Introduction
Glomerulonephritides are a group of rare diseases
that often affects younger individuals and can lead to end-stage
renal disease (ESRD) (1). Although
the clinical presentation of glomerulonephritides and their
outcomes are variable, well-established factors such as persistent
proteinuria, hypertension, diabetes and cardiovascular disease
increase the risk of ESRD progression (2). Podocyte injury is a hallmark of renal
diseases, presenting clinically with proteinuria,
glomerulosclerosis and kidney failure (3). Terminally differentiated podocytes have
a limited capacity to self-replicate; hence, direct cellular damage
contributes to the onset and progression of glomerular diseases
such as Focal Segmental Glomerulosclerosis (FSGS), Minimal Change
Disease and Diabetic Nephropathy (DN) (4). Additionally, immune-mediated damage is
responsible for the establishment and progression of secondary
glomerulonephritides, including lupus nephritis (LN) and
ANCA-associated vasculitis (AAV) (5).
The gold standard for confirming a diagnosis of
glomerulonephritides diagnosis is renal biopsy (6). However, biopsies are invasive, may have
complications and are hard to interpret due to the presence of
highly complex pathogenic mechanisms that translate into limited
histological responses of kidney injury (7). Thus, identifying non-invasive
biomarkers of glomerular disease may result in improved diagnosis
and guide therapeutic choices in the field of nephrology (8).
Long non-coding RNAs (lncRNAs) are RNA molecules
comprised of >200 nucleotides with no protein-coding capacity.
They are a class of RNAs comprised of heterogeneous intergenic
transcripts, sense or antisense transcripts that overlap with other
genes, or enhancer RNAs with a range of functions that includes the
regulation of gene expression, chromatin remodelling, microRNA
(miRNA/miR)-sponging and protein scaffolding (9,10).
LncRNAs modulate several biological processes, including
homeostasis, cellular metabolism, proliferation, apoptosis and
differentiation (10,11). Dysregulated expression of lncRNAs
such as metastasis-associated lung adenocarcinoma transcript 1,
LOC105374325, LOC105375913, X-inactive specific transcript and
RP11-2B6.2 contribute to the pathogenesis of various kidney
diseases (12-17).
Moreover, circulating lncRNAs are stable and easily detectable in
plasma, serum and urine, characteristics that make them convenient
for use diagnostically (18,19).
The lncRNA taurine upregulated gene 1 (TUG1),
located on chromosome 22q12, regulates podocyte health and
glomerulosclerosis by altering the expression of peroxisome
proliferator-activated receptor γ Coactivator 1α (PGC1A) (20-23).
PGC1A is a transcriptional coactivator that controls mitochondrial
biogenesis and is encoded by the PPARGC1A gene in humans
(24,25). A decrease in PGC1A expression
contributes to the onset of metabolic diseases such as DN, and
transgenic expression of PPARGC1A in tubular cells protects
mice from developing acute and chronic forms of kidney disease
(26,27). Long et al (28) found that podocyte-specific transgenic
expression of lncRNA TUG1 protects against DN in a mouse model, and
demonstrated that lower glomerular filtration rates (GFR) were
correlated with a decrease in TUG1 intrarenal expression in human
subjects. However, it is unknown if TUG1 can be detected in urine
and if its expression levels are correlated with histopathological
findings in kidney biopsies from patients diagnosed with
glomerulonephritides other than DN. In the present study, the
urinary expression of TUG1 in non-diabetic patients with
glomerulonephritides was characterized and it was shown that a
decrease in TUG1 expression was significantly associated with
FSGS.
Materials and methods
Ethical considerations
The present study complied with the ethical
principles for medical research specified in the Declaration of
Helsinki (29) and was approved by
the Local Ethics and Research Committee at the Hospital de
Especialidades NUM. 1, Bajio, Leon, Guanajuato, Instituto Mexicano
del Seguro Social (approval no. CLIEIS R-2018-1001-114). Each
participant provided written informed consent prior to enrolment in
the study.
Patient enrolment
A total of 11 patients with biopsy-confirmed
glomerulonephritides (7 females and 4 males; median age, 31 years;
range 19-59 years), and 10 healthy controls (6 females and 4 males;
median age 34 years; range 22-54) were enrolled in the present
study at UMAE-Hospital de Especialidades, CMNO in Guadalajara,
Mexico. Inclusion criteria were non-diabetic patients aged ≥18
years. Exclusion criteria were intake of non-steroid
anti-inflammatory drugs or platelet anti-aggregation medication a
week before sampling, administration of anticoagulant medication 96
h before the biopsy, blood pressure >160/90 mmHg, active urinary
infection or anaemia. Urine sample collection was taken shortly
before biopsy collection. Only 1 patient enrolled in the study was
unable to provide a urine sample and, therefore, was excluded from
further analysis.
