Introduction
Angiopoietin-like protein (ANGPTL) 8 is a member of
the ANGPTL family (1) and is also
known as RIFL (2), TD26(3), lipasin (4), PRO1185, PVPA599, C19orf80 and
betatrophin (5). ANGPTL8 is involved
in the partitioning of triglycerides to muscle and adipose tissues
in conjunction with ANGPTL3 and ANGPTL4 (6,7). They
regulate the activity of lipoprotein lipase (LPL) in organs
(7). ANGPTL8 interacts with ANGPTL3
and inhibits LPL in muscle tissue, whereas ANGPTL4 inhibits LPL in
adipose tissue.
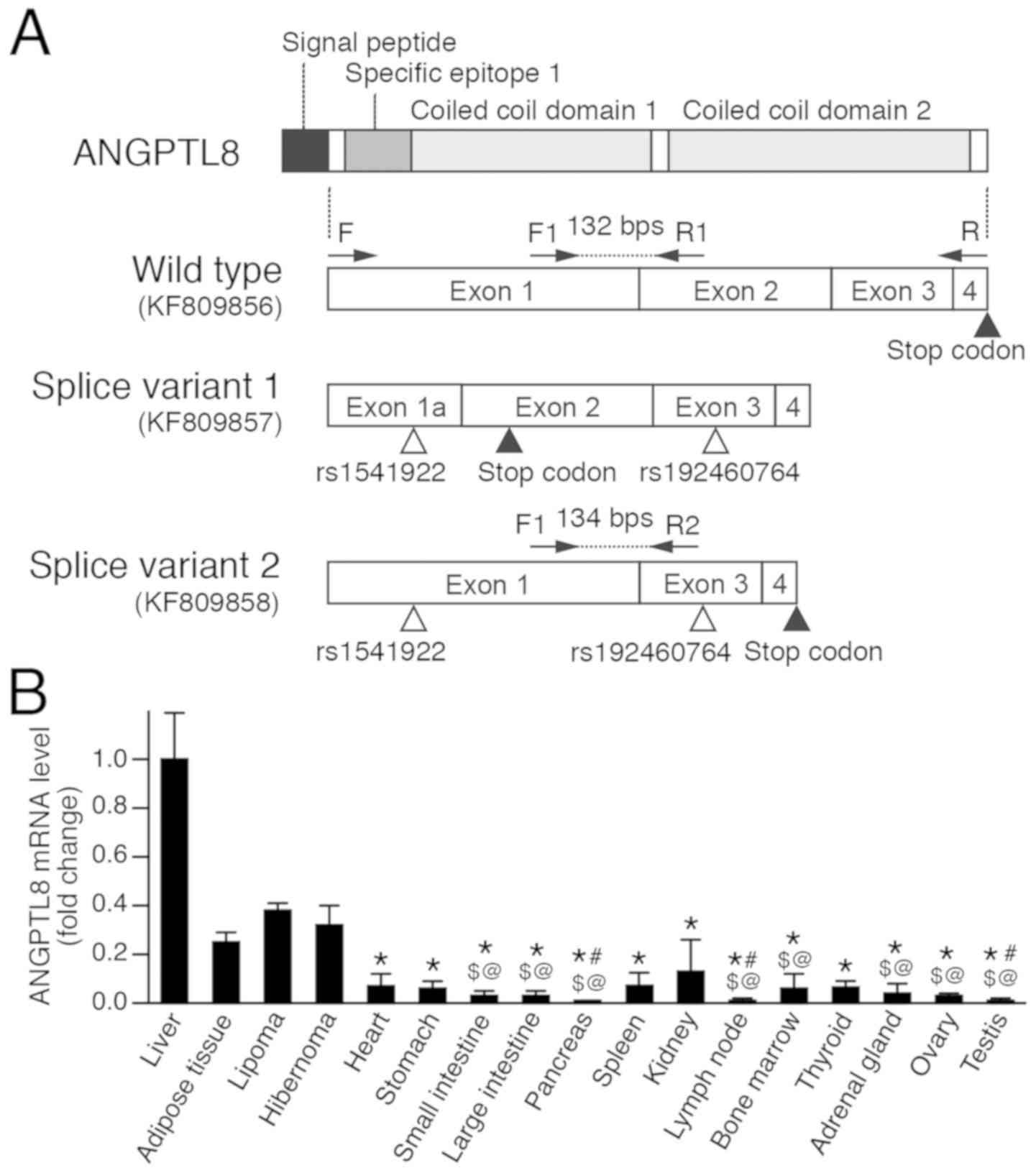
The ANGPTL8 gene is located on chromosome 19 and
consists of four exons that encode 198 amino acids. The protein
contains a signal peptide consisting of 20 amino acids at the
N-terminus, the specific epitope 1 domain and two coiled coil
domains (Fig. 1A). Specific epitope 1
domain is necessary for inhibition of LPL, and two coiled coil
domains are necessary for the complex formation with ANGPTL3 and
ANGPTL4 (6,7). The protein is processed to the mature
protein by cleavage of signal peptide and released into the
bloodstream (1).
It was reported that ANGPTL8 is highly expressed in
the liver, white and brown adipose tissue (BAT) (2,5,6). In vitro studies demonstrated that
the expression level of ANGPTL8 is altered depending on the
differentiation and functional state of the cells (2). However, the precise localization and
distribution of ANGPTL8-expressing cells in these organs remains
unclear. The elucidation of the localization of ANGPTL8 in these
organs will contribute to the understanding of the physiological
roles of ANGPTL8 and the association with local lipid metabolism.
