Introduction
Major depressive disorder (MDD) has a high worldwide
prevalence. Several hypotheses regarding the mechanism of
development of MDD have been proposed, including an immunological
mechanism involving cytokines (1,2). Increased
levels of proinflammatory cytokines, such as interleukin (IL)-6 and
tumor necrosis factor-α (TNF-α), have been identified in patients
with MDD by meta-analyses (3).
Clinical trials of a cyclooxygenase-2 inhibitor to suppress
inflammation in patients with MDD produced symptomatic
improvements; however, the immunological mechanisms underlying MDD
are not understood in detail.
IL-18 was initially identified as an interferon-γ
inducing factor (4). An inactive
24-kDa precursor form of IL-18 is cleaved by activated caspase-1 to
a mature 18-kDa active form (4-7).
In addition to its role as an inflammatory cytokine, IL-18 also
serves a role in energy metabolism, and in the central nervous
system it is involved in psychiatric disorders such as depression
(8-12).
Regarding energy homeostasis, IL-18 deficiency induces bulimia,
corpulency and insulin resistance which resembles type 2 diabetes
mellitus (8). IL-18-deficient mice
also exhibit dyslipidemia leading to nonalcoholic fatty liver
disease and steatohepatitis (9), and
hippocampal impairments that may cause depression-like behavioral
changes (12). Therefore, IL-18 is
closely related to energy homeostasis, central nervous system
function, and the occurrence of psychiatric disorders such as
depression. However, the biological mechanism by which Il-18
contributes to these processes remains unclear.
In our previous study (12), it was demonstrated that the
aforementioned behavioral changes were the result of hippocampal
impairments; however, other parts of the brain, including the
prefrontal cortex and amygdala, are also responsible for depression
(13,14). Therefore, to determine the biological
and molecular mechanisms responsible for depression caused by IL-18
deficiency, additional analyses of other brain regions are
required.
In the present study, candidate genes which may be
involved in the induction of depressive-like behavioral changes
were identified by comparing gene expression in the brains of
Il18+/+ and Il18-/- mice. At 12
weeks of age, a time at which Il18-/- mice start
to exhibit depression-like behavioral changes, the genes with a
|>1.5|-fold change were identified and analyzed using
Ingenuity® Pathway Analysis (IPA) as described
previously (10,11). Subsequently, to explore candidate
genes associated with depression at 12 weeks of age, microarray
analysis was performed to compare expression at 6 and 12 weeks of
age to screen for effector genes that induce depression. Finally,
to confirm the microarray results, reverse
transcription-quantitative PCR (RT-qPCR) was performed.
Materials and methods
Details regarding RNA purification, microarray
analysis, RT-qPCR, and IPA (Ingenuity® Systems;
ingenuity.com) have been described in our previous studies
(15-17).
Animals
Il18-/- C57BL/6 male mice were
used in the present study (18).
Littermate Il18+/+ male mice were used as the
controls. Mice were housed in groups of 3-5 in polycarbonate cages
in a controlled environment (temperature, 22±1˚C; humidity, 50-60%;
and a 12-h light/dark cycle) with free access to water and food
(MF; Oriental Yeast Co., Ltd.). A total of three
Il18+/+ and three Il18-/- mice
were used for microarray and RT-qPCR analysis.
Animal experiments performed in the present study
were performed in accordance with the Guide for the Care and Use of
Laboratory Animals published by the National Institutes of Health
and approved by the Animal Care Committee of Hyogo College of
Medicine (Hyogo, Japan; approval nos. #28041, #13-062, #14-020 and
#16-013) prior to performing the animal experiments.
Sample collection
Mice were euthanized by deep anesthesia using 5%
isoflurane inhalation at 10:00 a.m., and lack of ventilation was
used to confirm euthanasia. The brains were removed and immediately
placed in liquid nitrogen and subsequently kept at -80˚C until
required.
RNA purification
Total RNA was extracted and purified from mice
brains using a Sepasol-RNA I Super kit (Nacalai Tesque, Inc.)
according to the manufacturer's protocol. Genomic DNA contamination
was removed using 5 units RNase free DNase I at 37˚C for 30 min.
After phenol/chloroform extraction and ethanol precipitation, total
RNA was dissolved in de-ionized water. RNA concentrations were
measured using NanoDrop 1000 spectrophotometry (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.).
Microarray analysis
Expression profiling was performed as described
previously (9). Raw data were
imported into Subio platform version 1.18 (Subio, Inc.) and
normalized. Differentially expressed genes between
Il18+/+ and Il18-/- mouse
brains were defined by signal ratios of |>1.5|-fold. Data can be
accessed in Gene Expression Omnibus (accession no. GSE64307).
Heatmap design
Heatmaps were created using the regHeatmap function
of the Heatplus package (heatmaps with row and/or column covariates
and colored clusters; github.com/alexploner/Heatplus) in R version
2.30.0.
IPA
IPA (version spring 2019) was used to determine the
functionality of microarray results and for the interpreting the
gene profiling data. To investigate molecular mechanisms affected
by IL-18 expression, core analysis was performed with the following
settings: Tissue, Brain; all other settings, default. The network
explorer of IPA was used to detect relevant interactions among
genes extracted at 6 and 12 weeks of age and to reveal the shortest
direct pathways between genes without protein-protein
interactions.