Data collection
Biochemical parameters corresponding to each
glomerulonephritides patient at the time of biopsy were assayed by
routine laboratory techniques at UMAE-Hospital de Especialidades
and retrieved from patients' electronic medical records.
Reverse transcription-quantitative
(RT-q)PCR
Urine specimens from patients and healthy controls
were collected and centrifuged at 4,500 x g for 30 min at 4˚C. The
urinary sediments were recovered by discarding the supernatant, and
total RNA was extracted using RiboZol reagent (Amresco LLC)
according to the manufacturer's protocol. cDNA was synthesized by
reverse transcription using 1 µg total RNA and a QuantiTect Reverse
Transcription kit according to the manufacturer's protocol (Qiagen,
Inc.). The cDNA was used to perform quantitative PCR using EvaGreen
5x qPCR mix (qARTA Bio Inc.) and 10 pmol/µl each of the reverse and
forward primers in a Lightcycler 96 (Roche Diagnostics). The
sequences of the primers used to amplify human lncRNA TUG1(30), nephrin (NSPH1), podocin (NSPH2),
transcription factor A, mitochondrial (TFAM), cytochrome C oxidase
subunit 5A (COX5A), PPARGC1A and GAPDH are shown in Table I. The amplification conditions
consisted of 1 cycle of initial denaturation at 95˚C for 15 min;
followed by 40 cycles of denaturation at 95˚C for 15 sec, annealing
at 56˚C for 20 sec and extension at 72˚C for 20 sec. GAPDH was used
as the internal control to normalize gene expression data. The
relative mRNA expression was calculated using the 2-∆∆Cq
method (31).
 | Table ISequences of the primers. |
Table I
Sequences of the primers.
| Gene | Sequence,
5'-3' | Size, bp |
|---|
| TUG1 | | 150 |
|
Forward |
TAGCAGTTCCCCAATCCTTG | |
|
Reverse |
CACAAATTCCCATCATTCCC | |
| NPHS1 | | 124 |
|
Forward |
GTGCACTATGCTCCCACCAT | |
|
Reverse |
TCTCCCAGTTGAACATGCCC | |
| NPHS2 | | 160 |
|
Forward |
GAGGAAGGTACCAAATCCTCCG | |
|
Reverse |
GCAGATGTCCCAGTCGGAATA | |
| TFAM | | 105 |
|
Forward |
ATGGAGGCAGGAGTTTCGTT | |
|
Reverse |
CCTAGATGAGTTCTGCCTGCT | |
| COX5A | | 160 |
|
Forward |
AGATGCCTGGGAATTGCGTA | |
|
Reverse |
AGGTCCTGCTTTGTCCTTAACA | |
| PPARGC1A | | 158 |
|
Forward |
TGTGGAGTCCCTGGAATGGA | |
|
Reverse |
AAGATCGTGTTGGGCGAGAG | |
| GAPDH | | 153 |
|
Forward |
CCCACTCCTCCACCTTTGAC | |
|
Reverse |
TGGTCCAGGGGTCTTACTCC | |
In silico identification of
TUG1/miR-204-5p axis mRNA targets
The VarElect phenotype program from GeneAnalytics
(geneanalytics.genecards.org), a
pathway enrichment analysis tool, was used to identify predicted
mRNA targets of the TUG1/miR-204-5p axis involved in mitochondrial
biogenesis based on the genomic information stored in GeneCards
(32). In total, 453 predicted mRNA
targets of hsa-miR-204-5p downloaded from TargetScan version 7.2
(targetscan.org) (33) were profiled with GeneAnalytics. The
entries were scored and ranked using an algorithm that enables
matching of genomic expression, sequencing and microarray datasets
to tissues and cells.
Statistical analysis
Data are expressed as the mean ± standard error of
the mean. Non-parametric variables were compared using a
Mann-Whitney U-test or a Kruskal-Wallis test with a Dunn's post-hoc
test, as appropriate. A Spearman's rank correlation analysis was
used to analyse correlations. Statistical analysis was performed in
GraphPad Prism Version 6.01 (GraphPad Software, Inc.). P<0.05
was considered to indicate a statistically significant
difference.
