Introduction
Papillary thyroid carcinoma (PTC) is the main
histological type of thyroid cancer, accounting for >80% of all
thyroid malignancies (1). Although
most patients with PTC have low recurrence rate and exhibit optimal
prognosis as determined by long term follow-up, approximately 15%
of patients demonstrate aggressive behavior and poor outcome
(2,3).
To date, the pathogenesis of PTC has not been fully identified.
Therefore, detailed investigation of the biological processes
involved in PTC can provide insight to the diagnosis and
pathogenesis of this disease and to its potential treatment.
MicroRNAs (miRNAs) are a class of short
(approximately 22 nucleotides), non-coding and endogenous RNA
molecules that can suppress the expression of the corresponding
protein coding genes by binding to the 3'-untranslated region
(3'-UTR) of the target mRNAs (4,5). Previous
studies have suggested that miRNAs can function as regulators of
oncogenes or of tumor suppressor genes and are involved in the
occurrence and development of cancer (6-10).
However, due to the limitations of the traditional biomolecular
detection methods, most of the previous studies focused only on the
expression and function of single miRNAs in cancer. The application
of high-throughput detection technologies used for miRNA
expression, such as miRNA expression microarray and miRNA
sequencing has enabled the generation of an increasing number of
miRNA expression profiles and their deposition in the public
databases, such as Gene Expression Omnibus (GEO) and The Cancer
Genome Atlas (TCGA). By analyzing these miRNA expression profiles,
a comprehensive understanding of the expression levels of specific
miRNAs can be attained in a biological sample at a given
moment.
In the present study, high-throughput miRNA
expression profiles were obtained from the GEO database
(corresponding to PTC patients) and key miRNAs in PTC were
subsequently identified. Furthermore, the role of specific key
miRNAs in the development of PTC was further revealed by
investigating the potential function of their associated target
genes.
Materials and methods
miRNA expression profiles
Two independent microarray datasets (GSE73182 and
GSE113629) were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). GSE73182
included miRNA expression profiles of 19 primary PTCs and five
normal thyroid samples, which were generated using the Agilent
microarray platform (GPL20194) (11).
GSE113629 included miRNA expression profiles of five pairs of PTC
and normal thyroid tissues, which were generated using the Agilent
microarray platform (GPL24741) (12).
Data analysis
The GEO2R tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to
identify the differentially expressed miRNAs (DEmiRNAs) between PTC
and normal thyroid tissues. Multiple-testing corrections were
performed based on the Benjamini & Hochberg method in order to
correct for false-positive results. The miRNAs with the adjusted
P<0.05 and with a |logFC|>1 were selected as DEmiRNAs.
Identification of key miRNAs in
PTC
TheVennDiagram R package was used to identify common
DEmiRNAs in two independent datasets (13). These common DEmiRNAs were identified
as key miRNAs involved in PTC. Furthermore, the Human Cancer
Metastasis Database (HCMDB; http://hcmdb.i-sanger.com/index) was used to analyze
large-scale expression data of cancer metastasis. miRNA deep
sequencing data of three PTC and three matched normal tissues was
used to confirm the expression levels of the key miRNAs (14).
miRNA-target prediction
The miRWalk tool (http://mirwalk.umm.uni-heidelberg.de/) integrating
three prediction tools (miRDB, TargetScan and miRTarBase) was used
to predict the target genes of key miRNAs involved in PTC (15). The genes that were predicted
simultaneously by the three tools were identified as miRNA target
genes. The cytoscape software was used to construct the
miRNA-target gene interaction networks (16).
Functional annotation
To analyze the role of key miRNAs in PTC, Gene
Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analysis were performed using ClusterProfiler
package for the miRNA target genes (17). The adjusted P<0.05 was considered
to indicate statistically significant difference.