RT-qPCR
Brain samples from mice at 6 or 12 weeks of age were
used for analysis of expression. A total of 5 ng/reaction total RNA
was used, extracted as described above and was analyzed using
RNA-direct SYBR-Green Real-Time PCR Master mix and a One-step qPCR
kit (Toyobo Life Science). Samples were run in duplicate reactions
in 384-well plates. Median threshold cycle values were used to
calculate the fold changes between group samples. The fold change
values were normalized to Gapdh levels using the relative
standard curve method. qPCR was performed using QuantiStudio™ 12K
Flex (Thermo Fisher Scientific, Inc.). The reverse transcription
and thermocycling conditions were: 90˚C for 30 sec and 20 min at
61˚C for reverse transcription; followed by 45 cycles of 98˚C for 1
sec, 67˚C for 15 sec and 74˚C for 35 sec. The primer sequences for
RT-qPCR are shown in Table I.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Gene (GenBank
Accession no.) | Sequence,
5'-3' |
|---|
| Aqp4
(NM_009700) |
|
Forward |
CTTCCGCCCATCGAATGCTC |
|
Reverse |
CGACATTTGCAGCACATTGTCT |
| Bdnf
(NM_007540) |
|
Forward |
CGCCCATGAAAGAAGTAAACGTCCA |
|
Reverse |
GGCCCATTCACGCTCTCC |
| Btg1
(NM_007569) |
|
Forward |
GGCTTTGTCCGCATTTCCATAGCAG |
|
Reverse |
CTTTGCCCATGAGGTACACGTT |
| Btg2
(NM_007570) |
|
Forward |
CTGCCTCCTGGTCTCATGCTT |
|
Reverse |
ACAGTCCAGCTCTAGGGTTT |
| Cd46
(NM_010778) |
|
Forward |
GCCTACTCATCCTACCAAGCCT |
|
Reverse |
TTGGCTAAATATTCCTTCACGGGGAC |
| Chga
(NM_007693) |
|
Forward |
CCAATACCCAATCACCAACCAGC |
|
Reverse |
TCTCACTGTCTCCCGTGGC |
| Cplx1
(NM_007756) |
|
Forward |
TCTGCAAGGTTTGGCTTAAGAATTCCA |
|
Reverse |
ACCCGCTCCAAATCTATTGCT |
| Crh
(NM_205769) |
|
Forward |
AGAGAGCCTATATACCCCTTAATTAGCAT |
|
Reverse |
AGCATGGGCAATACAAATAACGC |
| Drp2
(NM_010078) |
|
Forward |
CCACAACAAGCAGCTCGAGT |
|
Reverse |
GAGCCATTGCCATCTGATTCACT |
| Eif6
(NM_010579) |
|
Forward |
CACCCTAAAACTTCTATCGAGGACCA |
|
Reverse |
CCTCGGTTCACAGTGCCT |
| Fgfr1
(NM_010206) |
|
Forward |
ATCATAATGGATTCTGTGGTGCCT |
|
Reverse |
CTCATTCTCCACGATGCAGGT |
| Gapdh
(NM_008084) |
|
Forward |
CCTTCCGTGTTCCTACCCCCAAT |
|
Reverse |
TTGATGTCATCATACTTGGCAGGTTTCTC |
| Hgs
(NM_001159328) |
|
Forward |
GACTCTCAGCCCATAACTCCCT |
|
Reverse |
CTCATGGCTCTCCTCCGACT |
| Htr2c
(NM_008312) |
|
Forward |
GCATACCAATGAACGTGTAGTTAGGAA |
|
Reverse |
GGCAGCTCTAAATTCTCTACCTGCATC |
| Kif5b
(NM_008448) |
|
Forward |
CTCACGGTTATGCAAGACAGACGA |
|
Reverse |
CACGGTCTCCTCCAAACCC |
| Ptpn1
(NM_011201) |
|
Forward |
ACACTGGAGCCTCACAACGG |
|
Reverse |
CCAGGCTGTCTTCATCCCC |
| Ucn3
(NM_031250) |
|
Forward |
GGGCACCAAGTTCACCCTT |
|
Reverse |
CGCAAATTCTTGGCCTTGTCG |
Statistical analysis
All results are expressed as the mean ± standard
deviation. Sigmaplot™ (version 11.0; Systat Software, Inc.) was
used for all statistical analyses. The microarray results were
analyzed using a Student's t-test and correlations between the
microarray and RT-qPCR results were analyzed using Spearman's rank
correlation tests. All analyses were performed >2 times to
confirm the results. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification and categorization of
differentially expressed genes in Il18-/- mice
The heatmaps of the microarray results at 6 and 12
weeks of age are shown in Fig. 1A and
B, respectively. A total of 699 and
2,805 genes showed a |>1.5|-fold-change
(Il18-/-/Il18+/+) in expression
at 6 and 12 weeks of age, respectively, and were extracted for
further analysis. The results of IPA core analysis are shown in
Table II. Based on the results of
IPA, 13 genes associated with ‘Major depression’ and ‘Depressive
disorder’ were identified: Aquaporin 4 (Aqp4), BTG
anti-proliferation factor (Btg)1, Btg2, CD46,
chromogranin A, complexin 1, corticotropin releasing hormone,
eukaryotic translation initiation factor 6, fibroblast growth
factor receptor 1 (Fgfr1), hepatocyte growth
factor-regulated tyrosine kinase substrate (Hgs), kinesin
family member 5B (Kif5b), protein tyrosine phosphatase,
non-receptor type 1 (Ptpn1) and urocortin 3 (Ucn3).
The heatmap of extracted genes is shown in Fig. 1C.
