Introduction
Oral cancer ranks as the 10th most common type of
cancer worldwide (1). Tongue squamous
cell carcinoma (TSCC) is the most prevalent and aggressive form of
oral cancer, accounting for 25–40% of all oral cancer cases
(2). Rapid local invasion, high rates
of lymph nodal metastasis and high recurrence rates are striking
features of TSCC, causing malfunction of mastication, speech and
deglutition, and leading to poor survival and quality of life
(3). In spite of the advances in
chemotherapy, radiotherapy and surgical therapy, the prognosis and
overall survival rates of patients with TSCC have not been
significantly improved over the last decade, with a current overall
five-year survival rate of <50% (4). Epidemiological study has revealed that
tongue cancer is associated with a variety of factors, including
age, geographic environment, family history, malnutrition,
infectious agents, and chronic alcohol, tobacco and betel nut
consumption (5). However, the
pathogenesis of tongue cancer is yet to be fully understood.
Mitochondria are organelles wherein aerobic
respiration is conducted to supply energy for a diverse range of
cellular events including fatty acid oxidation (6), calcium signaling (7), apoptosis (8), biosynthesis and biogenesis (9,10). Each
mitochondrion possesses its own 16,569 bp circular DNA (mtDNA)
(11), characteristically having 37
encoding genes for 13 subunits of respiratory chain complexes (I,
III, IV and V), 22 transfer (t)RNAs and 2 ribosomal (r)RNAs
(12), and a non-coding region
(D-loop) involved in regulation of mtDNA replication and
transcription (13–15). Unlike nuclear DNA, mtDNA lacks the
protection of histones and efficient repair mechanisms. Thus, mtDNA
is more vulnerable to oxidative damage by excessive reactive oxygen
species (ROS), a byproduct of oxidative phosphorylation in the
mitochondria (16). This may initiate
a vicious cycle: Damaged mtDNA can lead to an inefficient oxidative
phosphorylation system, which in turn gives rise to more ROS,
causing further damage to mtDNA and the genome. Importantly, the
resulting genomic instability directly contributes to
carcinogenesis.
Early on, it was proposed that injured mitochondria,
for instance when there is alteration of oxidative phosphorylation
or changes in the number, shape and function of mitochondria, may
be an important contributing factor in many cancer types (17). Recently, an increasing number of mtDNA
mutations, in the form of point mutations, deletions, insertions
and mitochondrial microsatellite instability (mtMSI), have been
reported in various cancers, including head and neck (18), pancreatic (19), colorectal (20,21) and
breast cancers (22–24). However, to date, the mtDNA alterations
in TSCC remain poorly defined. To profile the mtDNA alterations in
tongue cancer, the present study sequenced and analyzed the
mitochondrial genomes of tongue carcinoma, adjacent normal tissue
and matched peripheral blood from 8 patients with TSCC.
Materials and methods
Sample collection
A total of 8 patients with tongue cancer who
underwent primary surgery between January and December, 2015 at
Hunan Cancer Hospital, Changsha, China were enrolled. None of the
patients had a history of other known diseases potentially
associated with mitochondrial defects including diabetes and
hypertension (12). Cancer tissues
and adjacent normal tissues at a distance of 2 cm from the tumor
margin were divided by experienced surgeons and immediately frozen
at −80°C. The tumor node and metastasis staging for the tongue
cancer samples was classified by experienced pathologists. Matched
peripheral blood samples (2 ml venous blood per patient) were
collected. Information regarding age, sex and habits (smoking,
alcohol drinking and betel chewing) were also recorded (details are
given in Table I). Informed consent
was obtained from all subjects and the protocol of the study was
approved by the Medical Ethics Committees of the Hunan Cancer
Hospital and Family Planning Institute of Hunan Province (Changsha,
China).
 | Table I.Clinical data of patients with tongue
squamous cell carcinoma. |
Table I.