Results
Identification of DEmiRNAs
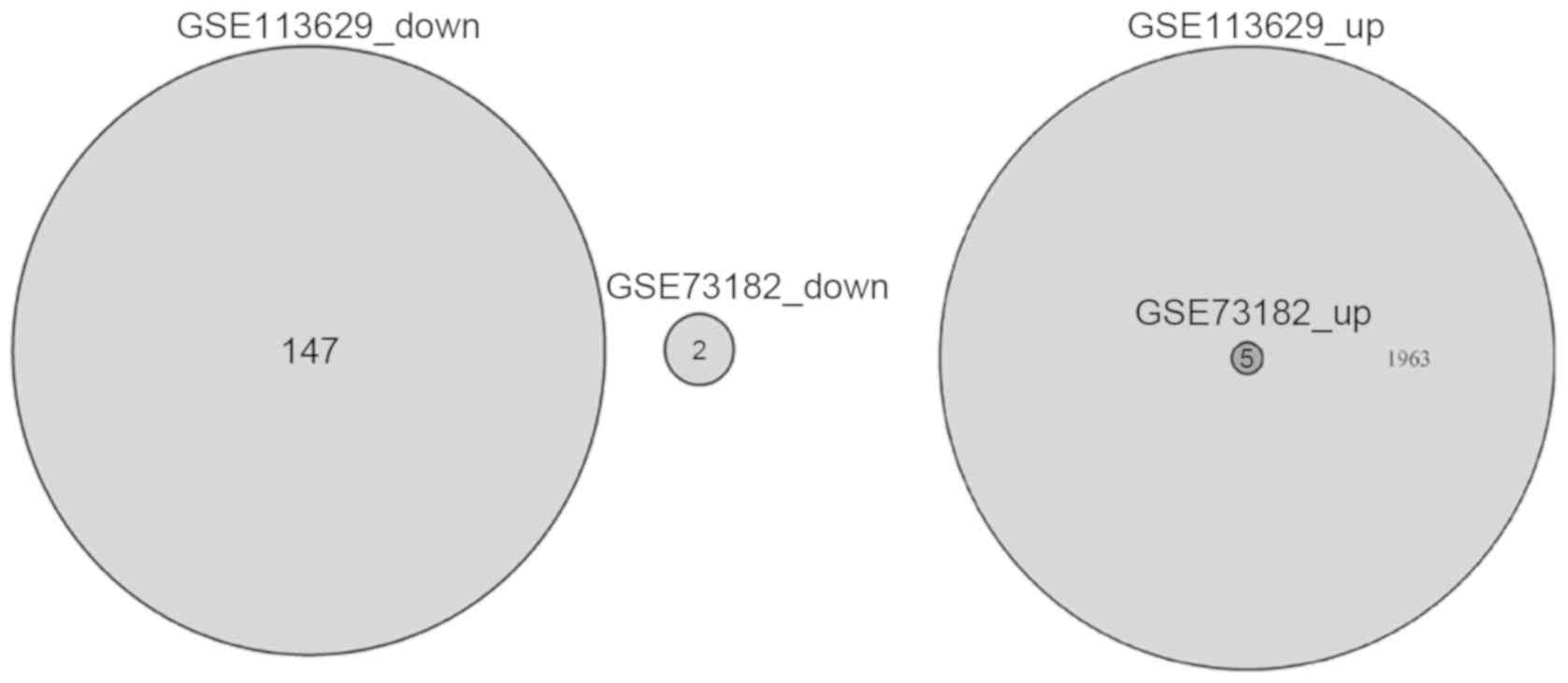
In the GSE113629 dataset 2,115 DEmiRNAs including
1,968 up- and 147 downregulated miRNAs were identified in PTC.
