Introduction
Immunothrombosis is a recently recognized
physiological process linking coagulation with innate immunity, of
which the deregulation may constitute a crucial event in the
development of thrombotic disorders (1). In this regard, this novel concept of
thrombosis implies that certain immune innate cells serve a key
role in thrombogenesis. Indeed, neutrophils are the first line of
defence against invading pathogens and are established to be an
essential component of the innate immune response; among the most
important mechanistic factors involved in the
hemostasis/inflammation crosstalk are neutrophil extracellular
traps (NETs) (2). NETs consist of
neutrophil web-like releasates formed by DNA, histones and other
nuclear and cytoplasmic components including elastase,
lactotransferrin, myeloperoxidase and calprotectin (3). To date, several stimuli able to induce
NET release have been described. The most recognized is infection
by pathogens (bacteria, fungi, viruses and protozoa), but other
factors have been described that depend on the host including
activated platelets, immune complexes and several inflammatory
stimuli (4). Thus, the presence of
NETs has been documented in several diseases with an elevated state
of inflammation, including autoimmune diseases, atherosclerosis,
vasculitis, cancer and several thrombotic disorders (5-9).
In particular, it has been demonstrated that NETs promote
thrombosis in vivo in mice models, and the implications of
several of their components including DNA and histones have been
widely studied (10,11). Notably, stimulation of platelets with
purified histones was sufficient to induce aggregation, and markers
of extracellular DNA can be detected in plasma following a deep
vein thrombosis episode (10,11). The pathogenic role of histones in
thrombosis was first proposed by showing that injection of histones
in mice induced thrombotic lesions similar to those observed in
severe sepsis (12). Indeed, histones
have been reported to induce tissue factor (TF; also known as
factor III) upregulation and thrombomodulin downregulation leading
to a procoagulant phenotype, hence they are considered as
damage-associated molecular patterns (DAMPs) (13). DAMPs are recognized as danger signals
by Toll-like receptors (TLRs), but the molecular mechanism remains
unclear (14-16).
DNA is also deemed to be a DAMP, and it has been demonstrated that
purified DNA may bind and activate proteins of the contact system,
which involves factor XI and factor XII, and boost thrombin
generation even in the absence of platelets (17). To date, the regulation of these
DAMP-induced processes has remained unclear and, moreover, there is
a lack of data on the effect of the different NET components on the
expression of other hemostatic factors besides TF. In the present
work, the aim was to evaluate the effect of DNA and histones on the
expression of the following hemostatic factors genes: Factor V
(F5), which is essential for thrombin generation catalysed
by the prothrombinase complex (18)
and necessary for the activation of prothrombin to thrombin by
factor Xa (F10) in the presence of Ca2+ (19); Factor VII (F7), a vitamin
K-dependent protease with a crucial role in the initiation of the
coagulation cascade (20); factor XII
(F12), involved in the activation of the intrinsic
coagulation pathway and the kallikrein-kinin system (21); PROC, which encodes protein C, a
fundamental factor involved in the cleavage of factors Va and VIIIa
(22); hepatocyte nuclear factor 4A
(HNF4A), a member of the nuclear receptor superfamily of
transcription factors and central to a complex regulatory network
controlling hepatocyte differentiation and metabolism (23); serpin family C member 1 (SERPINC1),
encoding antithrombin, the most important coagulation factor
inhibitor, which is a serine protease inhibitor with both
anticoagulant and anti-inflammatory action (24); serpin family F member 2
(SERPINF2), encoding the α2-antiplasmin, the primary
physiological inhibitor of plasmin that is also able to inhibit
other enzymes including elastase or trypsin (25); and finally, PROS1, which
encodes protein S, a vitamin K-dependent glycoprotein that has the
ability to inhibit factor Xa and act as a cofactor for the
activated protein C, taking part in the downregulation of
coagulation (26). Furthermore, our
group previously described the role of members of the microRNA
(miRNA/miR)-17/92 cluster and its paralog miR-106b-25 as modulators
of TF expression in a breast carcinoma cell line (MDA-MB-231) as
well as in a monocytic leukemia cell line (THP-1) (27). Moreover, miR-19a has been previously
described as an inhibitor of TF expression in breast cancer cells
(16). Therefore, the current study
also aimed to investigate if TF overexpression induced by histones
and DNA, acting as DAMPs, may be mediated by miRNA regulation. The
use of primary hepatocytes and monocytes would have been optimal to
study the effect of NETs in the expression of coagulation factors.
However, an inconvenience of these cells is the variability between
individuals that may impact on the interpretation as a first
approach (28). Thus, to minimize
variability, the experiments were performed with cell lines that
are well characterized and yield more homogeneous results (28).