Further, this may aid the understanding of the pathological role of
ANGPTL8 in metabolic diseases and the development of novel
therapies for these diseases.
The aim of the present study was to investigate the
expression and localization of ANGPTL8 in normal human tissues.
Using formalin-fixed paraffin-embedded (FFPE) specimens, ANGPTL8
expression levels and localization were examined by molecular
biological methods. During the cloning of ANGPTL8 mRNA from normal
liver, splice variants were identified. The structures of the
splice variants were also documented.
Materials and methods
Sequencing of ANGPTL8 mRNA from the
liver
The coding sequence of ANGPTL8 was amplified and
cloned from the total RNA of the liver (cat. no. K4000-1; lot no.
0111320; Thermo Fisher Scientific, Inc.). Total RNA (2 µg) was
treated with DNase I (Thermo Fisher Scientific, Inc.) at room
temperature (RT) for 15 min, and cDNA was synthesized by random
primer method using the SuperScript III First Strand Synthesis kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. PCR was performed with cDNA transcribed from 100 ng
total RNA using the AmpliTaq Gold PCR kit (Thermo Fisher
Scientific, Inc.). The primers used were: Forward (F),
5'-TAGGCGCCCCCATGGGCGGCCCAGA-3' and reverse (R),
5'-GGCTGGGAGCGCCGCTGTGT-3' (Fig. 1A).
The thermocycling conditions were as follows: 95˚C for 10 min,
followed by 40 cycles of 95˚C for 1 min, 55˚C for 1 min and 72˚C
for 1 min. The amplified PCR product was cloned into pCR-II TOPO
(Thermo Fisher Scientific, Inc.) using the TA-cloning method.
Cloned ANGPTL8 fragments were sequenced using the BigDye Terminator
v3.1 Cycle Sequencing kit (Thermo Fisher Scientific, Inc.).
Human tissue samples and histological
assessment
FFPE specimens of lipoma, hibernoma and normal human
tissues of the liver, adipose tissue, heart, stomach, small
intestine, large intestine, pancreas, spleen, kidney, lymph node,
bone marrow, thyroid, adrenal gland, ovary and testis were used in
the present study. The normal human tissues were collected adjacent
to malignant tumor tissues in resected organs. Lipomas and
hibernomas were resected tumor tissues. The tissues were fixed in
10% formalin at RT for 24 h and embedded in paraffin. Three
specimens of each normal tissue, lipoma and hibernoma were used for
the study. Tissues were obtained from the archives of pathological
specimens of Nippon Medical School Hospital (Tokyo, Japan) and were
originally obtained between January 2014 and December 2018. The
tissues obtained from 45 cases; median age, 65 (25-81) years. A
total of 21 male and 24 female patients were recruited. The
personal data were anonymized and only pathological diagnoses were
available for the study. This study was performed in accordance
with the principles of the Declaration of Helsinki, 2013 and the
Japanese Society of Pathology, Ethics Committee. The study was
approved by Ethics Committee of Nippon Medical School Hospital
(approval no. 30-11-1304). Written informed consent was obtained
from all patients at the time of hospitalization.
FFPE specimens of lipoma, hibernoma and normal human
tissues were stained with hematoxylin and eosin. Briefly,
4-µm-thick sections were stained with hematoxylin for 5 min and
eosin for 3 min at room temperature. The stained sections were
observed using a light microscope (magnifications, x100, x200 and
x400).
RNA extraction
Total RNA was extracted from FFPE specimens using
the RNeasy FFPE kit (Qiagen, Inc.) according to the manufacturer's
instructions. Briefly, five paraffin sections (4 µm) of a FFPE
specimen were deparaffinized in 1.5-ml tubes. After the sections
were dried, they were digested in PKD buffer with proteinase K at
56˚C until the tissues were completely dissolved. Total RNA was
extracted, and the concentration was measured.
Reverse transcription-quantitative
(RT-q)PCR of wild type ANGPTL8
The wild type ANGPTL8 mRNA (KF809856) was
quantitated in the normal human tissues, lipoma and hibernoma.
Total RNA (200 ng) were extracted from FFPE specimens and were
treated with DNase I (Thermo Fisher Scientific, Inc.) at RT for 15
min. cDNA was synthesized using the SuperScript VILO cDNA Synthesis
kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The primers for wild type ANGPTL8 mRNA
were: F1, 5'-CAGGAACAGCCTGGGTCTCTA-3' and R1,
5'-AGCTGCAGAATATCCTCCTCCAT-3' (Fig.
1A). For standardization, the expression level of 18S rRNA was
quantified using the following primers: Forward,
5'-TAGCCTTTGCCATCACTGCC-3' and reverse,
5'-CACCACAACTCCTTTCGTCTGTAA-3'. The qPCR was performed in 20 µl
reaction solution containing 1X PowerUp SYBR Green Master mix
(Thermo Fisher Scientific, Inc.), 500 nM forward and reverse
primers and cDNA transcribed from 100 ng total RNA as the template.
The reaction was initiated at 50˚C for 2 min and 95˚C for 2 min,
followed by 40 cycles of sequential incubations at 95˚C for 3 sec
and 60˚C for 30 sec. The changes in fluorescence were monitored
using a StepOne Plus Real-Time PCR system (Thermo Fisher
Scientific, Inc.) and quantitation cycles (Cq) were determined. The
expression level of wild type ANGPTL8 in the tissues was calculated
as follows: ΔCq = Cq(wild type)-Cq(18S rRNA); and ΔΔCq =
ΔCq(tissue)-ΔCq(liver). Expression levels were calculated following
the 2-ΔΔCq method (8).