Furthermore, the analysis indicated that 1,963 miRNAs were
upregulated only in the GSE113629 dataset. In the GSE73182 dataset,
seven DEmiRNAs including five up- and two downregulated miRNAs were
identified in PTC. Moreover, five miRNAs (hsa-miR-146b-5p,
hsa-miR-15a-5p, hsa-miR-21-5p, hsa-miR-221-3p and hsa-miR-222-3p)
were upregulated in both the GSE113629 and GSE73182 datasets, which
were identified as key miRNAs in PTC (Figs. 1 and 2
and Table I).
 | Table ISummary for key miRNAs in papillary
thyroid carcinoma. |
Table I
Summary for key miRNAs in papillary
thyroid carcinoma.
| miRBase | miRBase | miRBase | miRNA |
|---|
| hsa-miR-221-3p | hsa-miR-221 | MIMAT0000278 |
5'-AGCUACAUUGUCUGCUGGGUUUC-3' |
| hsa-miR-146b-5p | hsa-miR-146b | MIMAT0002809 |
5'-UGAGAACUGAAUUCCAUAGGCU-3' |
| hsa-miR-222-3p | hsa-miR-222 | MIMAT0000279 |
5'-AGCUACAUCUGGCUACUGGGU-3' |
| hsa-miR-21-5p | hsa-miR-21 | MIMAT0000076 |
5'-UAGCUUAUCAGACUGAUGUUGA-3' |
| hsa-miR-15a-5p | hsa-miR-15a | MIMAT0000068 |
5'-UAGCAGCACAUAAUGGUUUGUG-3' |
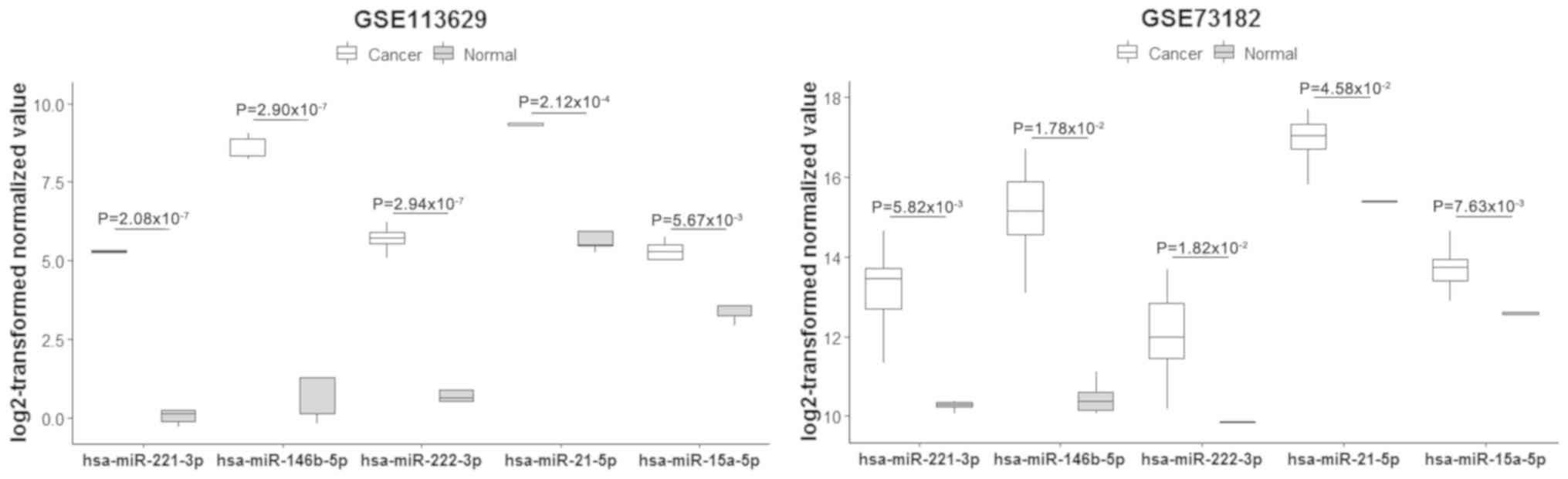
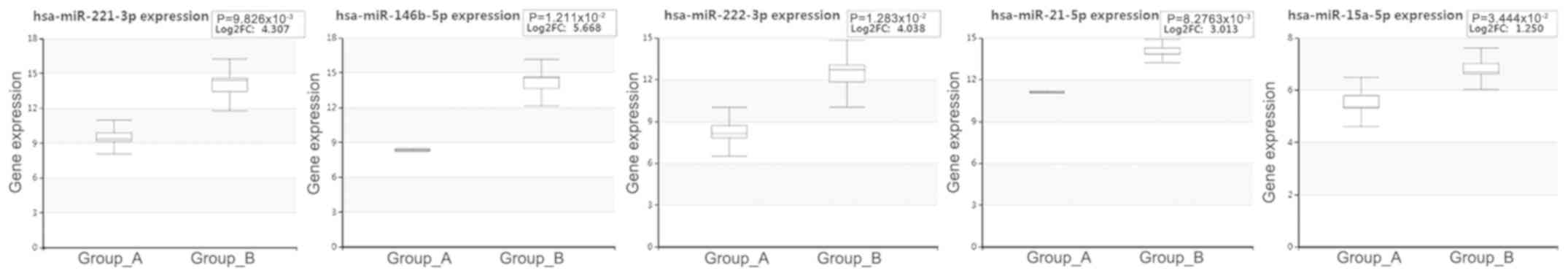
To validate the expression levels of these miRNAs in
PTC, the dataset (Exp ID: EXP00227) collected by HCMDB was analyzed