Clinical data of patients with tongue
squamous cell carcinoma.
|
|
|
|
|
| Carcinogen use |
|---|
|
|
|
|
|
|
|
|---|
| Patient no. | Sex | Age | TNM staging | Stage | Tobacco | Alcohol | Betel nut |
|---|
| 1 | M | 68 | T2N0M0 | II | Y | Y | N |
| 2 | M | 58 | T4N2M0 | IV | Y | Y | N |
| 3 | F | 44 | T2N0M0 | II | N | N | N |
| 4 | M | 51 | T2N1M0 | II | N | Y | Y |
| 5 | M | 59 | T4N1M0 | IV | Y | N | N |
| 6 | M | 50 | T2N1M0 | II | Y | Y | N |
| 7 | M | 52 | T2N0M0 | II | Y | N | N |
| 8 | M | 49 | T2N2M0 | IV | Y | Y | Y |
mtDNA amplification
Genomic DNA was isolated from tumor and adjacent
normal tissues with a High Pure Polymerase Chain Reaction (PCR)
Template Preparation kit (Roche Diagnostics, GmbH, Mannheim,
Germany). Genomic DNA from peripheral blood was extracted with a
GeneJET™ Whole Blood Genomic DNA Purification Mini kit (Thermo
Fisher Scientific, Inc., Waltham, MA, USA). The complete
mitochondrial genome was obtained using 24 pairs of primers as
previously described (25). PCR
amplification was performed using high-fidelity long PCR enzyme
PrimeSTAR® GXL DNA Polymerase (Takara Bio, Inc., Otsu,
Japan) on a Thermal Cycler 9700 machine (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The total reaction volume was 50 µl,
containing 10 µl of 5X PrimeSTAR GXL buffer (Mg2+ plus;
Takara Bio, Inc.), 4 µl of 2.5 mM dNTPs mix, 1.25 units polymerase
enzyme mix, 1.5 µl of 10 mM of each primer, and 50 ng template DNA,
and made up to 50 µl with nuclease-free water. The PCR
amplification protocol was as follows: Pre-denaturation at 95°C for
5 min, followed by 32 cycles of 94°C for 30 sec, 58°C for 30 sec
and 72°C for 9 min, and a final extension step at 72°C for 4 min.
PCR products were purified using a MiniBEST Agarose Gel DNA
Extraction kit Ver.4.0 (Takara Bio, Inc.).
Sequence analysis
Purified PCR products were sequenced with a BigDye
Terminator v3.1 Cycle Sequencing kit on a 3730 DNA sequencer (both
from Applied Biosystems; Thermo Fisher Scientific, Inc.). To detect
somatic variations and single nucleotide polymorphisms, the
original sequences of the tissues and matched blood samples were
aligned against the revised Cambridge Reference Sequence (rCRS;
GenBank access number: NC_012920.1; http://www.ncbi.nlm.nih.gov/gene/) using the online
software package CodonCode Aligner 6.0.2 (http://www.codoncode.com).
mtMSI analysis
A total of 10 microsatellite markers, namely C7TC5
(D310), (CA)5 (D514) and C5TC4 (D16184) in the D-loop, C6 in
NADH:ubiquinone oxidoreductase core subunit 1 (ND1), A7 in ND2, C6,
A8 and C3A3 in ND5, A7 in cytochrome C oxidase subunit 1 (CO1) and
T7 in CO3, were analyzed for length variations in cancer tissues
compared with adjacent normal tissues.
Results
mtDNA mutation in patients with tongue
cancer
To investigate the variance of mtDNA in patients
with TSCC, the mtDNA genomes of cancer tissues, adjacent normal
tissues and matched blood samples were sequenced and aligned to the
rCRS. Mutation was defined as a nucleotide variation occurring in
the mitochondrial genome of cancer tissues but not in those of
adjacent normal tissues and matched blood samples. A variation was
considered as a polymorphism when observed within all three samples
from a patient. In the present study, which involved 8 patients
(Table I), only one synonymous
mutation was observed in the ND5 gene in patient no. 2, which was a
transition of T-C at nucleotide position (np) 13,830 and caused no
substitution of an amino acid (not shown).
mtDNA polymorphisms in patients with
tongue cancer
When aligned with the rCRS, 21 nucleotide variations
of the mitochondrial genomes of TSCC were identified as
polymorphisms (Table II), as these
variations were present in not only the cancer tissues, but also
the adjacent normal tissues and matched blood samples. All the
polymorphisms exhibited a relatively high frequency in the study
population, as all were observed in at least 3/8 patients.