Expression ratio of splice variant
2/wild type ANGPTL8
The splice variant 2 of ANGPTL8 (KF809858) was
quantified in liver, adipose tissue and hibernoma. The splice
variant 2 was quantified using F1 and R2 primers (R2,
5'-TGTGGCTCTGCTTGTCA-3'; Fig. 1A).
RT-qPCR was performed as detailed in the previous section. The
expression ratio of splice variant 2/wild type ANGPTL8 was
determined as follows: ΔCq = Cq(splice variant 2)-Cq(wild type).
The expression ratio was obtained using the 2-ΔCq.
Immunohistochemistry (IHC)
IHC was performed on FFPE sections of normal human
tissues, lipoma and hibernoma. Paraffin sections (4 µm) were
deparaffinized in xylene and ethanol and hydrated in PBS. Sections
were then treated with 10 mM citrate buffer (pH 6.0) at 121˚C for
15 min. Endogenous peroxidase activity was blocked with 0.3%
hydrogen peroxide in methanol at RT for 30 min. After incubation
with 10% normal goat serum (Nichirei Biosciences, Inc.) at RT for
30 min, rabbit anti-ANGPTL8 antibody (cat. no. 7619; 1:500; ProSci,
Inc.) was applied on the sections. In the negative control without
an antibody, PBS was applied to the sections. Sections were
incubated at 4˚C overnight. Subsequently, slides were incubated
with a peroxidase-labeled anti-rabbit immunoglobulin antibody using
the Histofine Simple Stain MAX-PO® (cat. no. 424141;
prediluted; Nichirei Biosciences, Inc.) at RT for 1 h. Peroxidase
activity was detected by incubation with diaminobenzidine at RT for
2 min using Histofine DAB Substrate kit (Nichirei Biosciences,
Inc.) and the sections were counterstained with hematoxylin at RT
for 1 min. The immunostained sections were observed using a light
microscope (magnification, x200 and x400).
In situ hybridization (ISH)
ISH was performed with an ISH Reagent kit (cat. no.
SRK-02; GenoStaff, Co., Ltd.) using FFPE sections (4 µm) of normal
human tissues, lipoma, and hibernoma. Briefly, sections were
deparaffinized, rehydrated in PBS and subsequently incubated in 10%
neutral buffered formalin at RT for 15 min and treated with 20
µg/ml proteinase K (cat. no. S3004; Agilent Technologies, Inc.) in
Tris-HCl (pH 7.6) at 37˚C for 10 min. After incubation in 10%
neutral buffered formalin at RT for 15 min, samples were incubated
in 0.2 N HCl at RT for 10 min. G-Hybo hybridization solution (100
µl; provided with kit) mixed with a digoxigenin-labeled anti-sense
or sense probe was applied to the sections and incubated at 80˚C
for 10 min. The anti-sense and sense probes were synthesized from
the wild type ANGPTL8 using the T7/SP6 Digoxigenin Labeling kit
(Roche Diagnostics, K.K.) according to the manufacturer's
instructions. Following the initial incubation, sections were
incubated at 50˚C overnight and washed twice with 1X Wash Buffer
(provided with kit) at 50˚C for 10 min. Following blocking with
G-Block (provided with kit) at RT for 30 min, sections were further
incubated with an alkaline phosphatase-labeled anti-digoxigenin
antibody (anti-digoxigenin-AP, Fab fragment; cat. no. 11093274910;
1:1,000; Roche Diagnostics) and the enzymatic activity was detected
using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl
phosphate (NBT/BCIP Stock solution; cat. no. 11681451001; Roche
Diagnostics) according to the manufacturer's instructions. Two
types of negative control were used in the present study. In the
first type of negative control, the slides were treated with 100
µg/ml RNase A (Takara Bio, Inc.) in 1X standard saline citrate (150
mM NaCl/15 mM citrate; pH 7.0) at 37˚C for 1 h to remove all
endogenous RNA, and in the second type of negative control, the
slides were incubated with 100 µl G-Hybo without probes at 80˚C for
10 min. Slides were further processed in the same manner as
described. The stained sections were observed using a light
microscope (magnification, x200 and x400).
Statistical analysis
Data are expressed as the mean ± standard deviation
of three samples. The statistical analysis was performed using R
(version 3.6.1; https://www.r-project.org). The statistical
analysis among the organs was conducted using Kruskal Wallis
followed by Dunn's post-hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Sequencing of ANGPTL8 mRNA
Amplification and cloning of ANGPTL8 mRNA identified
three different fragments in the liver (Fig. 1A). One was KF809856, the wild type
mRNA, and KF809857 and KF809858 are two were alternatively spliced
mRNAs. The first splice variant (Splice Variant 1; KF809857) lacked
146 base pairs of exon 1. This deletion caused a frame shift
producing a truncated protein containing 63 amino acids due to a
premature stop codon in exon 2. The second splice variant (Splice
Variant 2; KF809858) lacked 162 base pairs, the entire exon 2, and
generated a short version of ANGPTL8. The splice variants contained
two single nucleotide polymorphisms (SNPs), rs1541922 and
rs192460764 (registered in dbSNP; available at https://www.ncbi.nlm.nih.gov/snp/). The SNP
rs192460764 caused the replacement of Arg to Trp (p.Arg172Trp). The
wild type ANGPTL8 did not contain these SNPs and no further SNPs
were identified.