Materials and methods
DNA purification
DNA was purified from the neutrophils of healthy
donors isolated using Histopaque 1077 (Sigma Aldrich; Merck KGaA,
Darmstadt, Germany). Briefly, neutrophils were purified from 12-ml
whole blood samples and pelleted at 150 x g for 15 min at room
temperature. The neutrophil pellet was resuspended in Hank's
balanced salt solution (supplemented with Ca2+ and
Mg2+; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
and cells were counted with an automatic cell counter (Bio Rad
Laboratories, Inc., Hercules, CA, USA). Purity was checked by flow
cytometry, considering a cluster of differentiation CD11b+/CD14-
population as neutrophils, and was always >90% (data not shown).
A BD Accuri™ C6 flow cytometer device (BD Biosciences, San Jose,
CA, USA) was used with CD11b+ fluorescein isothiocyanate-labelled
(cat. no. 11-0118-42) and CD14-phycoerythrin-conjugate antibodies
(cat. no. MHCD1417; Thermo Fisher Scientific, Inc.), applying 1 µl
(0.05 µg) antibody per 1x106 cells in a volume of 100 µl
[0.5% PBS-bovine serum albumin (Merck KGaA), 2 mM EDTA], following
the manufacturer's instructions. Neutrophils were acquired and
analysed with the Accuri C6 software. DNA was obtained using a
DNeasy kit (Qiagen GmbH, Hilden, Germany) following the
manufacturer's protocol.
The donor subjects met the eligibility criteria:
None of the subjects had a documented history of vascular disease,
or a personal history of thromboembolic/hemorrhagic disease or
cancer and all of them were <65 years old. All donors were
enrolled after providing written informed consent. A total of 100
healthy subjects were included in the study. Participants were
Caucasian, unrelated volunteers enrolled from the local blood donor
center (Centro Regional de Hemodonación, Murcia, Spain). Their
median age was 54 years (range, 46 to 63 years), and 33% were
female. Enrollment was performed from January to April, 2017.
The study was approved by the Ethical Committee of
Morales Meseguer University Hospital, Murcia, Spain (approval no.
20/14) and performed in accordance with the ethical standards laid
down in the 1964 Declaration of Helsinki and its subsequent
amendments.
Cell culture
The human liver carcinoma cell line HepG2 [American
Type Culture Collection (ATCC), Manassas, VA, USA] was cultured in
Eagle's minimum essential medium (MEM) supplemented with
non-essential amino acids at 0.1 mM and 10% fetal calf serum (FCS;
all from Thermo Fisher Scientific, Inc.). The monocytic leukemia
cell line THP-1 (ATCC) was cultured in ATCC-formulated RPMI-1640
medium with 10% FCS (Thermo Fisher Scientific, Madrid, Spain). Both
cell lines were maintained at 37˚C and 5% CO2.
It was preliminarily verified by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) that
DNA dose-dependently induced TF mRNA expression in the THP-1 cells
(tested at 0, 25 and 50 ng/µl; data not shown) and consequently the
highest concentration was used for subsequent experiments. THP-1
cells (500,000 cells per well in 24-well plates) were activated
with 50 ng/µl DNA from human neutrophils, or 20 µg/µl human
recombinant histone H4 (New England BioLabs, Inc., Ipswich, MA,
USA) as described by Kim et al (13); an equivalent volume of H2O
was used as a negative control and 1 µg/ml Escherichia coli
lipopolysaccharide (LPS; 0111:B4; Sigma-Aldrich) was used as a
positive control of cell activation, all of which were diluted in
Opti-MEM (Thermo Fisher Scientific, Inc.). After 2 h of incubation
at 37˚C and 5% CO2, the THP-1 cells were harvested.
HepG2 cells (1x106 cells per well in
12-well plates) were stimulated with DNA from human neutrophils or
human recombinant H4 via the same procedure as for THP-1 cells and
harvested after 24 h incubation at 37˚C and 5% CO2.
RNA isolation and detection of mRNA
expression by real-time RT-qPCR
Total RNA was isolated using RNAzol® RT
reagent (Molecular Research Center, Inc., Cincinnati, OH) according
to the manufacturer's protocol. Retrotranscription was performed
using 100 ng total RNA with MultiScribe Reverse Transcriptase and
Premix Ex Taq (Probe qPCR) Master Mix (Takara Biotechnology Co.,
Ltd., Dalian, China) for both mRNA and miRNA PCRs.