Furthermore, the polymorphisms were grouped on the basis of their
genomic location (Fig. 1). The
control region (D-loop) and Complex I gene harbored the most
polymorphisms, whereas there was no polymorphism identified in the
tRNA gene.
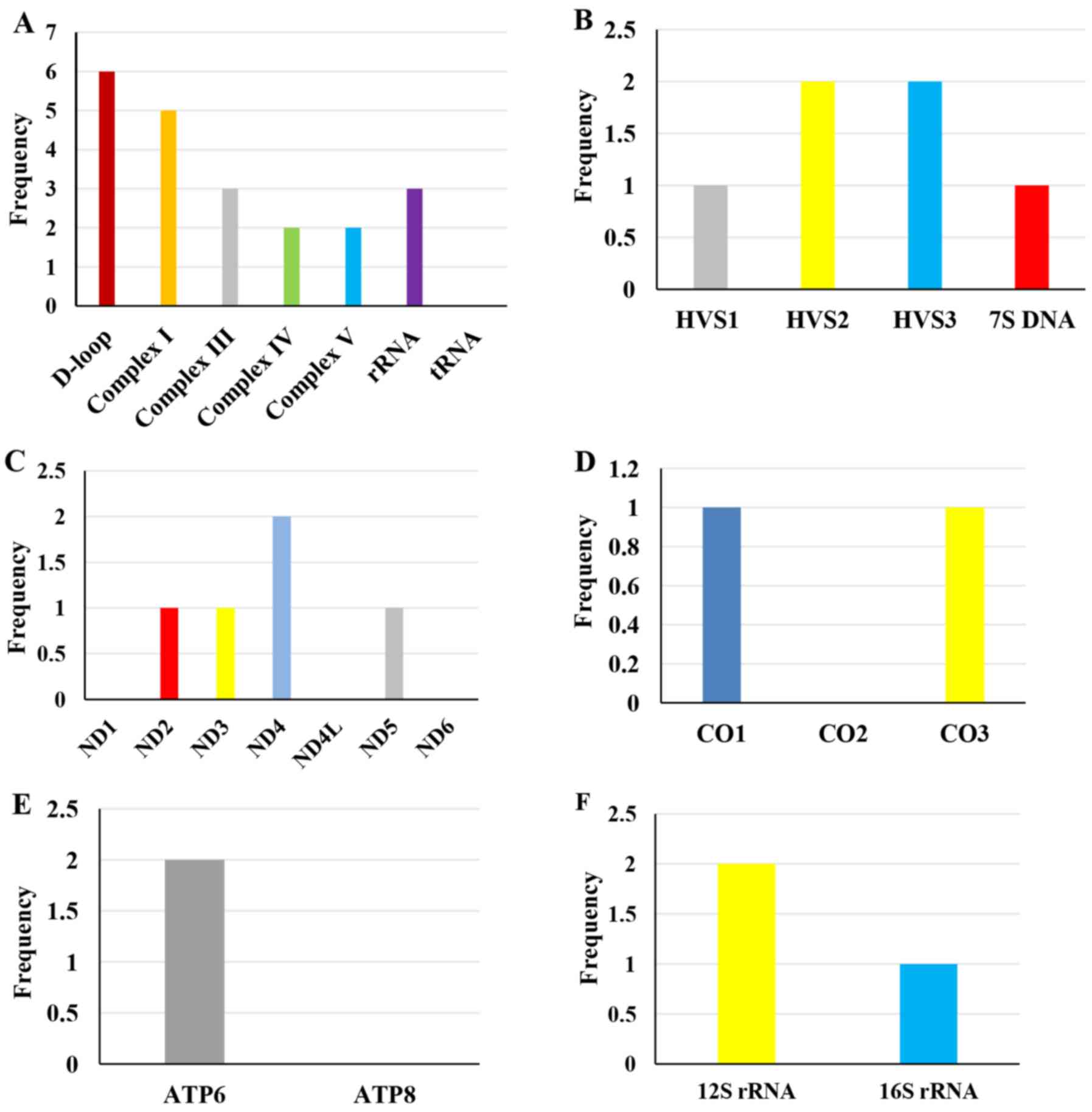 | Figure 1.Mitochondrial DNA polymorphisms
detected in the complete mitochondrial genome. (A) Overall
polymorphisms in the control region, four respiratory chain
complexes, rRNA and tRNA; the control region and Complex I harbored
the most polymorphisms while there was no polymorphism identified
in the tRNA gene. (B) The control region is divided into four
functional regions, including the HVS1, HVS2, HVS3 and 7S DNA
regions, among which six polymorphisms were mainly identified in
the HVS2 and HVS3 regions. (C-F) For the mitochondrial coding
regions, 15 polymorphisms were identified in the patients. (C)
Complex I: ND4 had the highest frequency of polymorphisms, while no
polymorphisms were found in the ND1, ND4L and ND6 genes. (D)
Complex IV: One polymorphism each in CO1 and CO3, and none in the
CO2 gene. (E) Complex V: Polymorphism was only identified in the
ATP6 gene, not in the ATP8 gene. (F) rRNA polymorphisms. HVS,
hyper-variable segment; rRNA, ribosomal RNA; tRNA, transfer RNA;
ND, NADH:ubiquinone oxidoreductase core subunit; CO, cytochrome C
oxidase subunit; ATP, ATP synthase membrane subunit. |
 | Table II.Mitochondrial DNA polymorphisms (in
the D-loop, Complexes I, III, IV and V, and rRNA) in tongue cancer
patients. |
Table II.
Mitochondrial DNA polymorphisms (in
the D-loop, Complexes I, III, IV and V, and rRNA) in tongue cancer
patients.