RT-qPCR of wild type ANGPTL8
The expression level of wild type ANGPTL8 mRNA was
determined in the FFPE tissue specimens. Results showed ANGPTL8
levels were highest in the liver, adipose tissue, lipoma and
hibernoma (Fig. 1B; Table I); levels in the other tissues were
low. The level of ANGPTL8 in the liver was significantly increased
compared with the other samples (P<0.05), except the adipose
tissue, lipoma and hibernoma where differences were not significant
(P>0.05). The levels in the adipose tissue, lipoma and hibernoma
were also higher than other tissues.
 | Table IANGPTL8 protein and mRNA expression in
human tissues. |
Table I
ANGPTL8 protein and mRNA expression in
human tissues.
| | ANGPTL8
expression |
|---|
| Tissue | RT-qPCR
(fold-change) | IHC | ISH |
|---|
| Liver | 1.00±0.01 | + | + |
| Adipose tissue | 0.25±0.04 | + | + |
| Lipoma | 0.38±0.03 | + | + |
| Hibernoma | 0.32±0.08 | + | + |
| Heart |
0.07±0.05a | - | - |
| Stomach |
0.06±0.03a | - | - |
| Small
intestine |
0.03±0.02a,c,d | - | - |
| Large
intestine |
0.03±0.02a,c,d | - | - |
| Pancreas |
0.01±0.01a-d | - | - |
| Spleen |
0.06±0.06a | - | - |
| Kidney |
0.13±0.13a | - | - |
| Lymph node |
0.01±0.01a-d | - | - |
| Bone marrow |
0.06±0.06a,c,d | - | - |
| Thyroid |
0.06±0.02a | - | - |
| Adrenal gland |
0.04±0.04a,c,d | - | - |
| Ovary |
0.03±0.01a,c,d | - | - |
| Testis |
0.01±0.01a-d | - | - |
Expression ratio of splice variant
2/wild type ANGPTL8
The level of ANGPTL8 splice variant 2 mRNA, which
generates a shorter form of ANGPTL8 due to partial lack of
coiled-coiled domains, was quantified in liver, adipose tissue and
hibernoma. The level of splice variant 1, which generates a
truncated form of ANGPTL8 and lacks almost entire coiled-coil
domains, was not quantitated in the present study. The expression
level of the splice variant 2 was below the detection limit in the
adipose tissue and hibernoma, and the variant was detected only in
the liver. The expression level of the splice variant 2 in the four
liver tissues was very low and the relative level was
0.0069±0.0024, compared with the wild type ANGPTL8.
IHC
In the liver, expression of ANGPTL8 was observed in
periportal zone 1 of hepatic acinus (Fig.
2A and B). In high magnification,
the cytoplasm in the hepatocytes showed a positive reaction
(Fig. 2C). The negative control
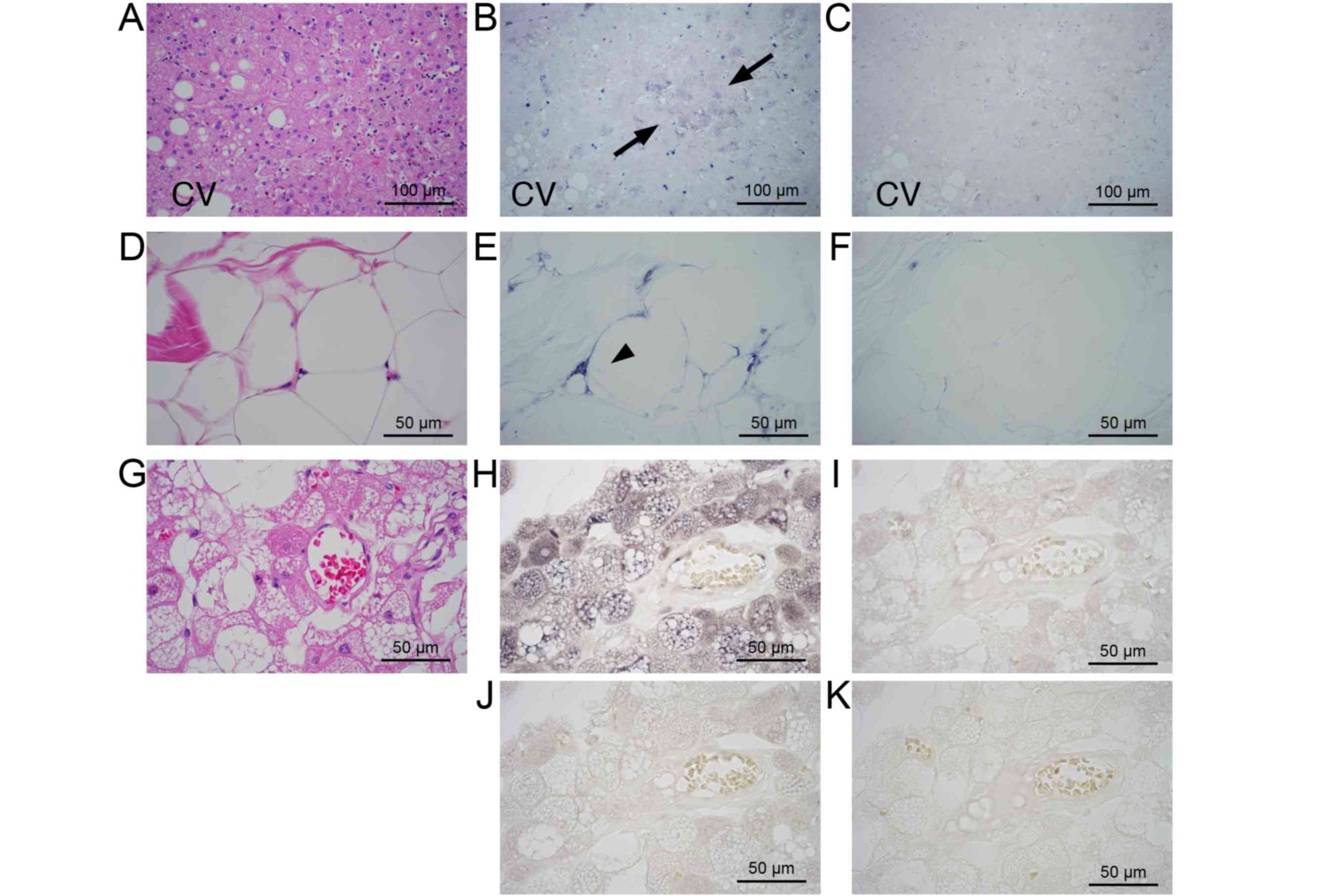
without the primary antibody showed no signal (Fig. 2D). In the adipose tissue, expression
of ANGPTL8 was observed in immature adipocytes (Fig. 2E; arrows). The cytoplasmic rims of
mature adipocytes in the adipose tissue and lipoma were also
stained (Fig. 2E and F). In hibernoma, the foamy cytoplasm of
tumor cells was strongly stained (Fig.