Quantitative real-time PCR was performed using
TaqMan Gene Expression Assays (Applied Biosystems; Thermo Fisher
Scientific) with gene-specific primers (for F5:
Hs00914120_m1; for F7: Hs01551992_m1; for F10:
Hs00984443_m1; for F12: Hs00166821; for PROC:
Hs00165584_m1; for PROS1: Hs00165590_m1; for
SERPINC1: Hs00166654_m1; for serpin family F member 2
(SERPINF2): Hs00168989_m1; for hepatocyte nuclear factor 4α
(HNF4A): Hs00230853_m1; and for β-actin (ACTB):
Hs99999903_m1; Thermo Fisher Scientific, Inc.). PCRs were performed
on a Light-Cycler 480 Real-Time PCR system (Roche Applied Science,
Penzberg, Germany) with the following thermocycling conditions: A
preincubation step at 95˚C for 30 sec, 40 cycles of amplification
with 3 sec at 95˚C and 20 sec at 60˚C, and finally, a cooling step
at 40˚C for 30 sec. Data were analysed using the comparative
threshold cycle method (2-ΔΔCq method) (29) with ACTB as an endogenous
reference control for quantification and normalization. To quantify
the levels of miRNAs, TaqMan Gene Expression Assays (Thermo Fisher
Scientific) were used with gene-specific primers for hsa-miR-17-5,
hsa-miR-18a, hsa-miR-19a, hsa-miR-20a and hsa-miR-106b. U6 snRNA
was used as an endogenous control (Thermo Fisher Scientific,
Inc.).
Statistical analysis
All data are presented as the mean ± standard error.
Experiments were performed in triplicate and repeated three times.
Statistical differences between groups were assessed by the
non-parametric Mann-Whitney U test using GraphPad Prism 6 software
(GraphPad Software Inc., La Jolla, CA). P<0.05 was considered to
indicate statistical significance.
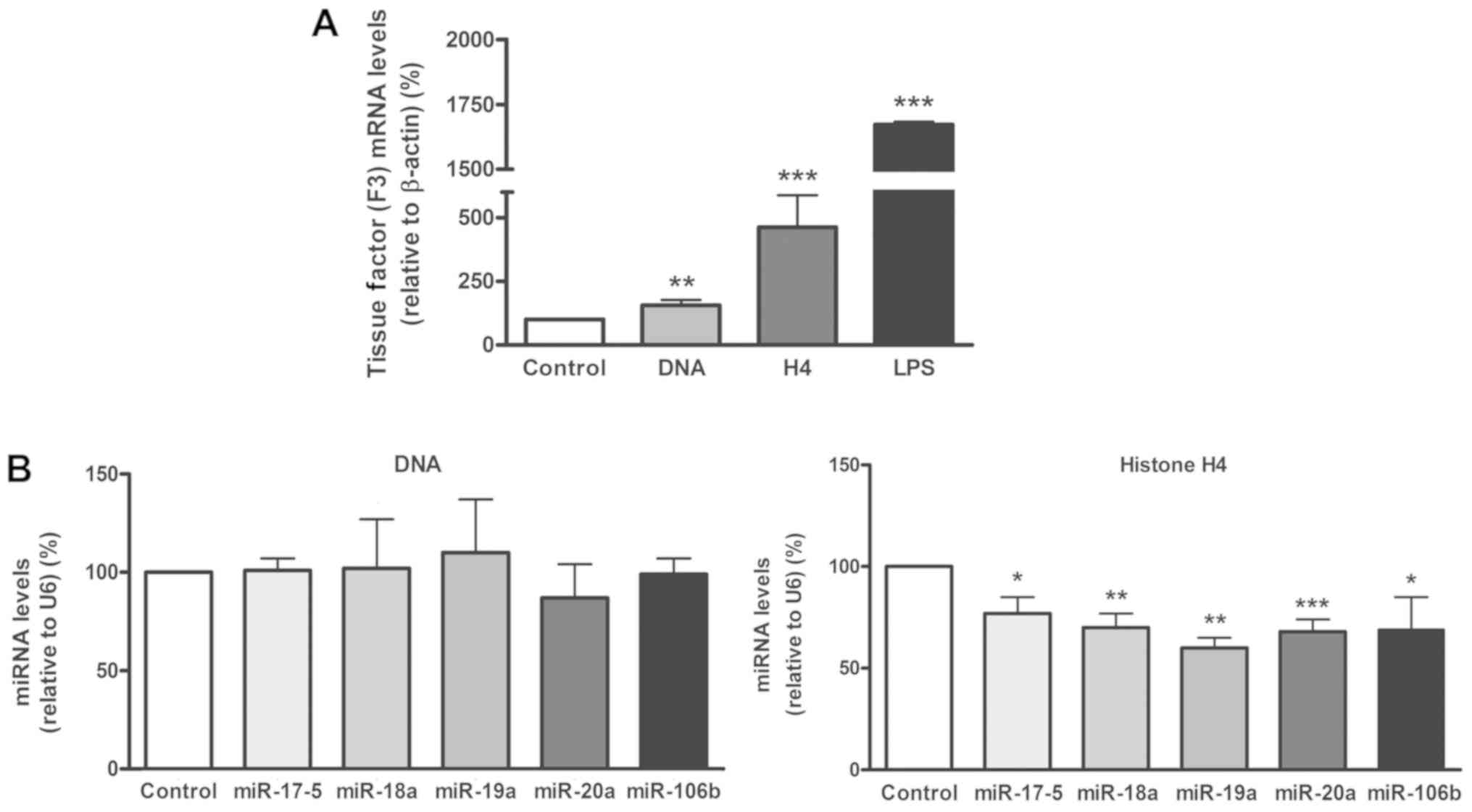
Results
H4 and DNA induce the expression of
coagulation factors
To study the influence of NET components on the
expression of coagulation factors in human liver, HepG2 cells were
incubated with H4 or DNA, and the mRNA levels of 8 genes related to
coagulation (F5, F7, F10, F12, PROC, PROS1, SERPINC1 and
SERPINF2) were quantified.