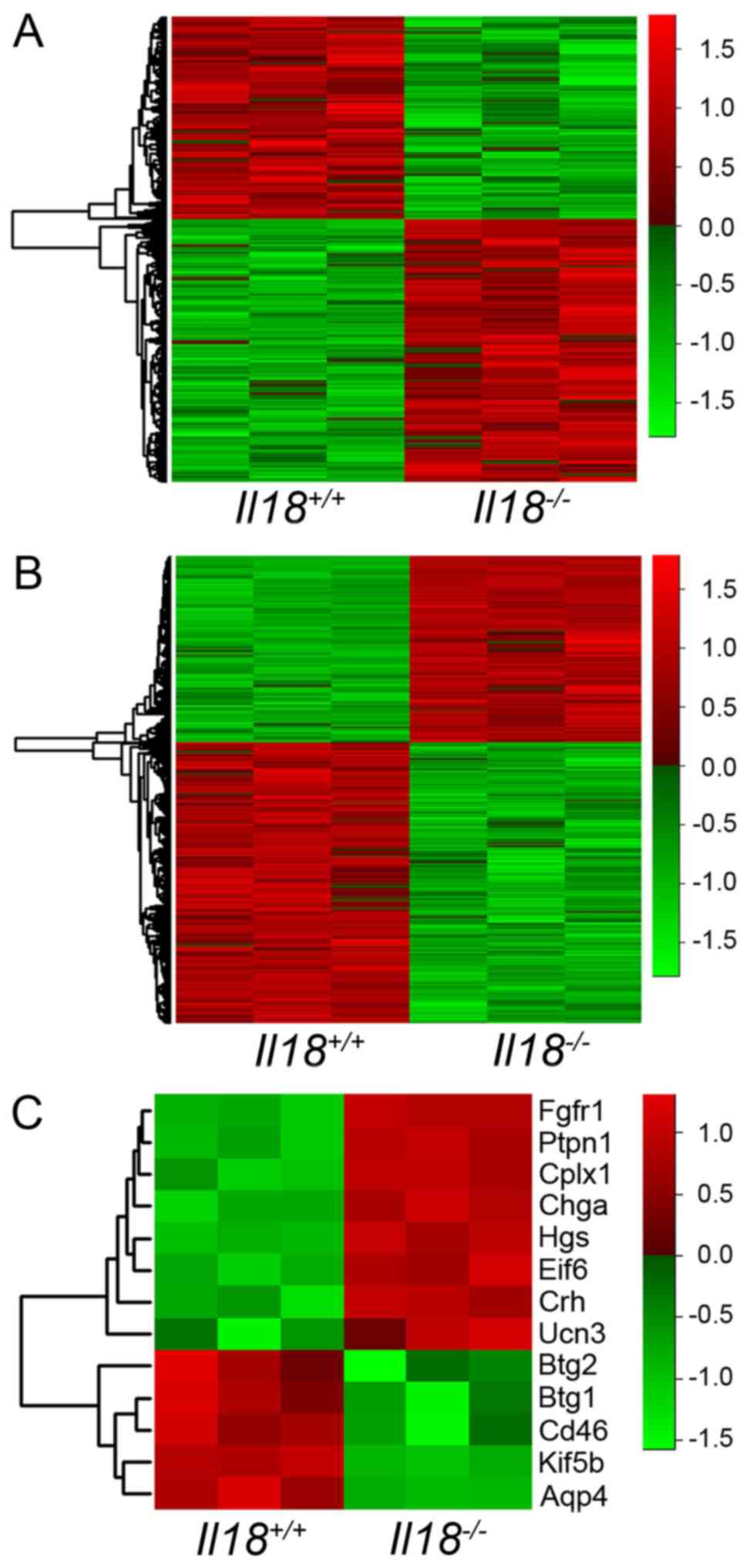 | Figure 1.Heatmap analyses of gene expression
profiles between Il18+/+ and
Il18-/- mice. Heatmap of microarray results at
(A) 6 and (B) 12 weeks of age. (C) Heatmap of 13 extracted genes
associated with depression at 12 weeks of age. The levels of gene
expression are shown in different colors, which transistion from
green to red with increasing expressions. IL, interleukin; Aqp4,
aquaporin 4; Btg, BTG anti-proliferation factor; Chga, chromogranin
A; Cplx1, complexin 1; Crh, corticotropin releasing hormone; Eif6,
eukaryotic translation 6; Hgs, hepatocyte growth factor-regulated
tyrosine kinase substrate; Ucn3, urocortin 3; Fgfr1, fibroblast
growth factor receptor 1; Ptpn1, protein tyrosine phosphatase,
non-receptor type 1; Kif5b, kinesin family member 5B. |
 | Table IIDisease association or functional
annotation of differentially expressed genes at 12 weeks of age
based on Ingenuity Pathway Analysis. |
Table II
Disease association or functional
annotation of differentially expressed genes at 12 weeks of age
based on Ingenuity Pathway Analysis.