(Fig. 3). The results indicated that
the expression levels of all these miRNAs were upregulated in PTC
compared with those of the normal tissues, which was consistent
with previous microarray results (11,12).
Potential function of of key
miRNAs
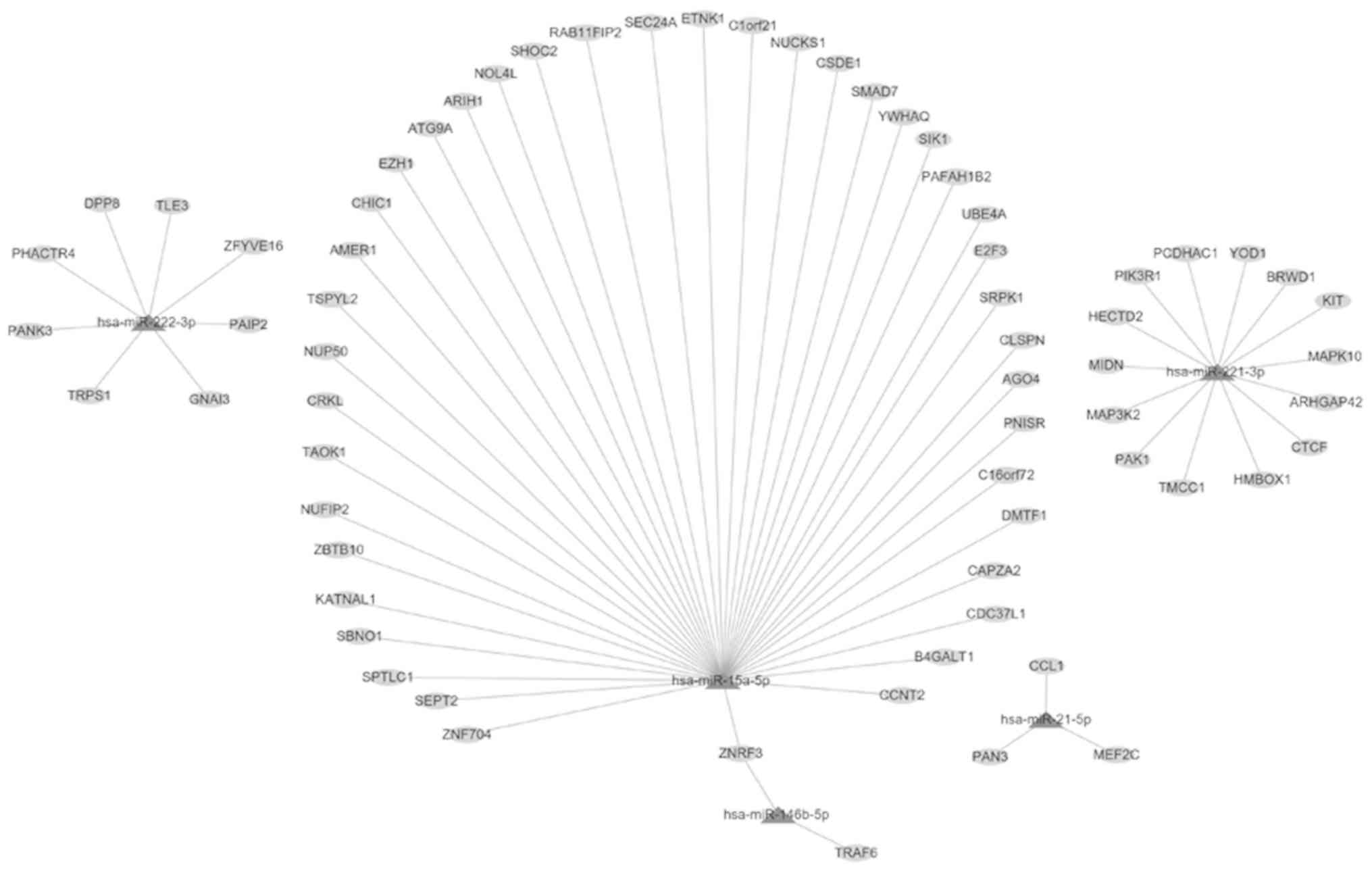
To reveal the potential function of the key miRNAs,
the prediction of their target genes was initially performed by the
miRWalk tool. The results indicated that hsa-miR-146b-5p exhibited
two target genes (ZNRF3 and TRAF6), whereas hsa-miR-15a-5p
exhibited 41 target genes (including ZNF704, SEPT2,
SPTLC1, SBNO1, KATNAL1, ZBTB10,
NUFIP2, TAOK1 and CRKL). hsa-miR-21-5p
exhibited three target genes (PAN3, CCL1 and
MEF2C), hsa-miR-221-3p 14 target genes (including
TMCC1, PAK1, MAP3K2, MIDN,
HECTD2, PIK3R1 and PCDHAC1) and hsa-miR-222-3p
eight target genes (including TRPS1, PANK3,
PHACTR4, DPP8, TLE3 and ZFYVE16)
(Fig. 4). Subsequently, GO analysis
demonstrated that the target genes of these key miRNAs were
significantly enriched in nine biological processes [regulation of
MAP kinase activity, Janus N-terminal kinase (JNK) cascade
signaling and regulation of protein serine/threonine kinase
activity] (Table II). KEGG pathway