|
|
Nucleotide
position in mitochondrial genome |
|---|
|
|
|
|---|
|
| D-loop | Complex I | Complex III | Complex IV | Complex V | rRNA |
|---|
|
|
|
|
|
|
|
|
|---|
|
| HVS1 | HVS2 | HVS3 | 7S DNA | ND2 | ND3 | ND4 | ND5 | CYTB | CO1 | CO3 | ATP6 | 12SrRNA | 16SrRNA |
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|---|
| Sample no. | 16,223 | 73 | 263 | 523 | 524 | 16,519 | 4,769 | 10,398 | 10,873 | 11,719 | 12,705 | 14,766 | 15,301 | 15,326 | 7,028 | 9,540 | 8,860 | 8,701 | 750 | 1,438 | 2,706 |
|---|
| 1 |
| A-G | A-G | Ad | Cd | T-C | A-G |
|
| G-A |
| C-T | A-G | C-T |
| A-G |
| A-G | A-G | A-G |
|
| 2 | C-T | A-G | A-G |
|
| T-C | A-G | A-G | T-C | G-A | C-T | C-T | G-A | A-G | C-T | T-C | A-G | A-G | A-G | A-G | A-G |
| 3 |
| A-G | A-G | Ad | Cd | T-C | A-G |
|
| G-A |
| C-T | A-G | C-T |
| A-G |
| A-G |
| A-G |
|
| 4 | C-T | A-G | A-G | Ad | Cd |
| A-G | A-G | T-C | G-A | C-T | C-T | G-A |
| C-T | T-C | A-G | A-G | A-G | A-G | A-G |
| 5 |
| A-G | A-G |
|
| T-C | A-G | A-G |
| G-A | C-T | C-T | A-G | C-T |
| A-G |
| A-G | A-G | A-G |
|
| 6 |
| A-G | A-G |
|
| T-C | A-G |
|
| G-A |
| C-T | A-G | C-T |
| A-G |
| A-G | A-G | A-G |
|
| 7 | C-T | A-G | A-G |
|
| T-C | A-G | A-G | T-C | G-A | C-T | C-T | G-A | A-G | C-T | T-C | A-G | A-G | A-G | A-G | A-G |
| 8 | C-T | A-G | A-G |
|
|
| A-G | A-G | T-C | G-A | C-T | C-T | G-A | A-G | C-T | T-C | A-G | A-G | A-G | A-G | A-G |
The control region is subdivided into HVS1
(hyper-variable segment 1, np 16024–16383), HVS2 (np 57–372) and
HVS3 (np 438–574). The current data exhibited a total of six
polymorphisms in the control region, and mainly in HVS2 and HVS3.