It was identified that each DAMP induced significant
increases in the expression of the different factors, both
coagulant (F5, F7, F12 and SERPINF2; Fig. 1A) and anticoagulant (PROS1 and
SERPINC1; Fig. 1B), though H4
consistently induced a marginally greater upregulation than DNA,
when compared with the cells without treatment. For F10 and
PROC, a trend towards an increase in expression was observed
with each DAMP but without statistical significance.
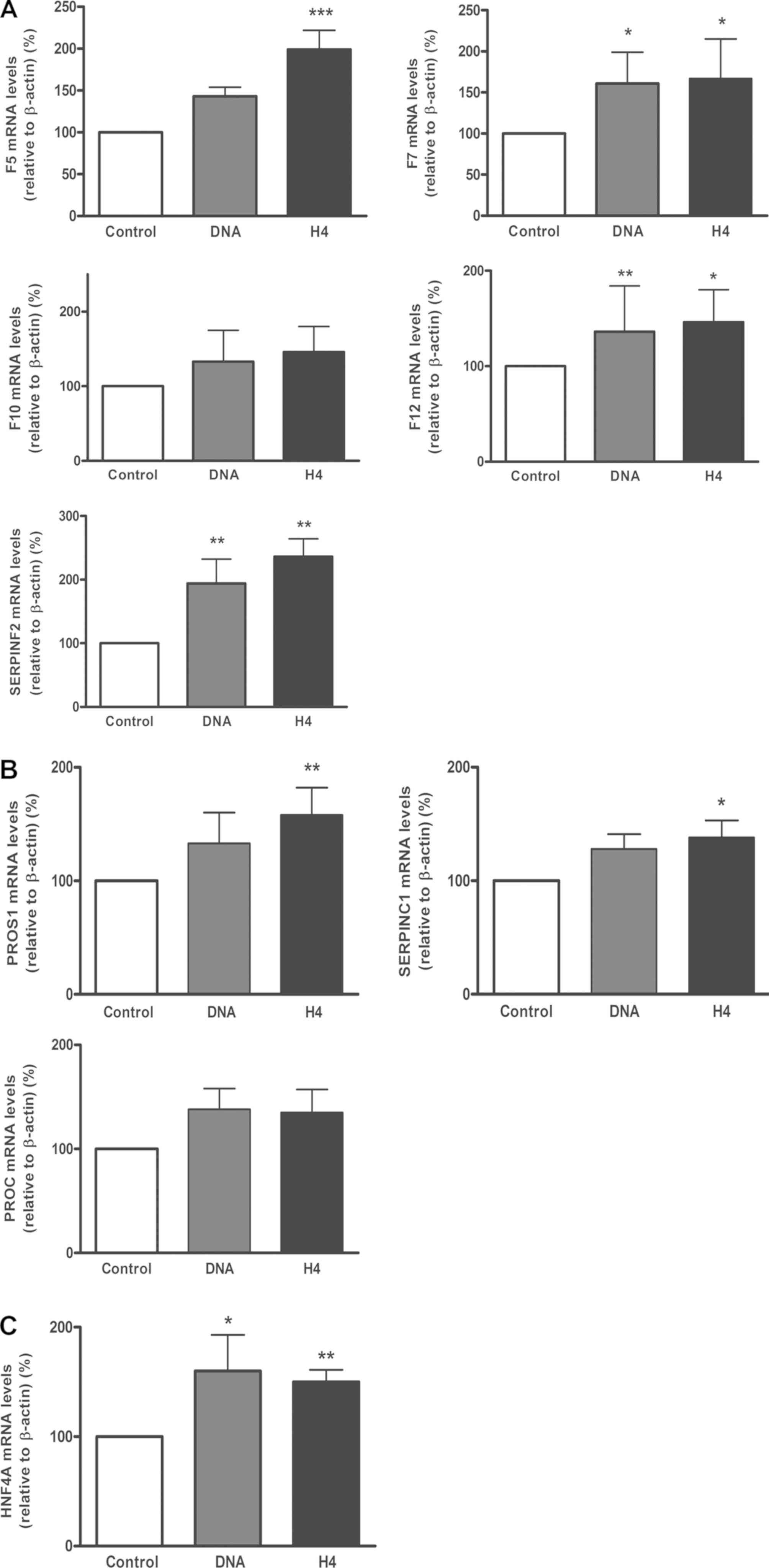 | Figure 1.Neutrophil extracellular trap
components increased coagulation gene expression in liver cells
in vitro. The expression of (A) coagulant genes F5,
F7, F10, F12, SERPINF2, (B)
anticoagulant genes PROS1, SERPINC1, PROC and
(C) HNF4A were measured in HepG2 cells incubated with DNA
(50 ng/ml) or H4 (20 µg/ml) for 24 h. All data are presented as the
mean ± standard error of the mean of 3 experiments performed in
triplicate. *P<0.05, **P<0.01 and
***P<0.001 vs. Control. F5/7/12, Factor
V/VII/XII; SERPINF2, serpin family F member 2; PROC,
protein C; PROS1, protein S; SERPINC1, serpin family
C member 1; HNF4A, hepatocyte nuclear factor 4α; H4, histone
H4. |
H4 and DNA upregulate the expression
of HNF4A
Since the association between HNF4A and the
expression of different coagulation factors, including Protein S,
Protein C and antithrombin, has previously been described (28-30),
the present study quantified the levels of HNF4A mRNA
following incubation with H4 or DNA. It was observed that each
agonist upregulated the expression of HNF4A to a significant
and similar extent (Fig. 1C).
H4 causes a decrease in miR-17/92
cluster expression
The next aim was to confirm the influence of NET
components on the expression of TF and the involvement of the
miR-17/92 cluster in this regulation. THP-1 cells were incubated
with DNA or H4 and the levels of TF mRNA, and of several of the
miRNAs included in the miR-17/92 cluster (miR-17-5, -18a, -19a and
-20a) and its paralog miR-106b, were assessed. LPS, was used as a
positive control only for TF, to ensure its expression (33).
It was confirmed that both DNA and H4 significantly
increased the expression of TF, with a higher increase
observed when using H4 (Fig. 2A).
Levels of the selected miRNA did not vary when incubating THP-1
cells with DNA (Fig. 2B, left panel),
while following the treatment with H4, all the studied miRNAs
significantly decreased in comparison with the negative control
(Fig. 2B, right panel).
Discussion
In recent years, research into the pathogenicity of
DAMPs has increased, and several reports have described a
prothrombotic role for histones and DNA (12,14,34,35).