| Diseases or
function annotation | P-value | No. of genes |
|---|
| Demyelination | 0.00817 | 5 |
| Neuropathy of
brain | 0.0130 | 3 |
| Despair
behavior | 0.0135 | 2 |
| Fear memory
acquisition | 0.0135 | 2 |
| Apoptosis of
anterior pituitary cells | 0.0135 | 2 |
| Childhood
adrenoleukodystrophy | 0.0135 | 2 |
| Feeding | 0.0219 | 8 |
| Astrocytoma | 0.0221 | 41 |
| Anorexia | 0.0238 | 3 |
| Grade 1-4
astrocytoma | 0.0255 | 40 |
| Abnormal morphology
of molecular layer of cerebellum | 0.0304 | 5 |
| Major
depression | 0.0357 | 12 |
| Brain
astrocytoma | 0.0367 | 39 |
| Depressive
disorder | 0.0375 | 13 |
| Apoptosis of neural
precursor cells | 0.0381 | 3 |
| Injury of cortical
neurons | 0.0381 | 3 |
| Movement
disorders | 0.0388 | 44 |
| Emotional
behavior | 0.0433 | 8 |
| Central nervous
system neuroepithelial tumor | 0.0434 | 48 |
| High grade
astrocytoma | 0.0444 | 38 |
| Glioma | 0.0454 | 48 |
| Abnormality of
cerebral cortex | 0.0467 | 38 |
| Glioma cancer | 0.0472 | 44 |
| Low grade
astrocytoma | 0.0491 | 5 |
Interactions between
depression-associated genes and Il18-/--specific genes
at 6 weeks of age
Interaction pathway analysis was used to identify
genes responsible for regulating depression-associated genes. As
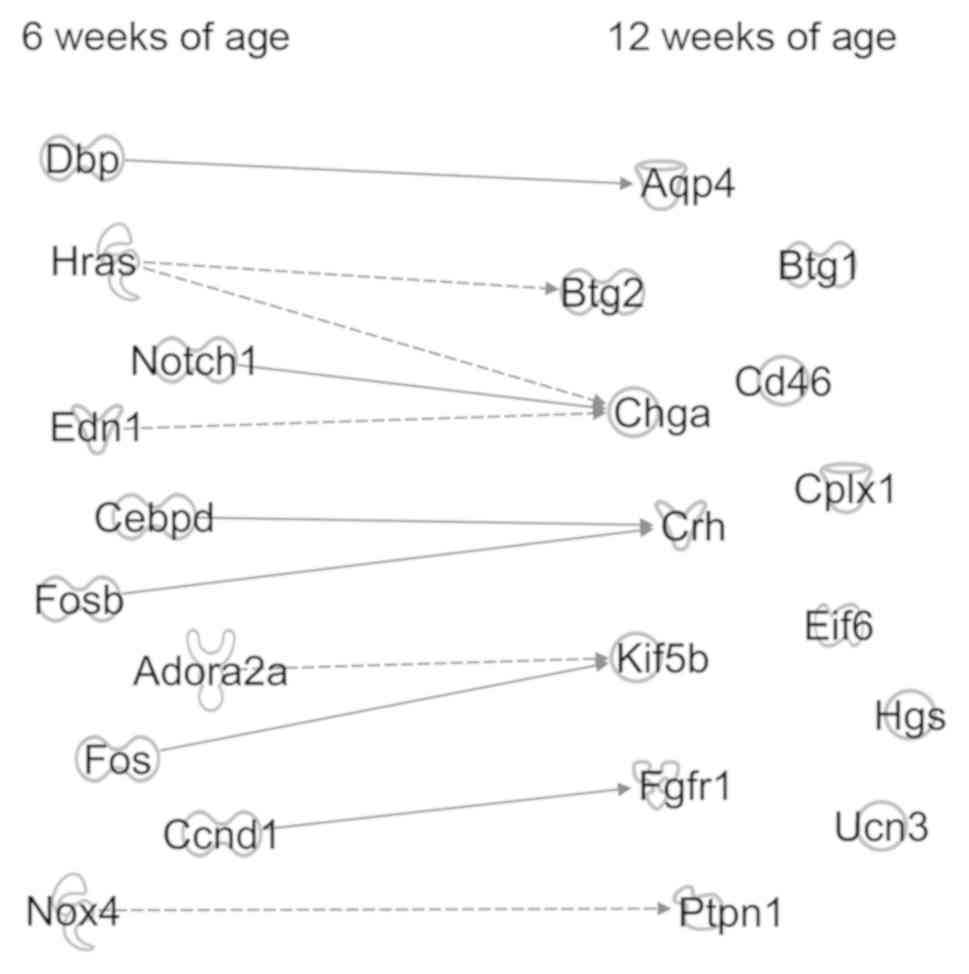
shown in Fig. 2, the interactions
deemed to be significant in the pathway analysis are shown for 10
genes: Adenosine A2a receptor (Adora2a), cyclin D1
(Ccnd1), CCAAT enhancer binding protein delta, D-box binding
PAR bZIP transcription factor, endothelin 1, Fos proto-oncogene,
AP-1 transcription factor subunit, FosB proto-oncogene, AP-1
transcription factor subunit, HRas proto-oncogene, GTPase,
Notch1 and NADPH oxidase 4 (Nox4).
Depression-inducing genes in
Il18-/- mice at 12 weeks of age
The 13 genes extracted in mice 12 weeks of age were
separated into depression-inducing and depression-suppressing genes
based on previous studies (19-21).
Among these 13 genes, Fgfr1, Ptpn1 and Ucn3
may be associated with the development of MDD
(depression-inducing), whereas the other 10 genes may be
depression-suppressing genes in MDD.
Interactions between
depression-inducing and -suppressing genes at 12 weeks of age with
the 10 extracted genes at 6 weeks of age
The interactions between the three
depression-inducing genes at 12 weeks of age, Fgfr1,
Ptpn1 and Ucn3, and 10 genes at 6 weeks of age were
examined and it was found that Ccnd1 and Nox4 may
affect development of depression. Ccnd1 inhibition increases
the expression of Fgfr1 (22),
and Nox4 decreases Ptpn1 expression (23).