Of the six polymorphisms, one was located at np 16223 in HVS1, two
at np 73 and np 263 in HVS2, two at np 523 and np 524 in HVS3, and
one at np 16519 in the 7S DNA region (not shown).
A total of 15 polymorphisms were identified in the
mitochondrial coding regions of the eight patients. Of these
polymorphisms, five (33.3%) belonged to the Complex I gene, with
ND4 having the most polymorphisms and no polymorphisms being
detected in the ND1, ND4L and ND6 genes. Three (20.0%) were located
in Complex III, and two (13.3%) in Complex IV, one each in CO1 and
CO3. Two (13.3%) were located in Complex V, both in the ATP6 gene.
The remaining three (20.0%) polymorphisms were seated in
mitochondrial rRNA genes (not shown).
mtMSI in patients with tongue
cancer
mtMSI refers to the change in length of short
repetitive sequences of mtDNA between normal and tumor tissues. The
present study observed alterations in the length of single
nucleotide short repeats within the D310 region in 2/8 patients
(Table III), whereas none of the
patients exhibited instability at the regions of D514 or D16184
(Table III), in the NADH
dehydrogenase subunits ND1, ND2 and ND5, or in the cytochrome
oxidase subunits COX1 and COX3 (not shown).
 | Table III.mtMSI in tongue cancer patients. |
Table III.
mtMSI in tongue cancer patients.
|
| D310 locus | D514 locus | D16184 locus |
|---|
|
|
|
|
|
|---|
| Sample no. | Tumor | Adjacent normal
tissue | Blood | mtMSI status | Tumor | Adjacent normal
tissue | Blood | mtMSI status | Tumor | Adjacent normal
tissue | Blood | mtMSI status |
|---|
| 1 | C13 | C13 | C13 | MSS |
(CA)4 |
(CA)4 |
(CA)4 | MSS | C13 | C13 | C13 | MSS |
| 2 |
C10TC6 |
C8TC6 |
C8TC6 | MSI |
(CA)6 |
(CA)6 |
(CA)6 | MSS |
C5TC4 |
C5TC4 |
C5TC4 | MSS |
| 3 |
C8TC6 |
C8TC6 |
C8TC6 | MSS |
(CA)4 |
(CA)4 |
(CA)4 | MSS |
C5TC4 |
C5TC4 |
C5TC4 | MSS |
| 4 |
C8TC6 |
C8TC6 |
C8TC6 | MSS |
(CA)4 |
(CA)4 |
(CA)4 | MSS |
CTC7 |
CTC7 |
CTC7 | MSS |
| 5 |
C8TC6 |
C8TC6 |
C8TC6 | MSS |
(CA)5 |
(CA)5 |
(CA)5 | MSS |
TC4TC4 |
TC4TC4 |
TC4TC4 | MSS |
| 6 |
C7TC6 |
C7TC6 |
C7TC6 | MSS |
(CA)5 |
(CA)5 |
(CA)5 | MSS |
C5TC4 |
C5TC4 |
C5TC4 | MSS |
| 7 |
C7TC6 |
C7TC6 |
C7TC6 | MSS |
(CA)5 |
(CA)5 |
(CA)5 | MSS |
C5TC4 |
C5TC4 |
C5TC4 | MSS |
| 8 |
C7TC6 |
C8TC6 |
C8TC6 | MSI |
(CA)5 |
(CA)5 |
(CA)5 | MSS |
C5TC4 |
C5TC4 |
C5TC4 | MSS |
Discussion
Somatic mutations in the mtDNA have been
increasingly observed in human cancers over recent years (21,23,24). To
contrast the many studies focusing on the control region (24,26), the
present study examined the entire mitochondrial genomes in cancer
tissues, adjacent normal tissues and matched peripheral blood
samples obtained from 8 patients with TSCC. As a result, synonymous
mutations, polymorphisms and MSI were identified in TSCC.