analysis indicated that the target genes of the key miRNAs
identified were significantly enriched in 28 pathways, including
the MAPK, the sphingolipid, the ErbB, the Ras and the C-type lectin
receptor signaling pathways (Table
III).
 | Table IIBiological processes enrichment
analysis of microRNA targets. |
Table II
Biological processes enrichment
analysis of microRNA targets.
| ID | Description | P.adjust |
|---|
| GO:0032147 | Activation of protein
kinase activity | 1.75x10 |
| GO:0043406 | Positive regulation
of MAP kinase activity | 1.85x10 |
| GO:0007254 | JNK cascade | 3.23x10 |
| GO:0071902 | Positive regulation
of protein serine/threonine kinase activity | 3.23x10 |
| GO:0043507 | Positive regulation
of JUN kinase activity | 3.23x10 |
| GO:0043405 | Regulation of MAP
kinase activity | 3.23x10 |
| GO:0000187 | Activation of MAPK
activity | 3.40x10 |
| GO:0071900 | Regulation of protein
serine/threonine kinase activity | 4.03x10 |
| GO:0043506 | Regulation of JUN
kinase activity | 4.03x10 |
 | Table IIIKyoto Encyclopedia of Genes and
Genomes pathway enrichment analysis of microRNA targets. |
Table III
Kyoto Encyclopedia of Genes and
Genomes pathway enrichment analysis of microRNA targets.