Results
Clinical and biochemical
characteristics of glomerulonephritides patients and healthy
controls
The present study included 10 patients with
biopsy-confirmed diagnosis, 4 males and 6 females, and 10
sex-matched healthy controls. The mean age at renal biopsy was
34.4±11.1 years old, the mean body mass index was 30.1±5.1, and the
diagnoses were as follows: 5 cases of FSGS, 3 cases of LN, 1 case
of AAV and 1 case of advanced glomerular sclerosis (AGS). Table II shows the biochemical parameters
of the glomerulonephritides patients. A total of 80% of the
patients had haemoglobin values within the normal range. All
glomerulonephritides patients had proteinuria, with the highest
levels present in the patient diagnosed with AGS. However, CKD
staging, classified according to the level of GFR (34) varied greatly. Only 2 patients had a
normal GFR of >90 ml/min/1.73 m2; 3 FSGS patients
were Stage 2; 1 AAV patient was Stage 3B; 3 patients were Stage 4
and only 1 LN patient was Stage 5 with a GFR <15
ml/min/1.73m2. The presence of glomerular damage in the
absence of renal failure highlights the importance of identifying
novel circulating markers able to detect glomerulonephritides
during the early stages of CKD.
 | Table IIBiochemical parameters of
glomerulonephritides patients. |
Table II
Biochemical parameters of
glomerulonephritides patients.
| |
Patient
no. |
|---|
| Factor | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|
| Diagnosis | FSGS | FSGS | AAV | LN | AGS | FSGS | LN | LN | FSGS | FSGS |
| Hemoglobin,
g/dl | 14.7 | 13.2 | 12.8 | 11.6 | 8.7 | 10.5 | 15.6 | 13.8 | 16.7 | 16.3 |
| sCr, mg/dl | 0.84 | 0.57 | 1.5 | 3.7 | 3.9 | 2.1 | 0.86 | 8.4 | 1.4 | 1.5 |
| GFR,