In previous studies, the T16519C substitution in the
control region has been demonstrated to be correlated with
increased risk of breast cancer (22)
and with worse prognosis in pancreatic cancer (19), and to exhibit a higher frequency in
patients with malignant melanoma and metastasis compared with
healthy controls (26). These
findings indicate a potential link between the T16519C polymorphism
and cancer progression. In addition to variation within the control
region, encoding sequence variation in the mitochondrial genome has
also been reported as relevant to cancer. The ND3 A10398G
non-synonymous polymorphism, which results in the substitution of
alanine by threonine within the NADH dehydrogenase subunit of
Complex I, has been shown to be associated with an increased risk
of breast cancer in European-American women (22). However, in another study, the 10398A
variant was reported to increase the risk of breast cancer in
African-American women but not in Caucasian women (27). A possible explanation for the
conflicting results may be that the A10398G variation was in
linkage disequilibrium with other causative polymorphisms in the
aforementioned study populations. In the present study, one
synonymous mutation and 21 polymorphisms were observed in 8
patients with TSCC. Of note, almost all the polymorphisms were
concentrated in the control region and in genes encoding Complexes
I, III and V, and rRNA. Theoretically, these variations may have
potential impact on mitochondrial function via affecting the
replication, transcription or translation events in
mitochondria.
In the current study, the MSI of the mitochondrial
genome in TSCC was analyzed. It appeared that mtMSI was most
frequently observed at the D310 locus (2/8 for the D310 locus vs.
undetected for the D514 or D16184 locus). This finding is in line
with previous reports (21,28). The D310 locus is considered to be the
most variable region in mtDNA, and alterations in this region,
including the 303-C insertion, T310C mutation or T deletion, 315-C
(CC) insertion and G316C mutation located within a poly-C stretch
of HVS II, may lead to MSI. The C-stretch structure is of
particular interest, since it is involved in the formation of the
persistent RNA-DNA hybrid that leads to initiation of mtDNA
heavy-strand replication (29).
Therefore, alterations of this region may have an impact on
replication and transcription of the mitochondrial genome. Studies
have also suggested that MSI at the non-coding D-loop region could
alter the rate of mtDNA replication, thereby disrupting
mitochondrion-induced apoptosis (30,31).
Regarding the biological significance of mtMSI in cancer, there are
studies having associated mtMSI with less differentiated
hepatocellular carcinoma, and late-stage progression and poor
prognosis of non-small cell lung cancer (32), as well as poor prognosis in colorectal
(20) and breast (24) cancer.
In summary, the current preliminary results suggest
a relatively high frequency of mtDNA polymorphisms and mtMSI in
TSCC. However, the generalization of this phenomenon requires
caution, since the present study used a relatively small sample
size and lacked healthy controls. Further large cohort studies are
needed to validate the findings and identify possible correlations
between the mtDNA variations and clinical parameters of patients
with TSCC. As increasing evidence supports that mitochondrial
genomic variations may serve a role in tumor development, further
investigations are expected to uncover the molecular mechanisms by
which mtDNA variations affect tumor progression, and their value in
cancer risk assessment and management.
Acknowledgements
The present study used the electronic medical record
database of the Department of Head and Neck Cancer (Oncoplastic
Surgery), Hunan Cancer Hospital (Changsha, China).