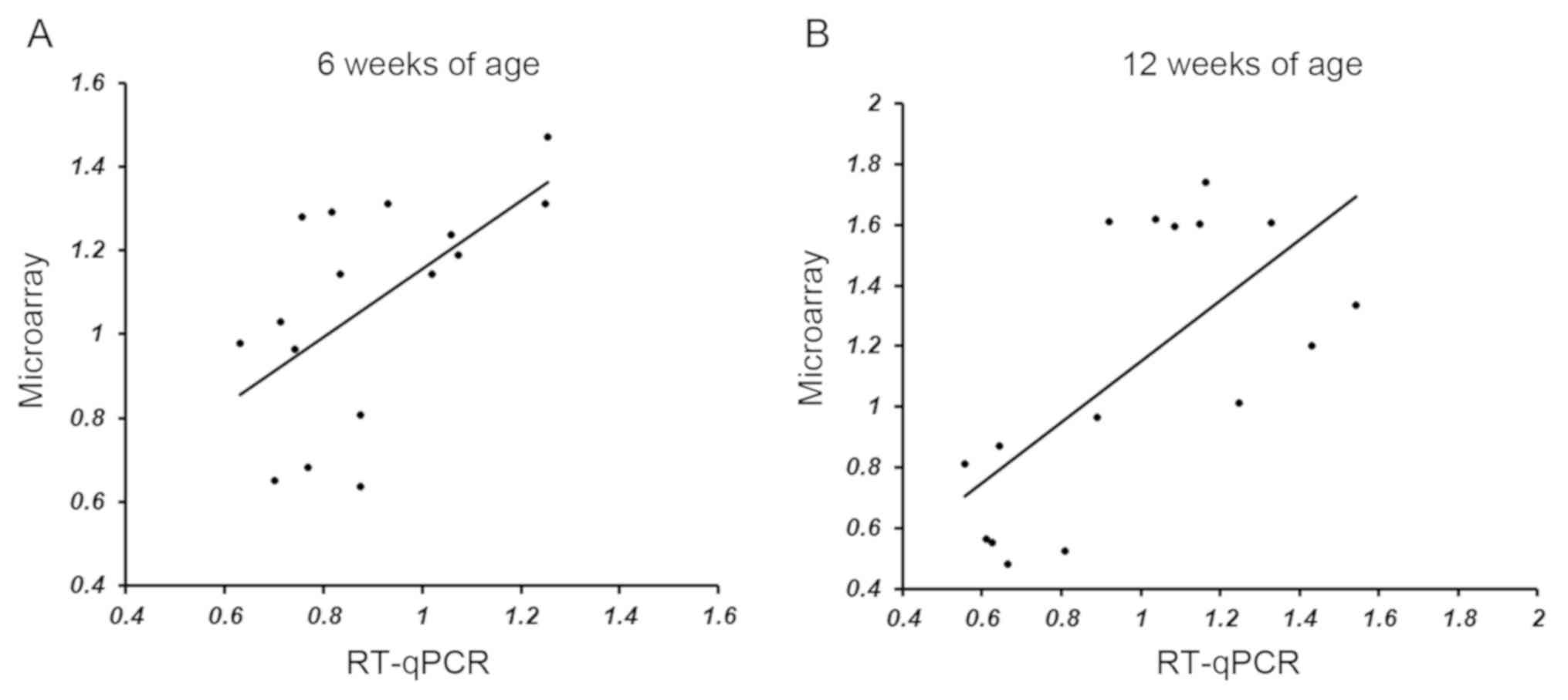
Correlation between microarray and
RT-qPCR results
To confirm the microarray analysis results, RT-qPCR
was performed, and a correlation test was performed between the
results of the microarray analysis and RT-qPCR (Table III). To determine the correlation
between microarray and RT-qPCR analysis, Spearman's rank
correlation coefficient analysis was performed for each group
(6-week-old group: P=0.014, ρ=0.60; 12-week-old group: P=0.007,
ρ=0.64; Fig. 3).
 | Table IIIComparison of microarray and RT-qPCR
gene expression data at 6 and 12 weeks of age. |
Table III
Comparison of microarray and RT-qPCR
gene expression data at 6 and 12 weeks of age.
| A, 6 weeks of
age |
|---|
| Gene symbol | FC (RT-qPCR) | FC
(Microarray) |
|---|
| Aqp4 | 0.875 | 0.808 |
| Bdnf | 1.074 | 1.190 |
| Btg1 | 0.702 | 0.651 |
| Btg2 | 1.255 | 1.470 |
| Cd46 | 1.249 | 1.312 |
| Chga | 0.631 | 0.980 |
| Cplx1 | 0.714 | 1.029 |
| Crh | 0.817 | 1.290 |
| Drp2 | 0.931 | 1.311 |
| Eif6 | 0.834 | 1.143 |
| Fgfr1 | 0.743 | 0.963 |
| Hgs | 1.019 | 1.145 |
| Htr2c | 0.768 | 0.684 |
| Kif5b | 0.875 | 0.636 |
| Ptpn1 | 1.059 | 1.237 |
| Ucn3 | 0.758 | 1.280 |
| B, 12 weeks of
age |
| Gene symbol | FC (RT-qPCR) | FC
(Microarray) |
| Aqp4 | 0.665 | 0.481 |
| Bdnf | 1.247 | 1.014 |
| Btg1 | 0.558 | 0.813 |
| Btg2 | 0.625 | 0.552 |
| Cd46 | 0.611 | 0.563 |
| Chga | 1.085 | 1.594 |
| Cplx1 | 1.148 | 1.604 |
| Crh | 1.165 | 1.742 |
| Drp2 | 1.431 | 1.202 |
| Eif6 | 0.890 | 0.965 |
| Fgfr1 | 0.922 | 1.610 |
| Hgs | 1.036 | 1.618 |
| Htr2c | 0.645 | 0.872 |
| Kif5b | 0.808 | 0.524 |
| Ptpn1 | 1.542 | 1.336 |
| Ucn3 | 1.328 | 1.609 |
Discussion
The present study identified potentially novel IL-18
pathways in the brain based on the core analysis, 13 genes were
identified to be associated with depression at 12 weeks of age; in
particular, Fgfr1, Ptpn1 and Ucn3 which may be
responsible for causing depression in Il18-/-
mice. Ccnd1 and Nox4 at 6 weeks of age interacted
with Fgfr1 and Ptpn1, indicating that Ccnd1
and Nox4 may be causative genes and the microarray and
RT-qPCR results were significantly correlated.