In the present study, this hypothesis was further supported by
demonstrating that H4 and DNA induced the transcription of several
coagulation factors. Specifically, the mRNA levels of F7,
F12 and SERPINF2 (encoding α2-antiplasmin) were
significantly upregulated by DNA and H4, while the mRNA levels of
F5, PROS1 and SERPINC1 (encoding antithrombin)
were upregulated only by H4. In the search of a possible mechanism
for this effect, HNF4α was selected as a suitable candidate. It has
been demonstrated that this hepatic transcription factor is
implicated in the transcription of numerous coagulation genes
including F5, F7, F12, SERPINF2 and SERPINC1
(31,32,36) among
others. In the current study, it was hypothesized that the observed
increase in HNF4A expression may be responsible for the increase in
the expression of certain coagulant factors induced by H4 and DNA,
although a direct effect of these DAMPs on the overexpression of
hemostatic factors cannot be discarded (37). Limited information is available
concerning the mechanisms implicated in the action of H4 and DNA on
different cells or tissues. As mentioned, normal primary
hepatocytes and primary monocytes would have been optimal models to
study the effect of NETs on the expression of coagulation factors,
but due to the variability between individuals that may affect the
interpretation of results, the experiments were performed with cell
lines that are well characterized and yield homogeneous results. It
should also be noted that in 2009 the phenotype of HepG2 was
described as hepatoblastoma instead of hepatocellular carcinoma
(38), but this misidentification is
not expected to affect interpretations in the current study.
To date, one of the sole proposed mechanisms
implicated in TF expression induced by histones in monocytes is the
activation of the TLR2-4/nuclear factor-κB pathway. In that report,
it was demonstrated that the selective blockade of TLR2 or TLR4
inhibited the overexpression of TF following activation with H4,
among other histones (13). Notably,
the expression of TLR4 has been reported in hepatocytes and it may
be implicated in the overexpression of hemostatic factors and/or
HNF4α as well as TF (39). Additional
studies using animal models may aid to define if these changes in
the coagulant state may occur in vivo and if overexpression
of hemostatic factors is also detectable in plasma following a
challenge with DNA or H4.
miRNAs have been recently associated with the
regulation of several hemostatic factors (40). In particular, coagulation factors
including fibrinogen, factor XI and tissue factor pathway inhibitor
are regulated by miRNAs (41-43).