Funding
The present study was supported by the Science and
Technology Foundation of the Health and Family Planning Commission
of Hunan province, China (grant no. B2017144).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HYZ and BN conceived and designed the study. HYS and
HCL were responsible for patient enrollment. HYZ, HCL and HYS
performed the data analyses. HYS and HYZ drafted the manuscript.
WQX and HYZ critically revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from all subjects and
the protocol of the study was approved by the Medical Ethics
Committees of the Hunan Cancer Hospital and Family Planning
Institute of Hunan Province (Changsha, China).
Patient consent for publication
Informed consent for the publication of patient data
was obtained from all patients analyzed in the present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: The impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rao Krishna SV, Mejia G, Roberts-Thomson K
and Logan R: Epidemiology of oral cancer in Asia in the past decade
- an update (2000–2012). Asian Pac J Cancer Prev. 14:5567–5577.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xie N, Wang C, Liu X, Li R, Hou J, Chen X
and Huang H: Tumor budding correlates with occult cervical lymph
node metastasis and poor prognosis in clinical early-stage tongue
squamous cell carcinoma. J Oral Pathol Med. 44:266–272. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chauhan SS, Kaur J, Kumar M, Matta A,
Srivastava G, Alyass A, Assi J, Leong I, MacMillan C, Witterick I,
et al: Prediction of recurrence-free survival using a protein
expression-based risk classifier for head and neck cancer.
Oncogenesis. 4:e1472015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Amtha R, Razak IA, Basuki B, Roeslan BO,
Gautama W, Puwanto DJ, Ghani WM and Zain RB: Tobacco (kretek)
smoking, betel quid chewing and risk of oral cancer in a selected
Jakarta population. Asian Pac J Cancer Prev. 15:8673–8678. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Djouadi F, Bonnefont JP, Munnich A and
Bastin J: Characterization of fatty acid oxidation in human muscle
mitochondria and myoblasts. Mol Genet Metab. 78:112–118. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Csordás G and Hajnóczky G: Sorting of
calcium signals at the junctions of endoplasmic reticulum and
mitochondria. Cell Calcium. 29:249–262. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Decuypere JP, Monaco G, Bultynck G,
Missiaen L, De Smedt H and Parys JB: The IP(3)
receptor-mitochondria connection in apoptosis and autophagy.
Biochim Biophys Acta. 1813:1003–1013. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Klinge CM: Estrogenic control of
mitochondrial function and biogenesis. J Cell Biochem.
105:1342–1351. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rossier MF: T channels and steroid
biosynthesis: In search of a link with mitochondria. Cell Calcium.
40:155–164. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Anderson S, Bankier AT, Barrell BG, de
Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA,
Sanger F, et al: Sequence and organization of the human
mitochondrial genome. Nature. 290:457–465. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chan DC: Mitochondria: Dynamic organelles
in disease, aging, and development. Cell. 125:1241–1252. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McFarland R, Taylor RW and Turnbull DM:
Mitochondrial disease - its impact, etiology, and pathology. Curr
Top Dev Biol. 77:113–155. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Penta JS, Johnson FM, Wachsman JT and
Copeland WC: Mitochondrial DNA in human malignancy. Mutat Res.
488:119–133. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stewart JB, Freyer C, Elson JL and Larsson
NG: Purifying selection of mtDNA and its implications for
understanding evolution and mitochondrial disease. Nat Rev Genet.
9:657–662. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dean OM, van den Buuse M, Berk M, Copolov
DL, Mavros C and Bush AI: N-acetyl cysteine restores brain
glutathione loss in combined 2-cyclohexene-1-one and
d-amphetamine-treated rats: Relevance to schizophrenia and bipolar
disorder. Neurosci Lett. 499:149–153. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pedersen PL: Tumor mitochondria and the
bioenergetics of cancer cells. Prog Exp Tumor Res. 22:190–274.