In our previous study, impaired learning and memory,
and depressive-like behavioral changes in Il18-/-
mice at 12 weeks of age were observed (12). To identify the genes responsible for
IL18 associated depression, microarray analysis of gene
expression in brains from Il18+/+ and
Il18-/- mice and IPA core analysis were performed
using previously described procedures (17). In the core analysis results, the
function and annotation of ‘Major depression’ and ‘Depressive
disorder’ were generated automatically. From the microarray
results, 13 genes at 12 weeks of age were associated with
‘depression’. Among these 13 genes, three, Fgfr1,
Ptpn1 and Ucn3, have previously been causally
associated with ‘Major depression’ and ‘Depressive disorder’
(19,21). Fgfr1 has been implicated in the
development of the brain and self-renewal of neural precursor cells
(24,25). In Il18-/- mice,
neurogenesis is decreased (12);
therefore, the increased expression of Fgfr1 may be a
compensatory response to the decrease in neurons during a
depressive state. Ptpn1 serves a role in negatively
regulating insulin signaling. Ptpn1 deficiency increases
insulin sensitivity and obesity resistance (26). In the present study, in the
Il18-/- mice, the expression of Ptpn1 was
increased, and it has previously been shown that
IL18-/- mice exhibit insulin resistance and
obesity (8). Ucn3 is
associated with stress-induced anxiety and depression, and with
energy homeostasis. Expression of Ucn3 in the rostral
perifornical area of the brain regulates not only anxiety-like
behaviors but also glucose metabolism of the body (27,28). The
expression of Ucn3 in Il18-/- mice at 12
weeks of age was increased; thus, a deficiency in IL-18 might
increase the expression of Ptpn1 and Ucn3, resulting
in not only depressive-like behavioral changes but also in an
energy imbalance. Additionally, in clinical studies on patients
with MDD, the mRNA expression levels of Fgfr1, Ptpn1
and Ucn3 were increased in the prefrontal cortex compared
with healthy controls (19,21). The expression of Fgfr1,
Ptpn1 and Ucn3 in Il18-/- mice at
12 weeks of age was higher compared with the
Il18+/+ mice based on the microarray results. The
localization of expression of these genes were determined using the
BrainStars database (brainstars.org) (29). Fgfr1 expression is upregulated
in the pontine nucleus and hippocampus, Ptpn1 is expressed
in the anterior olfactory bulb and hippocampus, and Ucn3 is
expressed in the paraventricular hypothalamic nucleus and amygdala.
Together, these results suggest that these three genes are
expressed in the brain and are involved in depression.
In contrast to these three genes, the other 10 genes
identified may have the opposite effect on depression, according to
previous findings (19,20,30). For
example, upregulated Aqp4 expression in the prefrontal
cortex was associated with MDD in patients. However, the expression
of Aqp4 in Il18-/- mice was decreased, and
the other nine genes also exhibited similar changes. Therefore,
these results suggest the existence of a negative feedback process
in MDD.
To analyze the molecular regulation of the three
genes at 12 weeks of age (Fgfr1, Ptpn1, and
Ucn3), interaction pathway analysis was performed, and based
on the results, Ccnd1 and Nox4 were extracted.
Inhibition of Ccnd1 expression increases Fgfr1 mRNA
levels in humans (31) and reduced
expression of Nox4-activated PTPN1 protein expression in
humans (23). These findings suggest
that Ccnd1 and Nox4 may regulate expression of
Fgfr1 and Ptpn1, and thus may be responsible for
inducing a depressive state in IL-18 deficiency.
In our previous study, it was shown that decreased
expression of Ccnd1 in the liver of
Il18-/- mice at 6 and 12 weeks of age may be
causally associated with energy imbalance through the Wnt signaling
pathway (9). In the present study, a
similar change was observed in Il18-/- mice at 6
weeks of age. Wnt signaling is indispensable for hippocampal memory
function (32) and is also associated
with depressive symptoms in an animal model of depression (33). Accordingly, Ccnd1 may regulate
energy levels and depression in Il18-/- mice.
Another gene extracted at 6 weeks of age,
Adora2a, also affected Kif5b expression at 12 weeks
of age. The expression of Adora2a was increased and
Kif5b was decreased, and Adora2a inhibition may have
decreased Kif5b expression in the present study, which
differs from the results of a previous study (34). Neuroinflammation is significantly
associated with MDD. In particular, inflammatory cytokines, such as
TNF-α, have been identified as mediators of MDD by impairing the
function of the blood brain barrier (35-37).
Increased Adora2a expression in cerebral endothelial cells
may induce blood-brain barrier leakage resulting in the impairment
of hippocampal memory function (38).
Therefore, these findings indicate that increased expression of
Adora2a at 6 weeks of age may be responsible for inducing
the behavioral phenotypes of Il18-/- mice through
neuroinflammation.
To assess the correlation between the microarray and
RT-qPCR results, Spearman's rank correlation coefficient analysis
was performed, and a significant correlation was detected between
the results.
A limitation of the present study was that the
molecular analysis was performed using whole brains; therefore,
genes affected by IL-18 only in select areas may not have been
detected, and further molecular studies using specific parts of the
brain are thus warranted. In addition, a more notable limitation is
that the present study did not analyze protein expression levels
encoded for by the identified genes. The identified genes may be
associated with depression; however the protein expression and
protein localization in the brain are required to determine
interactions between IL-18 and the proteins encoded for by the
extracted genes. Additionally, further studies using in
vitro and in vivo models are also required.
In conclusion, the gene expression profile of
Il18-/- mice brain was examined and an
association between the expression of certain genes and a
depressive phenotype in mice was identified. Fgfr1,
Ptpn1 and Ucn3 may be associated with depression, and
may be regulated by Ccnd1 and Nox4. Furthermore,
Adora2a and Ccnd1 may contribute to depression when
expression of IL-18 is downregulated.
In our previous study, it was hypothesized that
IL-18 may serve as a novel option for treatment of metabolic
disorders, such as diabetes mellitus and dyslipidemia, and for
neuropsychiatric disorders, such as MDD (9,11,12). The results of the present study
support the notion that IL-18 may serve as a novel treatment
strategy of metabolic and neuropsychiatric disorders, including
MDD.