ml/min/1.73m2 | 90 | 126 | 38 | 17 | 15 | 28 | 107 | 8 | 66 | 63 |
| Proteinuria, g/24
h | 1.28 | 2.68 | 1.9 | 2.24 | 5.8 | 3.68 | 1.59 | 2.9 | 2.72 | 1.79 |
| Albumin, g/dl | 3.7 | 2.1 | 4.1 | 3.6 | 2.5 | 2.3 | 2.9 | 3 | 3.5 | 4.3 |
| pANCA | + | - | + | +++ | - | + | +++ | +++ | - | - |
| cANCA | - | - | - | - | - | - | - | - | - | - |
| Antinuclear
antibodies | - | 0.09722 | 0.26389 | 0.48611 | 0.09722 | 0.09722 | 0.15278 | 0.15278 | 0.15278 | - |
| Anti-DNA
antibodies | - | - | - | +++ | - | - | ++ | ++ | - | - |
| C3 fraction,
mg/dl | 124 | 112 | 93.9 | 55 | 104 | 127 | 59.7 | 92.2 | 143 | 125 |
| C4 fraction,
mg/dl | 28.6 | 38.4 | 24.4 | 7 | 31.8 | 28.9 | 9.29 | 19.4 | 33.5 | 33.5 |
| CRP, mg/l | 3.75 | <3.23 | 5.53 | <3.23 | 15.8 | <3.23 | 16 | 11.1 | <3.23 | <3.23 |
| ESR, mm/h | 37 | 42 | 16 | 12 | 18 | - | 28 | 26 | - | 10 |
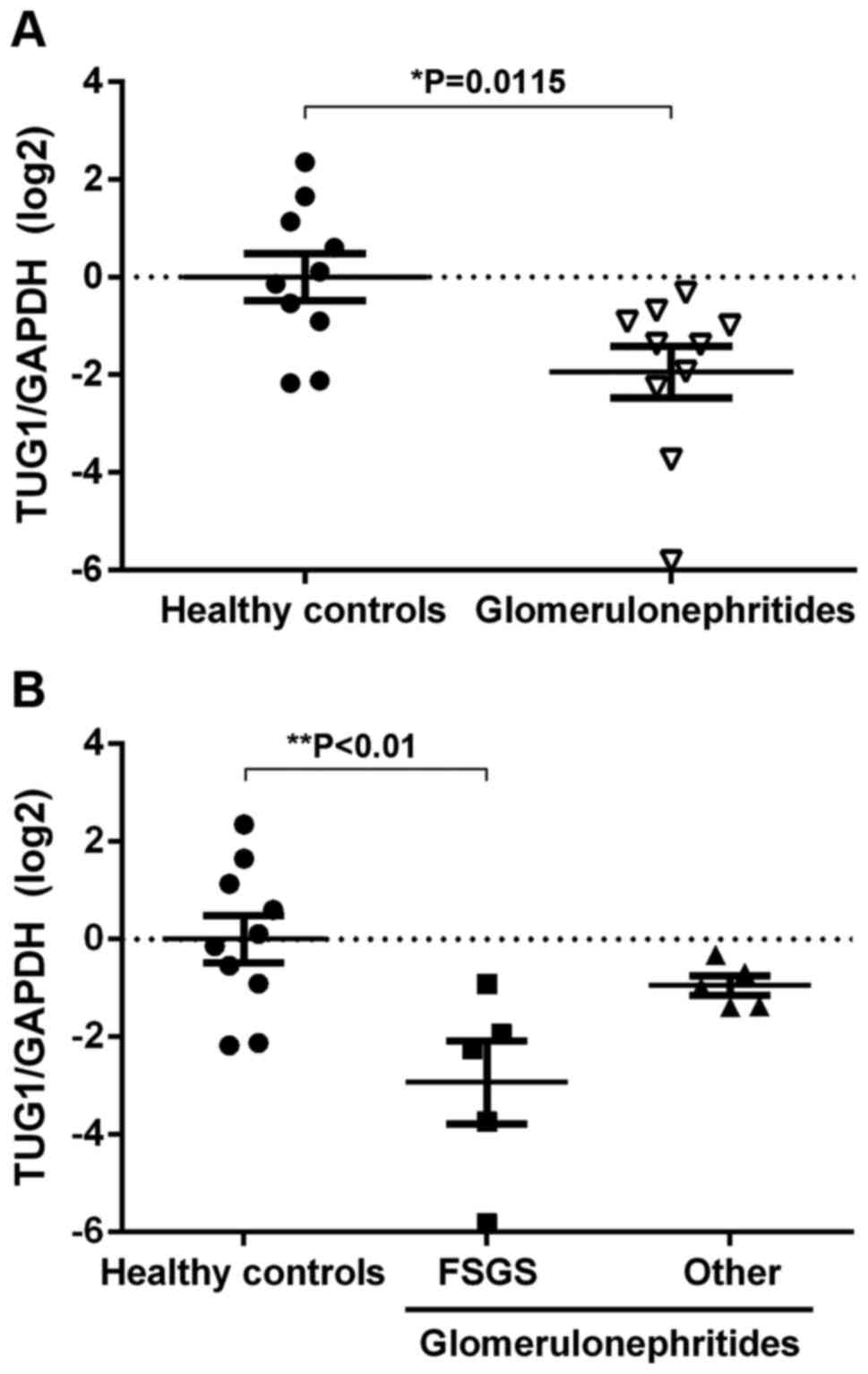
Glomerulonephritides patients have low
levels of urinary TUG1 expression
Cell-free lncRNAs are stable in urine and may
potentially be used as non-invasive biomarkers to identify
diseases, such as lupus nephritis and membranous nephropathy
(18,19). To assess alterations in TUG1
expression in the urinary sediment of patients with
glomerulonephritides, RT-qPCR was used. Urinary expression of TUG1
was reduced in glomerulonephritides patients compared with the
healthy controls (Fig. 1A).
Furthermore, significantly lower TUG1 expression levels in urinary
sediment was observed in patients with FSGS when compared with all
other diagnoses (Fig. 1B). These
results suggest that the urinary expression of TUG1 decreases with
podocytopathy.
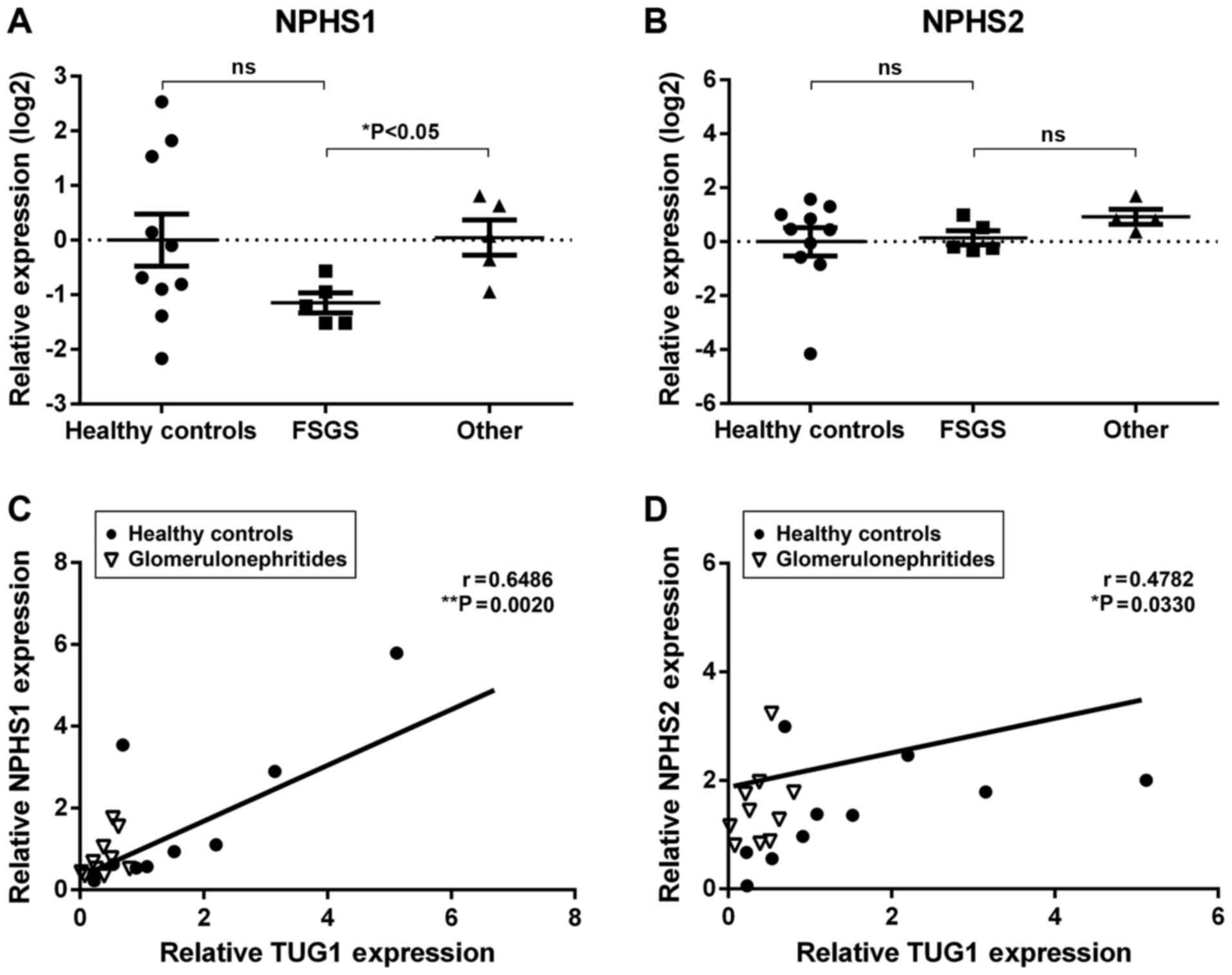
Urinary expression of TUG1 correlates