| ID | Description | P.adjust |
|---|
| hsa04010 | MAPK signaling
pathway | 1.12x10 |
| hsa04071 | Sphingolipid | 1.41x10 |
| hsa04012 | ErbB | 2.12x10 |
| hsa05170 | Human
immunodeficiency virus 1 infection | 3.7810 |
| hsa04014 | Ras | 4.80x10 |
| hsa04140 | Autophagy -
animal | 7.25x10 |
| hsa04625 | C-type | 1.47x10 |
| hsa05161 | Hepatitis B | 1.65x10 |
| hsa05142 | Chagas | 1.66x10 |
| hsa04137 | Mitophagy | 1.73x10 |
| hsa05120 | Epithelial cell
signaling in | 2.10x10 |
| hsa04722 | Neurotrophin | 2.26x10 |
| hsa05133 | Pertussis | 2.26x10 |
| hsa04062 | Chemokine signaling
pathway | 2.26x10 |
| hsa05211 | Renal cell
carcinoma | 2.35x10 |
| hsa05212 | Pancreatic
cancer | 2.55x10 |
| hsa05418 | Fluid shear stress
and atherosclerosis | 2.75x10 |
| hsa04510 | Focal adhesion | 3.02x10 |
| hsa04914 |
Progesterone-mediated oocyte
maturation | 3.23x10 |
| hsa04666 | Fc | 3.23x10 |
| hsa04024 | cAMP | 3.25x10 |
| hsa00600 | Sphingolipid | 3.25x10 |
| hsa04620 | Toll-like receptor
signaling pathway | 3.25x10 |
| hsa04912 | GnRH | 3.34x10 |
| hsa04930 | Type II diabetes
mellitus | 3.60x10 |
| hsa01522 | Endocrine
resistance | 3.70x10 |
| hsa04141 | Protein processing
in endoplasmic reticulum | 3.70x10 |
| hsa04622 | RIG-I-like receptor
signaling pathway | 4.47x10 |
Discussion
Bioinformatic analysis for microarray and RNA-seq
has been widely used recently to identify potential targets for the
diagnosis and therapy of different cancer types (18-20).
Xia et al (12) demonstrated
that promoter DNA methylation caused silencing of miR-204 that
could serve as a potential diagnostic biomarker of PTC based on
miRNA expression profiling (GSE113629). Minna et al
(11) highlighted that miR-451a
expression was downregulated and that it targeted the protein
kinase B/mammalian target of rapamycin pathway in PTC based on
miRNA expression profiling (GSE73182). In the present study, the
two sets of data were analyzed by bioinformatic methods and five
key miRNAs (hsa-miR-146b-5p, hsa-miR-15a-5p, hsa-miR-21-5p,
hsa-miR-221-3p and hsa-miR-222-3p) were identified in PTC. The
expression levels of these key miRNAs were increased in PTC
compared with those noted in normal tissues, which was also
confirmed by an RNA-seq dataset. By reviewing the relevant
literature, the data demonstrated the carcinogenic or
cancer-promoting effects of hsa-miR-146b-5p and hsa-miR-222-3p in
PTC. For instance, hsa-miR-146b-5p expression was associated with
advanced PTC stage and promoted the development of this disease by
targeting CCDC6 in vitro and in vivo, which may serve
as a promising target for PTC treatment (21). The enhanced expression of miR-222-3p
promoted the proliferation of PTC cells, while miR-222-3p knockdown
inhibited it (22). Although no
direct functional study has been conducted to date on
hsa-miR-15a-5p or hsa-miR-21-5p in PTC, the findings by Jiang et
al (23) and Zhang et al
(24) indicated that these two miRNAs
may exert tumor-suppressive effects in PTC. These results are
contradictory to the present findings possibly due to tumor
heterogeneity or due to the opposing function of these two miRNAs
in the development of PTC. The upregulated expression of
hsa-miR-221-3p in PTC has also been reported in a previous study
(25). However, no relevant
functional experiment was performed previously to the best of our
knowledge. Therefore, further studies are required to explore the
function of these key miRNAs in PTC, notably of hsa-miR-15a-5p,
hsa-miR-21-5p and hsa-miR-221-3p. Chen et al (26) explored several types of RNAs involved
in the development of PTC based on TCGA and identified 30
differentially expressed miRNAs between PTC and normal samples.