As another key element, TF has also been demonstrated to be
regulated by miRNAs (17). Thus, in
the present study it was hypothesized that another possible effect
explaining the increased expression of TF by DAMPs in monocytes may
be an alteration of miRNA expression. In this regard, it was
identified that, following the treatment with H4, the decrease of
miRNAs targeting TF was significant. Philippe et al
demonstrated that activation of TLR2 in synoviocytes by bacterial
lipoproteins (44) or of TLR4 by LPS
(45) decreased the expression of
miR-19a/b and miR-20a, respectively. These results are in accord
with the present findings, although the mechanism by which these
miRNAs are decreased by DAMPs is still unknown. Meanwhile, DNA had
no effect on the expression of miRNAs in the miR-17-92 cluster,
suggesting that the upregulation of TF by DNA is independent of
miR-17-92 and other unknown mechanisms are involved.
In conclusion, the present results indicated that H4
and, to a lesser extent, DNA cause an increase in the mRNA
expression of various coagulation factors. This upregulation may
alter the hemostatic balance and may explain the prothrombotic
effect of histones and DNA, although this requires further
confirmation. Additionally, both DNA and H4 triggered an increase
in HNF4A mRNA expression that may indicate its partial involvement
in the regulation of clotting factors mediated by DAMPs. Finally,
it was determined that H4 could induce a decrease in the expression
of certain miRNAs of the miR-17/92 cluster, which may partially
explain the upregulation of the expression of TF induced by H4.
Acknowledgements
Not applicable.
Funding
The current study was supported by research grants
from the Instituto de Salud Carlos III and Fondo Europeo de
Desarrollo Regional (FEDER; grant no. PI17/00051) and the Fundación
Séneca (grant no. 19873/GERM/15). ABA has a research fellowship
from Sociedad Española de Trombosis y Hemostasia (SETH).
Availability of data and materials
Data and material related to this manuscript are
available from the corresponding authors on reasonable request.
Authors' contributions
RGC and CM conceptualized the study. AMDLRG, RGC and
CM were responsible for data acquisition. AMDR, AAC and ABA
performed the analyses of the data. AMDLRG, AA, ABA, NGB, VV, RGC
and CM performed the experiments. NGB provided technical support in
data acquisition. AMDLRG, AA and ABA drafted the manuscript. RGC,
VV and CM reviewed and edited the manuscript.
Ethics approval and consent to
participate
All subjects who met eligibility criteria were
enrolled after providing written informed consent. The study was
approved by the Ethical Committee of Morales Meseguer University
Hospital, Murcia, Spain (approval no., 20/14) and performed in
accordance with the ethical standards laid down in the 1964
Declaration of Helsinki and its subsequent amendments.
Patient consent for publication
All subjects gave their consent for the publication
of associated data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Engelmann B and Massberg S: Thrombosis as
an intravascular effector of innate immunity. Nat Rev Immunol.
13:34–45. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Swystun LL and Liaw PC: The role of
leukocytes in thrombosis. Blood. 128:753–762. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Urban CF, Ermert D, Schmid M, Abu-Abed U,
Goosmann C, Nacken W, Brinkmann V, Jungblut PR and Zychlinsky A:
Neutrophil extracellular traps contain calprotectin, a cytosolic
protein complex involved in host defense against Candida
albicans. PLoS Pathog. 5(e1000639)2009.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zawrotniak M and Rapala-Kozik M:
Neutrophil extracellular traps (NETs) - formation and implications.
Acta Biochim Pol. 60:277–284. 2013.PubMed/NCBI
|
|
5
|
Rao AN, Kazzaz NM and Knight JS: Do
neutrophil extracellular traps contribute to the heightened risk of
thrombosis in inflammatory diseases? World J Cardiol. 7:829–842.
2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lee KH, Kronbichler A, Park DD, Park Y,
Moon H, Kim H, Choi JH, Choi Y, Shim S, Lyu IS, et al: Neutrophil
extracellular traps (NETs) in autoimmune diseases: A comprehensive
review. Autoimmun Rev. 16:1160–1173. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Knight JS, Luo W, O'Dell AA, Yalavarthi S,
Zhao W, Subramanian V, Guo C, Grenn RC, Thompson PR, Eitzman DT, et
al: Peptidylarginine deiminase inhibition reduces vascular damage
and modulates innate immune responses in murine models of
atherosclerosis. Circ Res. 114:947–956. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tang S, Zhang Y, Yin SW, Gao XJ, Shi WW,
Wang Y, Huang X, Wang L, Zou LY, Zhao JH, et al: Neutrophil
extracellular trap formation is associated with autophagy-related
signalling in ANCA-associated vasculitis. Clin Exp Immunol.