with podocyte marker expression
Glomerular injury is associated with podocyte loss,
a phenomenon that can be monitored by detecting podocyte-specific
mRNAs in the urine (35). In the
present study, two podocyte-specific mRNAs, NPHS1 and NPHS2, were
detected using RT-qPCR. Although the expression levels of urinary
NPHS1 was not significantly different between glomerulonephritides
patients and healthy controls, NPHS1 urinary expression was
significantly lower in the FSGS patients when compared with other
diagnoses (Fig. 2A). In contrast,
NPHS2 mRNA did not differ significantly amongst the groups
(Fig. 2B). Interestingly, a positive
correlation was observed between TUG1 urinary expression and both
podocyte-specific mRNAs (NPHS1, r=0.6486, **P<0.01;
NPHS2, r=0.4782, *P<0.05; Fig. 2C and D). These results further suggest an
association between TUG1 urinary expression and podocyte
damage.
Urinary expression of mitochondrial
biogenesis markers is correlated with TUG1 expression levels
Several studies have validated the functional role
of TUG1 as a lncRNA in directly binding to specific miRNAs to
regulate post-transcriptional processing via a competing endogenous
RNA mechanism (21,23,28,30). One
such target is miR-204-5p (30).
Thus, the VarElect phenotype program from Gene Analytics was used
to predict mRNA targets of the TUG1/miR-204-5p axis involved in
mitochondrial biogenesis. Through gene data profiling, a total of
27 genes significantly associated with mitochondrial biogenesis
were identified (Table III).
 | Table IIIPredicted targets of TUG1/miR-204-5p
axis involved in mitochondrial biogenesis. |
Table III
Predicted targets of TUG1/miR-204-5p
axis involved in mitochondrial biogenesis.