However, hsa-miR-15a-5p, hsa-miR-21-5p, hsa-miR-221-3p and
hsa-miR-222-3p were not reported in that study (26).
Considering that the miRNA function is mainly
dependent on its target gene, the target genes of the key miRNAs
investigated in the present study were predicted by miRWalk. A
total of 68 genes were identified as targets of these key miRNAs.
Subsequently, GO and KEGG pathway enrichment analysis based on
these target genes was performed to indicate the function of these
key miRNAs in PTC. The results indicated that the key miRNAs could
significantly influence nine biological processes and 28 signaling
pathways, such as the regulation of mitogen associated protein
kinase (MAPK) activity, JNK cascade signaling, the MAPK signaling
pathway, the sphingolipid signaling pathway, the ErbB signaling
pathway and the Ras signaling pathway. These observations can aid
the understanding of the pathogenesis of PTC.
Although the present study identified key miRNAs in
PTC based on data mining and bioinformatic methods, the following
limitation must be highlighted: Functional experiments are required
in future studies to elucidate the mechanism of action of the key
miRNAs identified in PTC.
In conclusion, findings of the present study have
revealed a group of miRNAs associated with PTC. Therefore,
hsa-miR-146b-5p, hsa-miR-15a-5p, hsa-miR-21-5p, hsa-miR-221-3p as
well as hsa-miR-222-3p may be regarded as promising diagnostic
biomarkers and therapeutic targets for PTC.
Acknowledgements
Not applicable.
Funding
The present study was supported by Public Projects
of Zhejiang Province (grant no. GF18H040021).
Availability of data and materials
The datasets analyzed in the present study are all
available on NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
Authors' contributions
JW, SL and XL designed the study. JW and LW wrote
the manuscript. JW, LW and YJ performed the bioinformatics
analysis. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gimm O: Thyroid cancer. Cancer Lett.
163:143–156. 2001.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Maia FF, Vassallo J, Pinto GA, Pavin EJ,
Matos PS and Zantut-Wittmann DE: Expression of Mcl-1 and Ki-67 in
papillary thyroid carcinomas. Exp Clin Endocrinol Diabetes.
124:209–214. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wen Q, Zhao L, Wang T, Lv N, Cheng X,
Zhang G and Bai L: LncRNA SNHG16 drives proliferation and invasion
of papillary thyroid cancer through modulation of miR-497. Onco
Targets Ther. 12:699–708. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Feng B, Dong TT, Wang LL, Zhou HM, Zhao
HC, Dong F and Zheng MH: Colorectal cancer migration and invasion
initiated by microRNA-106a. PLoS One. 7(e43452)2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhu Z, Yang Q, Zhang B, Wu W, Yuan F and
Zhu Z: miR-106b promotes metastasis of early gastric cancer by
targeting ALEX1 in vitro and in vivo. Cell Physiol Biochem.