2G). The negative control without the primary antibody showed
no positive staining (Fig. 2H).
Adipose tissue in the bone marrow showed a positive staining
(Fig. 2I). The positive staining was
not observed in the negative control without primary antibody
(Fig. 2J). Positive staining was not
observed in the epithelial cells of the small intestine (Fig. 2K), exocrine tissue and the islets of
the pancreas (Fig. 2L; arrow heads).
Other tissues did not show a positive reaction for ANGPTL8 (data
not shown). The results of IHC are summarized in Table I.
 | Figure 2.ANGPTL8 expression in human tissues by
immunohistochemistry. (A) Hematoxylin & eosin staining of liver
tissues (magnification, x40). ANGPTL8 expression in portal
hepatocytes (B) at magnification, x40 and (C) at higher
magnification of the portal area (magnification, x200). (D)
Negative control liver tissues without primary antibody
(magnification, x200). (E) ANGPTL8 expression in the adipose
tissue; positive staining of small immature adipocytes (arrows;
magnification, x200). (F) ANGPTL8 expression in lipoma
(magnification, x200). (G) ANGPTL8 expression in tumor cells in
hibernoma (magnification, x200). (H) Negative control of hibernoma
without primary antibody (magnification, x200). (I) ANGPTL8
expression in bone marrow (magnification, x200). (J) Negative
control of the bone marrow without primary antibody (magnification,
x200). ANGPTL8 expression in (K) the small intestine or (L)
pancreatic islets (arrow heads; magnification, x100). ANGPTL8,
angiopoietin-like protein 8; CV, central vein; PV, portal vein. |
ISH
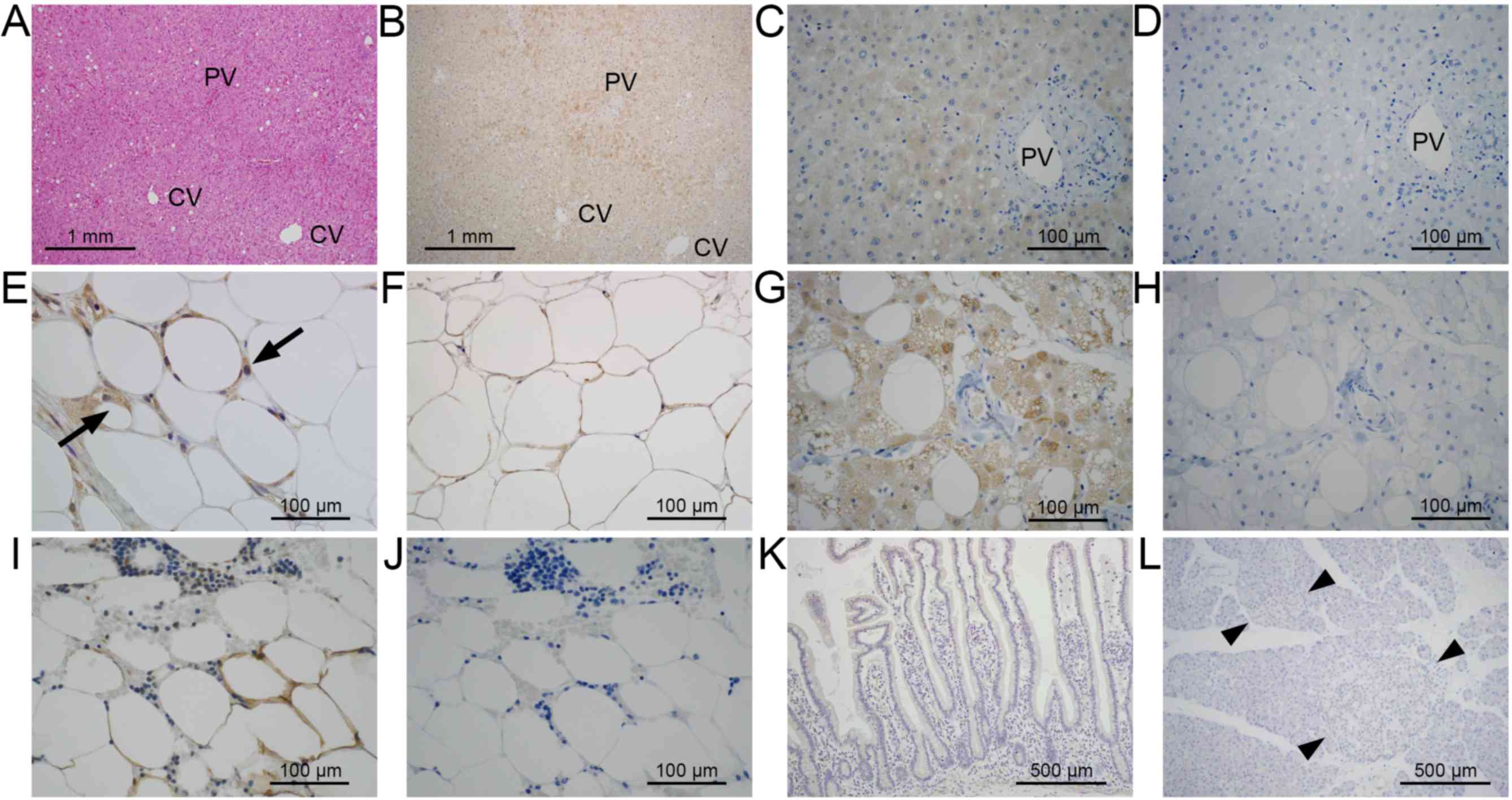
In the liver (Fig.
3A), the cytoplasm of hepatocytes in zone 1 showed a positive
signal with the anti-sense probe (Fig.
3B; arrows), whereas no signal was detected with sense probe
(Fig. 3C). In the adipose tissue
(Fig. 3D), the immature adipocytes
showed a strong signal (Fig. 3E;
arrowheads), and a weak signal was observed in mature adipocytes
(Fig. 3F). In hibernoma (Fig. 3G), the tumor cells showed a positive
reaction in the cytoplasm (Fig. 3H).
No signal was detected in slides hybridized with the sense probe
(Fig. 3I). In the section treated
with RNase and hybridized with anti-sense probe (Fig. 3J) or in the section hybridized with no
probe (Fig. 3K), no signals were
detected. In lipoma, a weak signal was observed in mature tumor
cells with the anti-sense probe, and other tissues did not show a
positive reaction (data not shown). The results of ISH are
summarized in Table I.
Discussion
In the present study, RT-qPCR showed that ANGPTL8
mRNA was expressed in the liver, normal adipose tissue, lipoma and
hibernoma. These results are consistent with previous findings,
which suggest ANGPTL8 is abundantly expressed in the liver, adipose
tissue and BAT (2,5,6). The
localization data of ANGPTL8-positive cells by IHC and ISH in these