1978. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou S, Kachhap S, Sun W, Wu G, Chuang A,
Poeta L, Grumbine L, Mithani SK, Chatterjee A, Koch W, et al:
Frequency and phenotypic implications of mitochondrial DNA
mutations in human squamous cell cancers of the head and neck. Proc
Natl Acad Sci USA. 104:7540–7545. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Navaglia F, Basso D, Fogar P, Sperti C,
Greco E, Zambon CF, Stranges A, Falda A, Pizzi S, Parenti A, et al:
Mitochondrial DNA D-loop in pancreatic cancer: Somatic mutations
are epiphenomena while the germline 16519 T variant worsens
metabolism and outcome. Am J Clin Pathol. 126:593–601. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lièvre A, Chapusot C, Bouvier AM,
Zinzindohoué F, Piard F, Roignot P, Arnould L, Beaune P, Faivre J
and Laurent-Puig P: Clinical value of mitochondrial mutations in
colorectal cancer. J Clin Oncol. 23:3517–3525. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lim SW, Kim HR, Kim HY, Huh JW, Kim YJ,
Shin JH, Suh SP, Ryang DW, Kim HR and Shin MG: High-frequency
minisatellite instability of the mitochondrial genome in colorectal
cancer tissue associated with clinicopathological values. Int J
Cancer. 131:1332–1341. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bai RK, Leal SM, Covarrubias D, Liu A and
Wong LJ: Mitochondrial genetic background modifies breast cancer
risk. Cancer Res. 67:4687–4694. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tipirisetti NR, Govatati S, Pullari P,
Malempati S, Thupurani MK, Perugu S, Guruvaiah P, Rao K L,
Digumarti RR, Nallanchakravarthula V, et al: Mitochondrial control
region alterations and breast cancer risk: A study in South Indian
population. PLoS One. 9:e853632014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW,
Lee LM, Wei YH and Lee HC: Mitochondrial DNA mutations and
mitochondrial DNA depletion in breast cancer. Genes Chromosomes
Cancer. 45:629–638. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rieder MJ, Taylor SL, Tobe VO and
Nickerson DA: Automating the identification of DNA variations using
quality-based fluorescence re-sequencing: Analysis of the human
mitochondrial genome. Nucleic Acids Res. 26:967–973. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ebner S, Lang R, Mueller EE, Eder W,
Oeller M, Moser A, Koller J, Paulweber B, Mayr JA, Sperl W, et al:
Mitochondrial haplogroups, control region polymorphisms and
malignant melanoma: A study in middle European Caucasians. PLoS
One. 6:e271922011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Canter JA, Kallianpur AR, Parl FF and
Millikan RC: Mitochondrial DNA G10398A polymorphism and invasive
breast cancer in African-American women. Cancer Res. 65:8028–8033.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pinheiro M, Veiga I, Pinto C, Afonso L,
Sousa O, Fragoso M, Santos L, Lopes P, Pais I, Lopes C, et al:
Mitochondrial genome alterations in rectal and sigmoid carcinomas.
Cancer Lett. 280:38–43. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kang D, Miyako K, Kai Y, Irie T and
Takeshige K: In vivo determination of replication origins of human
mitochondrial DNA by ligation-mediated polymerase chain reaction. J
Biol Chem. 272:15275–15279. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mambo E, Chatterjee A, Xing M, Tallini G,
Haugen BR, Yeung SC, Sukumar S and Sidransky D: Tumor-specific
changes in mtDNA content in human cancer. Int J Cancer.
116:920–924. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yin PH, Lee HC, Chau GY, Wu YT, Li SH, Lui
WY, Wei YH, Liu TY and Chi CW: Alteration of the copy number and
deletion of mitochondrial DNA in human hepatocellular carcinoma. Br
J Cancer. 90:2390–2396. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Matsuyama W, Nakagawa M, Wakimoto J,
Hirotsu Y, Kawabata M and Osame M: Mitochondrial DNA mutation
correlates with stage progression and prognosis in non-small cell
lung cancer. Hum Mutat. 21:441–443. 2003. View Article : Google Scholar : PubMed/NCBI
|