Acknowledgements
The authors would like to thank Mr. Nobutaka
Okamura and Mrs. Naomi Gamachi for their technical support, Mr.
Nobutaka Okamura for his assistance with animal care and the
collection of samples, and Mrs. Naomi Gamachi for assistance with
the experiments.
Funding
The present study was supported by JSPS KAKENHI
(grant no. JP17K16404) and by the Takeda Science Foundation
(2019).
Availability of data and materials
The datasets shown and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
KY and HM designed the study. TH, MM, KM, KI, NU,
TI, and DO performed experiments. YW, HO, and HY analyzed the data.
KY and HM wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Animal experiments were approved by the Animal Care
Committee of Hyogo College of Medicine (Hyogo, Japan; approval nos.
#28041, #13-062, #14-020, and #16-013) prior to beginning animal
experiments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Maes M: Major depression and activation of
the inflammatory response system. Adv Exp Med Biol. 461:25–46.
1999.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yirmiya R, Weidenfeld J, Pollak Y, Morag
M, Morag A, Avitsur R, Barak O, Reichenberg A, Cohen E, Shavit Y
and Ovadia H: Cytokines, ‘depression due to a general medical
condition’ and antidepressant drugs. Adv Exp Med Biol. 461:283–316.
1999.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Dowlati Y, Herrmann N, Swardfager W, Liu
H, Sham L, Reim EK and Lanctôt KL: A meta-analysis of cytokines in
major depression. Biol Psychiatry. 67:446–457. 2010.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Okamura H, Tsutsi H, Komatsu T, Yutsudo M,
Hakura A, Tanimoto T, Torigoe K, Okura T, Nukada Y, Hattori K, et
al: Cloning of a new cytokine that induces IFN-gamma production by
T cells. Nature. 378:88–91. 1995.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Okamura H, Tsutsui H, Kashiwamura S,
Yoshimoto T and Nakanishi K: Interleukin-18: A novel cytokine that
augments both innate and acquired immunity. Adv Immunol.
70:281–312. 1998.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tsutsui H, Kayagaki N, Kuida K, Nakano H,
Hayashi N, Takeda K, Matsui K, Kashiwamura S, Hada T, Akira S, et
al: Caspase-1-independent, Fas/Fas ligand-mediated IL-18 secretion
from macrophages causes acute liver injury in mice. Immunity.
11:359–367. 1999.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ghayur T, Banerjee S, Hugunin M, Butler D,
Herzog L, Carter A, Quintal L, Sekut L, Talanian R, Paskind M, et
al: Caspase-1 processes IFN-gamma-inducing factor and regulates
LPS-induced IFN-gamma production. Nature. 386:619–623.
1997.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Netea MG, Joosten LA, Lewis E, Jensen DR,
Voshol PJ, Kullberg BJ, Tack CJ, van Krieken H, Kim SH, Stalenhoef
AF, et al: Deficiency of interleukin-18 in mice leads to
hyperphagia, obesity and insulin resistance. Nat Med. 12:650–656.
2006.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Yamanishi K, Maeda S, Kuwahara-Otani S,
Watanabe Y, Yoshida M, Ikubo K, Okuzaki D, El-Darawish Y, Li W,
Nakasho K, et al: Interleukin-18-deficient mice develop
dyslipidemia resulting in nonalcoholic fatty liver disease and
steatohepatitis. Transl Res. 173:101–114.e7. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yamanishi K, Mukai K, Hashimoto T, Ikubo
K, Nakasho K, El-Darawish Y, Li W, Okuzaki D, Watanabe Y, Hayakawa
T, et al: Physiological and molecular effects of interleukin-18
administration on the mouse kidney. J Transl Med.
16(51)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yamanishi K, Maeda S, Kuwahara-Otani S,
Hashimoto T, Ikubo K, Mukai K, Nakasho K, Gamachi N, El-Darawish Y,
Li W, et al: Deficiency in interleukin-18 promotes differentiation
of brown adipose tissue resulting in fat accumulation despite
dyslipidemia. J Transl Med. 16(314)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yamanishi K, Doe N, Mukai K, Ikubo K,
Hashimoto T, Uwa N, Sumida M, El-Darawish Y, Gamachi N, Li W, et
al: Interleukin-18-deficient mice develop hippocampal abnormalities
related to possible depressive-like behaviors. Neuroscience.
408:147–160. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Amat J, Baratta MV, Paul E, Bland ST,
Watkins LR and Maier SF: Medial prefrontal cortex determines how
stressor controllability affects behavior and dorsal raphe nucleus.
Nat Neurosci. 8:365–371. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Hamilton JP, Siemer M and Gotlib IH:
Amygdala volume in major depressive disorder: A meta-analysis of
magnetic resonance imaging studies. Mol Psychiatry. 13:993–1000.
2008.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yamanishi K, Doe N, Sumida M, Watanabe Y,
Yoshida M, Yamamoto H, Xu Y, Li W, Yamanishi H, Okamura H and
Matsunaga H: Hepatocyte nuclear factor 4 alpha is a key factor
related to depression and physiological homeostasis in the mouse
brain. PLoS One. 10(e0119021)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ikubo K, Yamanishi K, Doe N, Hashimoto T,
Sumida M, Watanabe Y, El-Darawish Y, Li W, Okamura H, Yamanishi H
and Matsunaga H: Molecular analysis of the mouse brain exposed to
chronic mild stress: The influence of hepatocyte nuclear factor 4α
on physiological homeostasis. Mol Med Rep. 16:301–309.