tissues were comparable, suggesting the specific reactivity of the
antibody used in the current study. The present study revealed the
precise localization of ANGPTL8-expressing cells in the liver and
adipose tissue. The expression of ANGPTL8 was homogenous in
hibernoma. The distribution of ANGPTL8-expressing cells in the
liver and adipose tissue was heterogeneous; there was a difference
in the expression levels among the cells in the organs.
In the liver, ANGPTL8 expression was observed in the
periportal hepatocytes of the hepatic acinus. The hepatic acinus is
separated into three functional zones, and hepatocytes in the
periportal zone 1 actively uptake triglycerides and utilize them
through β-oxidation (9). ANGPTL8 is
expressed in hepatocytes, which actively metabolize triglycerides
under physiological conditions. Increased expression of ANGPTL8 has
previously been detected in cultured cells of hepatocellular
carcinoma (3). The expression of
ANGPTL8 may reflect the metabolic state of carcinoma cells. Serum
levels of ANGPTL8 have been shown to correlate with liver steatosis
(10) and non-alcoholic fatty liver
disease (11). The association of the
precise localization of ANGPTL8-expressing hepatocytes with lipid
deposition requires to be elucidated.
In adipose tissue, ANGPTL8 was more strongly
expressed in immature adipocytes compared with mature adipocytes.
This is consistent with previous findings on the expression of
ANGPTL8 in preadipocytes, which differentiate into mature
adipocytes (2). ANGPTL8 expression
may be necessary for the storage of triglycerides. Previous studies
reported that the knockdown of ANGPTL8 by siRNA in cultured
adipocytes does not affect differentiation, but alters the
expression levels of enzymes involved in lipolysis and fatty acid
oxidation, resulting in a reduction in triglyceride levels
(2,12). Decreases in body weight and fatty
tissue have been reported in ANGPTL8 knockout mice (7). The results in the present study further
suggested an important role for ANGPTL8 in lipid metabolism by
immature adipocytes.
A treatment with an ANGPTL8 antisense
oligonucleotide has been reported to promote the uptake and storage
of triglycerides in adipose tissue and eventually prevent liver
steatosis, and this has been associated with improvements in
glucose intolerance in rodents (13).
In this situation, immature adipocytes may be a therapeutic target
of antisense oligonucleotides against ANGPTL8. Immature adipocytes
release various metabolic factors, such as leptin and adiponectin
(14), and the expression of enzymes
associated with the lipid metabolism and leptin are upregulated in
adipocytes with ANGPTL8 knockdown (12). Therefore, it is conceivable that the
prevention of steatosis and improvements in glucose metabolism was,
in part, attributed to the modulation of metabolic factors by
ANGPTL8 knockdown in immature adipocytes.