52:606–616. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yin Y, Hong S, Yu S, Huang Y, Chen S, Liu
Y, Zhang Q, Li Y and Xiao H: MiR-195 inhibits tumor growth and
metastasis in papillary thyroid carcinoma cell lines by targeting
CCND1 and FGF2. Int J Endocrinol. 2017(6180425)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang D, Wang H, Li Y and Li Q: MiR-362-3p
functions as a tumor suppressor through targeting MCM5 in cervical
adenocarcinoma. Biosci Rep. 38(pii: BSR20180668)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Park YR, Seo SY, Kim SL, Zhu SM, Chun S,
Oh JM, Lee MR, Kim SH, Kim IH, Lee SO, et al: MiRNA-206 suppresses
PGE2-induced colorectal cancer cell proliferation, migration, and
invasion by targetting TM4SF1. Biosci Rep. 38:2018.pii:
BSR20180664. PubMed/NCBI View Article : Google Scholar
|
|
11
|
Minna E, Romeo P, Dugo M, De Cecco L,
Todoerti K, Pilotti S, Perrone F, Seregni E, Agnelli L, Neri A, et
al: miR-451a is underexpressed and targets AKT/mTOR pathway in
papillary thyroid carcinoma. Oncotarget. 7:12731–12747.
2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xia F, Wang W, Jiang B, Chen Y and Li X:
DNA methylation-mediated silencing of miR-204 is a potential
prognostic marker for papillary thyroid carcinoma. Cancer Manag
Res. 11:1249–1262. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Chen H and Boutros PC: VennDiagram: A
package for the generation of highly-customizable Venn and Euler
diagrams in R. BMC Bioinformatics. 12(35)2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Saiselet M, Gacquer D, Spinette A, Craciun
L, Decaussin-Petrucci M, Andry G, Detours V and Maenhaut C: New
global analysis of the microRNA transcriptome of primary tumors and
lymph node metastases of papillary thyroid cancer. BMC Genomics.
16(828)2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Sticht C, De La Torre C, Parveen A and
Gretz N: miRWalk: An online resource for prediction of microRNA
binding sites. PLoS One. 13(e0206239)2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gao X, Wang J and Zhang S: Integrated
bioinformatics analysis of hub genes and pathways in anaplastic
thyroid carcinomas. Int J Endocrinol. 2019(9651380)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gao X, Wang X and Zhang S: Bioinformatics
identification of crucial genes and pathways associated with
hepatocellular carcinoma. Biosci Rep. 38(pii:
BSR20181441)2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang S, Wang Q, Han Q, Han H and Lu P:
Identification and analysis of genes associated with papillary
thyroid carcinoma by bioinformatics methods. Biosci Rep. 39(pii:
BSR20190083)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jia M, Shi Y, Li Z, Lu X and Wang J:
MicroRNA-146b-5p as an oncomiR promotes papillary thyroid carcinoma
development by targeting CCDC6. Cancer Lett. 443:145–156.
2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang XF, Ye Y and Zhao SJ: LncRNA Gas5
acts as a ceRNA to regulate PTEN expression by sponging miR-222-3p
in papillary thyroid carcinoma. Oncotarget. 9:3519–3530.
2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jiang L, Wu Z, Meng X, Chu X, Huang H and
Xu C: LncRNA HOXA-AS2 facilitates tumorigenesis and progression of
papillary thyroid cancer by modulating the miR-15a-5p/HOXA3 axis.
Hum Gene Ther. 30:618–631. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang H, Cai Y, Zheng L, Zhang Z, Lin X
and Jiang N: LncRNA BISPR promotes the progression of thyroid
papillary carcinoma by regulating miR-21-5p. Int J Immunopathol
Pharmacol.
32(2058738418772652)2018.https://doi.org/10.1177/2058738418772652.
PubMed/NCBI View Article : Google Scholar
|
|
25
|
Swierniak M, Wojcicka A, Czetwertynska M,
Stachlewska E, Maciag M, Wiechno W, Gornicka B, Bogdanska M,
Koperski L, de la Chapelle A, et al: In-depth characterization of
the microRNA transcriptome in normal thyroid and papillary thyroid
carcinoma. J Clin Endocrinol Metab. 98:E1401–1409. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chen S, Fan X, Gu H, Zhang L and Zhao W:
Competing endogenous RNA regulatory network in papillary thyroid
carcinoma. Mol Med Rep. 18:695–704. 2018.PubMed/NCBI View Article : Google Scholar
|