| Symbol | Description | Global
Ranka | Scorea,b |
|---|
| PPARGC1A | PPARG Coactivator
1α | 1 | 18.76 |
| TFAM | Transcription
Factor A, Mitochondrial | 4 | 11.67 |
| CREB1 | CAMP Responsive
Element Binding Protein 1 | 25 | 4.98 |
| SIRT1 | Sirtuin 1 | 26 | 4.80 |
| COX5A | Cytochrome C
Oxidase Subunit 5A | 39 | 3.13 |
| ATF2 | Activating
Transcription Factor 2 | 52 | 2.42 |
| MAPK1 | Mitogen-Activated
Protein Kinase 1 | 56 | 2.32 |
| ESR1 | Estrogen Receptor
1 | 61 | 1.96 |
| CAMK2D | Calcium/Calmodulin
Dependent Protein Kinase II δ | 68 | 1.68 |
| NFATC3 | Nuclear Factor of
Activated T Cells 3 | 68 | 1.68 |
| OGT | O-Linked
N-Acetylglucosamine (GlcNAc) Transferase | 69 | 1.59 |
| MXI1 | MAX Interactor 1,
Dimerization Protein | 69 | 1.59 |
| ESRRG | Estrogen Related
Receptor γ | 75 | 0.68 |
| CPOX | Coproporphyrinogen
Oxidase | 75 | 0.68 |
| ESR2 | Estrogen Receptor
2 | 80 | 0.36 |
| TFAP2A | Transcription
Factor AP-2α | 80 | 0.36 |
| ZBTB20 | Zinc Finger and BTB
Domain Containing 20 | 81 | 0.26 |
| NPAS3 | Neuronal PAS Domain
Protein 3 | 81 | 0.26 |
| NR4A2 | Nuclear Receptor
Subfamily 4 Group A Member 2 | 81 | 0.26 |
| RICTOR | RPTOR Independent
Companion of mTOR Complex 2 | 81 | 0.26 |
| CAMK1 | Calcium/Calmodulin
Dependent Protein Kinase I | 81 | 0.26 |
| TET2 | Tet Methylcytosine
Dioxygenase 2 | 81 | 0.26 |
| ATXN1 | Ataxin 1 | 81 | 0.26 |
| VHL | Von Hippel-Lindau
Tumor Suppressor | 81 | 0.26 |
| HMGCR |
3-Hydroxy-3-Methylglutaryl-CoA
Reductase | 81 | 0.26 |
| ZNF521 | Zinc Finger Protein
521 | 81 | 0.26 |
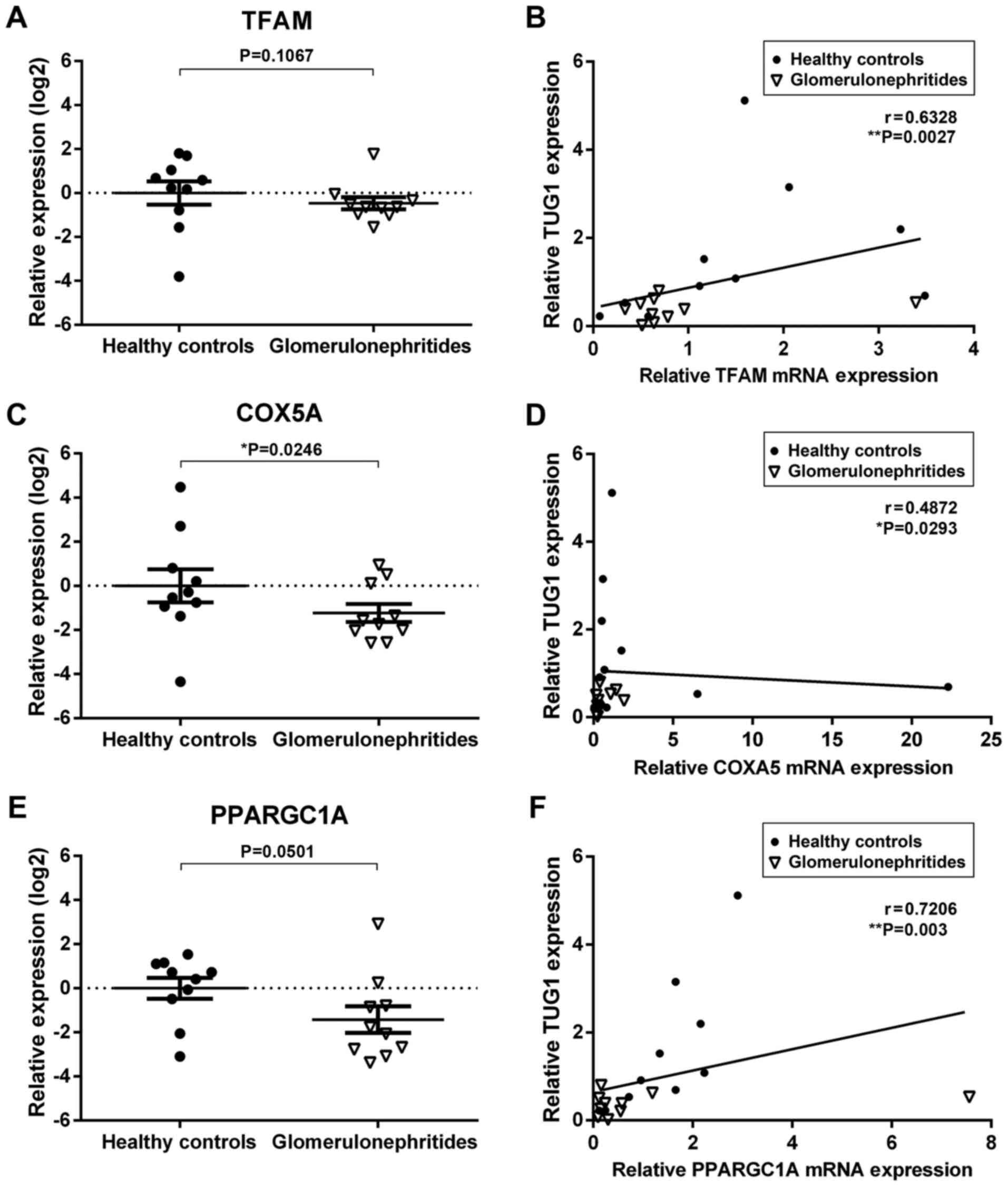
Next, the urinary expression of two predicted
TUG1/miR-204 mRNA targets significantly associated with
mitochondrial biogenesis, TFAM and COX5A, as well as one validated
target, PPARGC1A (36), were
quantified (Fig. 3). The relative
expression levels of COXA5 and PPARGC1A were lower in the urinary
sediment of glomerulonephritides patients when compared with
healthy controls (Fig. 3C and
E). Additionally, there was a
significant correlation between TUG1 urinary expression and all
three mitochondrial biogenesis mRNAs quantified in the present
study (TFAM, r=0.6328, **P<0.01; COXA5, r=0.4872,
*P<0.05; PPARGC1A, r=0.7206, **P<0.01;
Fig. 3B-F). These results suggest
that urinary RNAs may reflect molecular processes involved in the
pathophysiology of renal dysfunction.
Discussion
The diagnosis of glomerulonephritides in current
clinical practice is primarily dependent on the histological
analysis of renal biopsies (6).