In the present study, we identified two splice
variants of ANGPTL8 in the liver, which resulted in production of
truncated forms of ANGPTL8 protein. These splice variants contain a
conserved region of specific epitope 1, which is necessary for the
inhibition of LPL. However, the coiled-coil domains, which are
necessary for binding with ANGPTL3 (15,16), were
deleted. The inhibitory effect of LPL by ANGPTL8 requires complex
formation with ANGPTL3 (17-19).
ANGPTL8 further forms a complex with ANGPTL4, inactivating the
inhibitory effect of ANGPTL4 on LPL (19). The almost complete deletion of
C-terminal coiled-coil domains of ANGPTL8, like splice variant 1,
may loose the ability to form complex. The partial deletion of the
C-terminal coiled-coil domain of ANGPTL8, like splice variant 2,
may interfere the complex formation with ANGPTL3 and ANGPTL4 and
affect the partitioning of lipids by modulating the inhibitory
effect on LPL. Although the expression level of splice variant 2
was very low in the normal liver, the expression level of ANGPTL8
in metabolic and liver diseases needs to be elucidated.
The splice variants assessed in the present study
contained two SNPs (rs1541922 and rs192460764). SNP rs192460764
caused the replacement of an amino acid (p.Arg172Trp). A previous
study reported that another SNP rs2278426, causing the same
replacement (p.Arg59Trp), is associated with ethnic and
gender-specific differences in lipid levels in the Chinese
population (20). The frequency of
this SNP is high in patients with type 2 diabetes in Japan
(21). The biological significance of
SNPs of ANGPTL8 in lipid metabolism and metabolic diseases needs to
be clarified.
An increase in serum levels of ANGPTL8 was reported
in patients with type 1 and type 2 diabetes (22,23), as
well as gestational diabetes (24).
It was reported that ANGPTL8 stimulates the proliferation of
β-cells in the pancreatic islet (5);
however, the biological activity is not reproduced (25,26). The
increase in ANGPTL8 in the serum is considered to be associated
with lipid dysmetabolism in diabetes (27). A direct association between ANGPTL8
expression and β-cell proliferation was not evident in the present
study.
The present study demonstrated high expression
levels of ANGPTL8 and detailed the precise localization of the
cells expressing ANGPTL8 in the liver and adipose tissue. The
expression was associated with the activity of lipid metabolism and
differentiation state in these organs. The modulation of metabolism
and partitioning of triglyceride by the alteration of ANGPTL8 by
anti-sense oligonucleotide may be a novel therapy for lipid
dysmetabolism or steatohepatitis. The localization of
ANGPTL8-expressing cells in the liver and adipose tissue may aid
the understanding of partitioning of lipids and development of
novel therapies targeting the lipid metabolism.
Acknowledgements
The authors would like to acknowledge the excellent
assistance of Ms. Kiyoko Kawahara and Mr. Takenori Fujii for help
with ISH, Mr. Kiyoshi Teduka and Ms. Yoko Kawamoto for help with
IHC and Ms. Taeko Kitamura for help with the subcloning and RT-qPCR
(Department of Integrated Diagnostic Pathology, Nippon Medical
School, Tokyo, Japan).
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
available from the corresponding author on reasonable request.
Authors' contributions
NA, RW and ZN designed the study and wrote the
manuscript. NA performed histological examinations. NA and RW
conducted biochemical examinations, data analyses and statistical
analyses. NA prepared the figures and table. NA and KI provided
clinical data of the patients and assisted with revising the
manuscript. KI and ZN supervised the experimental design and
manuscript writing. All authors read and approved the final
manuscript. All authors agree to be accountable for all aspects of
the work in ensuring that questions related to the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the Nippon Medical School Hospital (Tokyo, Japan; approval no.
30-11-1304). Informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li Y and Teng C: Angiopoietin-like
proteins 3, 4 and 8 regulating lipid metabolism and providing new
hope for metabolic syndrome. J Drug Target. 22:679–687.
2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ren G, Kim JY and Smas CM: Identification
of RIFL, a novel adipocyte-enriched insulin target gene with a role
in lipid metabolism. Am J Physiol Endocrinol Metab. 303:E334–E351.
2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Dong XY, Pang XW, Yu ST, Su YR, Wang HC,
Yin YH, Wang YD and Chen WF: Identification of genes differentially
expressed in human hepatocellular carcinoma by a modified
suppression subtractive hybridization method. Int J Cancer.
112:239–248. 2004.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Fu Z, Yao F, Abou-Samra AB and Zhang R:
Lipasin, thermoregulated in brown fat, is a novel but atypical
member of the angiopoietin-like protein family. Biochem Biophys Res
Commun. 430:1126–1131. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yi P, Park JS and Melton DA: Betatrophin:
a hormone that controls pancreatic β cell proliferation. Cell.
153:747–758. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Quagliarini F, Wang Y, Kozlitina J,
Grishin NV, Hyde R, Boerwinkle E, Valenzuela DM, Murphy AJ, Cohen
JC and Hobbs HH: Atypical angiopoietin-like protein that regulates
ANGPTL3. Proc Natl Acad Sci USA. 109:19751–19756. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wang Y, Quagliarini F, Gusarova V, Gromada
J, Valenzuela DM, Cohen JC and Hobbs HH: Mice lacking ANGPTL8
(Betatrophin) manifest disrupted triglyceride metabolism without
impaired glucose homeostasis. Proc Natl Acad Sci USA.