180:408–418. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Berger-Achituv S, Brinkmann V, Abed UA,
Kühn LI, Ben-Ezra J, Elhasid R and Zychlinsky A: A proposed role
for neutrophil extracellular traps in cancer immunoediting. Front
Immunol. 4(48)2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Fuchs TA, Brill A, Duerschmied D,
Schatzberg D, Monestier M, Myers DD Jr, Wrobleski SK, Wakefield TW,
Hartwig JH and Wagner DD: Extracellular DNA traps promote
thrombosis. Proc Natl Acad Sci USA. 107:15880–15885.
2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Brill A, Fuchs TA, Savchenko AS, Thomas
GM, Martinod K, De Meyer SF, Bhandari AA and Wagner DD: Neutrophil
extracellular traps promote deep vein thrombosis in mice. J Thromb
Haemost. 10:136–144. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Semeraro F, Ammollo CT, Morrissey JH, Dale
GL, Friese P, Esmon NL and Esmon CT: Extracellular histones promote
thrombin generation through platelet-dependent mechanisms:
Involvement of platelet TLR2 and TLR4. Blood. 118:1952–1961.
2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kim JE, Yoo HJ, Gu JY and Kim HK: Histones
Induce the Procoagulant Phenotype of Endothelial Cells through
Tissue Factor Up-Regulation and Thrombomodulin Down-Regulation.
PLoS One. 11(e0156763)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Ammollo CT, Semeraro F, Xu J, Esmon NL and
Esmon CT: Extracellular histones increase plasma thrombin
generation by impairing thrombomodulin-dependent protein C
activation. J Thromb Haemost. 9:1795–1803. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yang X, Li L, Liu J, Lv B and Chen F:
Extracellular histones induce tissue factor expression in vascular
endothelial cells via TLR and activation of NF-κB and AP-1. Thromb
Res. 137:211–218. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Gould TJ, Lysov Z, Swystun LL, Dwivedi DJ,
Zarychanski R, Fox-Robichaud AE and Liaw PC: Canadian Critical Care
Translational Biology Group: Extracellular Histones Increase Tissue
Factor Activity and Enhance Thrombin Generation by Human Blood
Monocytes. Shock. 46:655–662. 2016.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bhagirath VC, Dwivedi DJ and Liaw PC:
Comparison of the Proinflammatory and Procoagulant Properties of
Nuclear, Mitochondrial, and Bacterial DNA. Shock. 44:265–271.
2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Mann KG, Nesheim ME, Tracy PB, Hibbard LS
and Bloom JW: Assembly of the prothrombinase complex. Biophys J.
37:106–107. 1982.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Krishnaswamy S: The transition of
prothrombin to thrombin. J Thromb Haemost. 11 (Suppl 1):265–276.
2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
O'Hara PJ, Grant FJ, Haldeman BA, Gray CL,
Insley MY, Hagen FS and Murray MJ: Nucleotide sequence of the gene
coding for human factor VII, a vitamin K-dependent protein
participating in blood coagulation. Proc Natl Acad Sci USA.
84:5158–5162. 1987.PubMed/NCBI
|
|
21
|
Didiasova M, Wujak L, Schaefer L and
Wygrecka M: Factor XII in coagulation, inflammation and beyond.
Cell Signal. 51:257–265. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Esmon NL, Owen WG and Esmon CT: Isolation
of a membrane-bound cofactor for thrombin-catalyzed activation of
protein C. J Biol Chem. 257:859–864. 1982.PubMed/NCBI
|
|
23
|
Hayhurst GP, Lee Y-H, Lambert G, Ward JM
and Gonzalez FJ: Hepatocyte nuclear factor 4alpha (nuclear receptor
2A1) is essential for maintenance of hepatic gene expression and
lipid homeostasis. Mol Cell Biol. 21:1393–1403. 2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Björk I and Olson ST: Antithrombin. A
bloody important serpin. Adv Exp Med Biol. 425:17–33.
1997.PubMed/NCBI
|
|
25
|
Kettle P and Mayne EE: A bleeding disorder
due to deficiency of alpha 2-antiplasmin. J Clin Pathol.
38:428–429. 1985.PubMed/NCBIPubMed/NCBI
|
|
26
|
Furie B and Furie BC: The molecular basis
of blood coagulation. Cell. 53:505–518. 1988.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Teruel R, Pérez-Sánchez C, Corral J,
Herranz MT, Pérez-Andreu V, Saiz E, García-Barberá N,
Martínez-Martínez I, Roldán V, Vicente V, et al: Identification of
miRNAs as potential modulators of tissue factor expression in
patients with systemic lupus erythematosus and antiphospholipid
syndrome. J Thromb Haemost. 9:1985–1992. 2011.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ramboer E, De Craene B, De Kock J,
Vanhaecke T, Berx G, Rogiers V and Vinken M: Strategies for
immortalization of primary hepatocytes. J Hepatol. 61:925–943.