2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yoshida M, Watanabe Y, Yamanishi K,
Yamashita A, Yamamoto H, Okuzaki D, Shimada K, Nojima H, Yasunaga
T, Okamura H, et al: Analysis of genes causing hypertension and
stroke in spontaneously hypertensive rats: Gene expression profiles
in the brain. Int J Mol Med. 33:887–896. 2014.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Takeda K, Tsutsui H, Yoshimoto T, Adachi
O, Yoshida N, Kishimoto T, Okamura H, Nakanishi K and Akira S:
Defective NK cell activity and Th1 response in IL-18-deficient
mice. Immunity. 8:383–390. 1998.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tochigi M, Iwamoto K, Bundo M, Sasaki T,
Kato N and Kato T: Gene expression profiling of major depression
and suicide in the prefrontal cortex of postmortem brains. Neurosci
Res. 60:184–191. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tezval H, Jahn O, Todorovic C, Sasse A,
Eckart K and Spiess J: Cortagine, a specific agonist of
corticotropin-releasing factor receptor subtype 1, is anxiogenic
and antidepressive in the mouse model. Proc Natl Acad Sci USA.
101:9468–9473. 2004.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kang HJ, Adams DH, Simen A, Simen BB,
Rajkowska G, Stockmeier CA, Overholser JC, Meltzer HY, Jurjus GJ,
Konick LC, et al: Gene expression profiling in postmortem
prefrontal cortex of major depressive disorder. J Neurosci.
27:13329–13340. 2007.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Tashiro E, Maruki H, Minato Y, Doki Y,
Weinstein IB and Imoto M: Overexpression of cyclin D1 contributes
to malignancy by up-regulation of fibroblast growth factor receptor
1 via the pRB/E2F pathway. Cancer Res. 63:424–431. 2003.PubMed/NCBI
|
|
23
|
Mahadev K, Motoshima H, Wu X, Ruddy JM,
Arnold RS, Cheng G, Lambeth JD and Goldstein BJ: The NAD(P)H
oxidase homolog Nox4 modulates insulin-stimulated generation of
H2O2 and plays an integral role in insulin signal transduction. Mol
Cell Biol. 24:1844–1854. 2004.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Trokovic R, Trokovic N, Hernesniemi S,
Pirvola U, Vogt Weisenhorn DM, Rossant J, McMahon AP, Wurst W and
Partanen J: FGFR1 is independently required in both developing mid-
and hindbrain for sustained response to isthmic signals. EMBO J.
22:1811–1823. 2003.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Stevens HE, Smith KM, Maragnoli ME, Fagel
D, Borok E, Shanabrough M, Horvath TL and Vaccarino FM: Fgfr2 is
required for the development of the medial prefrontal cortex and
its connections with limbic circuits. J Neurosci. 30:5590–5602.
2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Elchebly M, Payette P, Michaliszyn E,
Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J,
Chan CC, et al: Increased insulin sensitivity and obesity
resistance in mice lacking the protein tyrosine phosphatase-1B
gene. Science. 283:1544–1548. 1999.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kuperman Y, Issler O, Regev L, Musseri I,
Navon I, Neufeld-Cohen A, Gil S and Chen A: Perifornical
Urocortin-3 mediates the link between stress-induced anxiety and
energy homeostasis. Proc Natl Acad Sci USA. 107:8393–8398.
2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li C, Chen P, Vaughan J, Lee KF and Vale
W: Urocortin 3 regulates glucose-stimulated insulin secretion and
energy homeostasis. Proc Natl Acad Sci USA. 104:4206–4211.
2007.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kasukawa T, Masumoto KH, Nikaido I, Nagano
M, Uno KD, Tsujino K, Hanashima C, Shigeyoshi Y and Ueda H:
Quantitative expression profile of distinct functional regions in
the adult mouse brain. PLoS One. 6(e23228)2011.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sawada K, Young CE, Barr AM, Longworth K,
Takahashi S, Arango V, Mann JJ, Dwork AJ, Falkai P, Phillips AG and
Honer WG: Altered immunoreactivity of complexin protein in
prefrontal cortex in severe mental illness. Mol Psychiatry.
7:484–492. 2002.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chen G, Wang J, Liu Z and Kornmann M: Exon
III splicing of fibroblast growth factor receptor 1 is modulated by
growth factors and cyclin D1. Pancreas. 37:159–164. 2008.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Fortress AM, Schram SL, Tuscher JJ and
Frick KM: Canonical Wnt signaling is necessary for object
recognition memory consolidation. J Neurosci. 33:12619–12626.
2013.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Hui J, Zhang J, Pu M, Zhou X, Dong L, Mao
X, Shi G, Zou J, Wu J, Jiang D and Xi G: Modulation of
GSK-3β/β-catenin signaling contributes to learning and memory
impairment in a rat model of depression. Int J
Neuropsychopharmacol. 21:858–870. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Yu L, Haverty PM, Mariani J, Wang Y, Shen
HY, Schwarzschild MA, Weng Z and Chen JF: Genetic and
pharmacological inactivation of adenosine A2A receptor reveals an
Egr-2-mediated transcriptional regulatory network in the mouse
striatum. Physiol Genomics. 23:89–102. 2005.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Krishnadas R and Cavanagh J: Depression:
An inflammatory illness? J Neurol Neurosurg Psychiatry. 83:495–502.
2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Khairova RA, Machado-Vieira R, Du J and
Manji HK: A potential role for pro-inflammatory cytokines in
regulating synaptic plasticity in major depressive disorder. Int J
Neuropsychopharmacol. 12:561–578. 2009.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liu H, Luiten PG, Eisel UL, Dejongste MJ
and Schoemaker RG: Depression after myocardial infarction:
TNF-α-induced alterations of the blood-brain barrier and its
putative therapeutic implications. Neurosci Biobehav Rev.
37:561–572. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yamamoto M, Guo DH, Hernandez CM and
Stranahan AM: Endothelial Adora2a activation promotes blood-brain
barrier breakdown and cognitive impairment in mice with
diet-induced insulin resistance. J Neurosci. 39:4179–4192.
2019.PubMed/NCBI View Article : Google Scholar
|