However, this traditional approach is being challenged by molecular
techniques that go beyond description and examine disease
mechanisms (37,38). Multiple studies have shown the
association between dysregulated expression of certain lncRNAs and
the development and progression of pathological disease states that
affect the kidney (12-17).
Urinary non-coding RNAs are a promising non-invasive tool able to
reflect renal disease, aid in its appropriate diagnosis, and guide
therapeutic choices (12,39,40). In
the present study, it was shown that lncRNA TUG1 was present in the
urinary sediment, and its expression was significantly reduced in
patients with biopsy-confirmed glomerulonephritides, particularly
those diagnosed with FSGS. Moreover, there was a positive
correlation between urinary TUG1 expression and mRNAs known to be
involved in pathogenic mechanisms associated with podocyte
loss.
Mutations in NPHS1 or NPHS2 lead to proteinuria and
rapid ESRD progression, highlighting the critical role of these
podocyte-specific structural proteins in glomerular filtration
barrier function (41,42). Although NPHS1 and NPHS2 are
detectable in the urine of glomerulonephritides patients and have
been described as clinically valuable biomarkers of podocyte
injury, the results between studies are inconsistent and raise
questions regarding the reasons behind the contrasting findings
(43). In the present study, a
decrease in urinary NPHS1 expression in FSGS was observed when
compared to other diagnoses, but no differences in NPHS2 levels
within the groups was found. These results partially agree with
previous studies that showed urinary NPHS1 and NPHS2 mRNA levels
were reduced in patients with MCN and FSGS, a feature that
correlates with the degree of proteinuria (44,45).
However, TUG1 expression levels showed no correlation with GFR,
proteinuria or albuminuria in the present study (data not shown).
Although it has been proposed that these podocyte-specific
biomarkers can detect nephropathy before the development of
albuminuria (46-48),
a larger patient cohort is required to assess the relationship
between TUG1 urinary expression and clinical markers of renal
dysfunction traditionally used in the diagnosis of
glomerulonephritides. In the present study, a positive correlation
between NPHS1 and NPHS2 mRNA levels and TUG1 expression was
observed, which suggests that the molecular mechanisms described
in vitro and in animal models of DN regarding the
relationship between TUG1 and podocyte loss are reflected in the
urine of non-diabetic patients (28).