110:16109–16114. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hijmans BS, Grefhorst A, Oosterveer MH and
Groen AK: Zonation of glucose and fatty acid metabolism in the
liver Mechanism and metabolic consequences. Biochimie. 96:121–129.
2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
von Loeffelholz, Pfeiffer AFH, Lock JF,
Lieske S, Docke S, Murahovschi V, Kriebel J, de Las Heras Gala,
Grallert H, Rudovich N, et al: ANGPTL8 (Betatrophin) is expressed
in visceral adipose tissue and relates to human hepatic steatosis
in two independent clinical collectives. Horm Metab Res.
49:343–349. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lee YH, Lee SG, Lee CJ, Kim SH, Song YM,
Yoon MR, Jeon BH, Lee JH, Lee BW, Kang ES, et al: Association
between betatrophin/ANGPTL8 and non-alcoholic fatty liver disease
Animal and human studies. Sci Rep. 6(24013)2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mysore R, Liebisch G, Zhou Y, Olkkonen VM
and Nidhina Haridas PA: Angiopoietin-like 8 (Angptl8) controls
adipocyte lipolysis and phospholipid composition. Chem Phys Lipids.
207:246–252. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vatner DF, Goedeke L, Camporez JG, Lyu K,
Nasiri AR, Zhang D, Bhanot S, Murray SF, Still CD, Gerhard GS, et
al: Angptl8 antisense oligonucleotide improves adipose lipid
metabolism and prevents diet-induced NAFLD and hepatic insulin
resistance in rodents. Diabetologia. 61:1435–1446. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yu YH and Zhu H: Chronological changes in
metabolism and functions of cultured adipocytes A hypothesis for
cell aging in mature adipocytes. Am J Physiol Endocrinol Metab.
286:E402–E410. 2004.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Siddiqa A, Ahmad J, Ali A, Paracha RZ,
Bibi Z and Aslam B: Structural characterization of ANGPTL8
(betatrophin) with its interacting partner lipoprotein lipase.
Comput Biol Chem. 61:210–220. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Tseng YH, Yeh YH, Chen WJ and Lin KH:
Emerging regulation and function of betatrophin. Int J Mol Sci.
15:23640–23657. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Haller JF, Mintah IJ, Shihanian LM, Stevis
P, Buckler D, Alexa-Braun CA, Kleiner S, Banfi S, Cohen JC, Hobbs
HH, et al: ANGPTL8 requires ANGPTL3 to inhibit lipoprotein lipase
and plasma triglyceride clearance. J Lipid Res. 58:1166–1173.
2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Nidhina Haridas PA, Soronen J, Sadevirta
S, Mysore R, Quagliarini F, Pasternack A, Metso J, Perttila J,
Leivonen M, Smas CM, et al: Regulation of angiopoietin-like
proteins (ANGPTLs) 3 and 8 by insulin. J Clin Endocrinol Metab.
100:E1299–E1307. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Kovrov O, Kristensen KK, Larsson E, Ploug
M and Olivecrona G: On the mechanism of angiopoietin-like protein 8
for control of lipoprotein lipase activity. J Lipid Res.
60:783–793. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Guo T, Yin RX, Wu J, Lin QZ, Shi GY, Shen
SW, Sun JQ, Li H, Lin WX and Yan DZ: Association of the
angiopoietin-like protein 8 rs2278426 polymorphism and several
environmental factors with serum lipid levels. Mol Med Rep.
12:3285–3296. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liu J, Yagi K, Nohara A, Chujo D, Ohbatake
A, Fujimoto A, Miyamoto Y, Kobayashi J and Yamagishi M: High
frequency of type 2 diabetes and impaired glucose tolerance in
Japanese subjects with the angiopoietin-like protein 8 R59W
variant. J Clin Lipidol. 12:331–337. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Espes D, Lau J and Carlsson PO: Increased
circulating levels of betatrophin in individuals with long-standing
type 1 diabetes. Diabetologia. 57:50–53. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hu H, Sun W, Yu S, Hong X, Qian W, Tang B,
Wang D, Yang L, Wang J, Mao C, et al: Increased circulating levels
of betatrophin in newly diagnosed type 2 diabetic patients.
Diabetes Care. 37:2718–2722. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ebert T, Kralisch S, Wurst U, Lössner U,
Kratzsch J, Blüher M, Stumvoll M, Tönjes A and Fasshauer M:
Betatrophin levels are increased in women with gestational diabetes
mellitus compared to healthy pregnant controls. Eur J Endocrinol.
173:1–7. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gusarova V, Alexa CA, Na E, Stevis PE, Xin
Y, Bonner-Weir S, Cohen JC, Hobbs HH, Murphy AJ, Yancopoulos GD and
Gromada J: ANGPTL8/betatrophin does not control pancreatic beta
cell expansion. Cell. 159:691–696. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yi P, Park JS and Melton DA: Retraction
notice to Betatrophin A hormone that controls pancreatic β cell
proliferation. Cell. 168(326)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Fenzl A, Itariu BK, Kosi L,
Fritzer-Szekeres M, Kautzky-Willer A, Stulnig TM and Kiefer FW:
Circulating betatrophin correlates with atherogenic lipid profiles
but not with glucose and insulin levels in insulin-resistant
individuals. Diabetologia. 57:1204–1208. 2014.PubMed/NCBI View Article : Google Scholar
|