2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Δ Δ C(T)) method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Salloum-Asfar S, Arroyo AB, Teruel-Montoya
R, García-Barberá N, Roldán V, Vicente V, Martínez C and
González-Conejero R: miRNA-Based Regulation of Hemostatic Factors
through Hepatic Nuclear Factor-4 Alpha. PLoS One.
11(e0154751)2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Erdmann D and Heim J: Orphan nuclear
receptor HNF-4 binds to the human coagulation factor VII promoter.
J Biol Chem. 270:22988–22996. 1995.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tarumi T, Kravtsov DV, Zhao M, Williams SM
and Gailani D: Cloning and characterization of the human factor XI
gene promoter: Transcription factor hepatocyte nuclear factor
4alpha (HNF-4alpha ) is required for hepatocyte-specific expression
of factor XI. J Biol Chem. 277:18510–18516. 2002.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Brand K, Fowler BJ, Edgington TS and
Mackman N: Tissue factor mRNA in THP-1 monocytic cells is regulated
at both transcriptional and posttranscriptional levels in response
to lipopolysaccharide. Mol Cell Biol. 11:4732–4738. 1991.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Xu J, Zhang X, Pelayo R, Monestier M,
Ammollo CT, Semeraro F, Taylor FB, Esmon NL, Lupu F and Esmon CT:
Extracellular histones are major mediators of death in sepsis. Nat
Med. 15:1318–1321. 2009.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Noubouossie DF, Whelihan MF, Yu YB,
Sparkenbaugh E, Pawlinski R, Monroe DM and Key NS: In vitro
activation of coagulation by human neutrophil DNA and histone
proteins but not neutrophil extracellular traps. Blood.
129:1021–1029. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Farsetti A, Moretti F, Narducci M, Misiti
S, Nanni S, Andreoli M, Sacchi A and Pontecorvi A: Orphan receptor
hepatocyte nuclear factor-4 antagonizes estrogen receptor
alpha-mediated induction of human coagulation factor XII gene.
Endocrinology. 139:4581–4589. 1998.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tremp GL, Duchange N, Branellec D,
Cereghini S, Tailleux A, Berthou L, Fievet C, Touchet N, Schombert
B, Fruchart JC, et al: A 700-bp fragment of the human antithrombin
III promoter is sufficient to confer high, tissue-specific
expression on human apolipoprotein A-II in transgenic mice. Gene.
156:199–205. 1995.PubMed/NCBI View Article : Google Scholar
|
|
38
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Lee YS, Kim YH, Jung YS, Kim KS, Kim DK,
Na SY, Lee JM, Lee CH and Choi HS: Hepatocyte Toll-like receptor 4
mediates lipopolysaccharide-induced hepcidin expression. Exp Mol
Med. 49(e408)2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Teruel-Montoya R, Rosendaal FR and
Martínez C: MicroRNAs in hemostasis. J Thromb Haemost. 13:170–181.
2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Fish RJ and Neerman-Arbez M: Fibrinogen
gene regulation. Thromb Haemost. 108:419–426. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Salloum-Asfar S, Teruel-Montoya R, Arroyo
AB, García-Barberá N, Chaudhry A, Schuetz E, Luengo-Gil G, Vicente
V, González-Conejero R and Martínez C: Regulation of coagulation
factor XI expression by microRNAs in the human liver. PLoS One.
9(e111713)2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
B Arroyo A, Salloum-Asfar S, Pérez-Sánchez
C, Teruel-Montoya R, Navarro S, García-Barberá N, Luengo-Gil G,
Roldán V, Hansen JB, López-Pedrera C, et al: Regulation of TFPIα
expression by miR-27a/b-3p in human endothelial cells under normal
conditions and in response to androgens. Sci Rep.
7(43500)2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Philippe L, Alsaleh G, Suffert G, Meyer A,
Georgel P, Sibilia J, Wachsmann D and Pfeffer S: TLR2 expression is
regulated by microRNA miR-19 in rheumatoid fibroblast-like
synoviocytes. J Immunol. 188:454–461. 2012.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Philippe L, Alsaleh G, Pichot A, Ostermann
E, Zuber G, Frisch B, Sibilia J, Pfeffer S, Bahram S, Wachsmann D,
et al: miR-20a regulates ASK1 expression and TLR4-dependent
cytokine release in rheumatoid fibroblast-like synoviocytes. Ann
Rheum Dis. 72:1071–1079. 2013.PubMed/NCBI View Article : Google Scholar
|