A decrease in PGC1A, a transcriptional coactivator
that regulates energy homeostasis and mitochondrial biogenesis, has
been implicated in the development of acute kidney injury, DN and
renal fibrosis (49). The expression
levels and activity of PGC1A depend on multiple transcriptional and
post-transcriptional mechanisms, some mediated by non-coding RNAs
(22,28). Interactions between TUG1 and the
promoter of PGC1A enhances its transcription, and increases
mitochondrial content and cellular ATP levels whilst reducing
mitochondrial reactive oxygen species levels. In addition, TUG1
acts as a competitive endogenous RNA for miRNAs such as miR-145,
miR-144 and miR-204-5p (30,50,51).
miRNA miR-204-5p is involved in multiple cellular processes,
including angiogenesis, vascular disease, metabolism and glucose
homeostasis (30,52). Through in silico analysis,
targets of the TUG1/miR-204-5p axis involved in mitochondrial
biogenesis were predicted and it was subsequently shown that PGC1A,
COX5A and TFAM were also detectable in the urine and were
correlated with TUG1 expression. PGC1A is a known
posttranscriptional target of miR-204-5p (53,54);
however, COX5A and TFAM still require experimental validation.
Nonetheless, the results suggest that the TUG1/miR-204 axis and
their mitochondrial biogenesis targets are relevant in FSGS
pathogenesis and may serve as potential biomarkers.
The present study has some limitations, including
the small number of glomerulonephritides patients enrolled and the
variability of diagnosis, which included primary and secondary
glomerulonephritides. Nonetheless, our findings support a role for
lncRNA TUG1 in the development of glomerulonephritides, and justify
further exploration of urinary TUG1 as a potential biomarker of
FSGS. More extensive studies, with carefully selected subjects are
required to confirm the relationship between urinary TUG1
expression, clinical markers of renal dysfunction and glomerular
damage observed in kidney biopsies; and to validate lncRNA TUG1 as
a biomarker of FSGS. Additionally, more in vitro and animal
experiments focused on the TUG1/miR-204-5p axis should be performed
to increase our understanding of these non-coding RNAs in
mitochondrial bioenergetics and podocyte loss.
In conclusion, the results of the present study
highlight the potential of urinary non-coding RNAs, such as TUG1,
in the diagnosis of renal disease, and suggests that this lncRNA
and its mitochondrial-associated pathways are relevant in
glomerulonephritides other than DN. Further studies are required to
evaluate urinary TUG1 as a potential biomarker of podocytopathy,
and to determine its association with kidney dysfunction and
patient prognosis.
Acknowledgements
We would like to thank Claudia Susana Meza Calvillo
for her technical assistance as part of the Scientific Research
Summer Program of the Mexican Academy of Science 2019.
Funding
This study was supported by Consejo Nacional de
Ciencia y Tecnología (CONACyT), Mexico (grant no.
SALUD-2018-02-B-S-42687).
Availability of data and materials
The datasets used and/or analysed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FJS-T, MM-P and RE conceived the study. FJS-T and
MM-P collected and analysed the data. ZM performed the experiments.
CM-C performed the histopathological analysis of renal biopsies. RE
drafted the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study complied with the ethical
principles for medical research specified in the Declaration of
Helsinki and was approved by the Local Ethics and Research
Committee at the Hospital de Especialidades NUM. 1, Bajio, Leon,
Guanajuato, Instituto Mexicano del Seguro Social (approval no.
CLIEIS R-2018-1001-114). Each participant provided written informed
consent prior to enrolment in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Floege J and Amann K: Primary
glomerulonephritides. Lancet. 387:2036–2048. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Caliskan Y and Kiryluk K: Novel biomarkers
in glomerular disease. Adv Chronic Kidney Dis. 21:205–216.
2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Matovinović MS: Podocyte injury in
glomerular diseases. EJIFCC. 20:21–27. 2009.PubMed/NCBI
|
|
4
|
Mallipattu SK and He JC: The podocyte as a
direct target for treatment of glomerular disease? Am J Physiol
Renal Physiol. 311:F46–F51. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bagavant H and Fu SM: Pathogenesis of
kidney disease in systemic lupus erythematosus. Curr Opin
Rheumatol. 21:489–494. 2009.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hogan J, Mohan P and Appel GB: Diagnostic
tests and treatment options in glomerular disease: 2014 update. Am
J Kidney Dis. 63:656–666. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Parikh SV, Ayoub I and Rovin BH: The
kidney biopsy in lupus nephritis: Time to move beyond histology.
Nephrol Dial Transplant. 30:3–6. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Rovin BH, Almaani S and Malvar A:
Reimagining the kidney biopsy in the era of diagnostic biomarkers
of glomerular disease. Kidney Int. 95:265–267. 2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Mongelli A, Martelli F, Farsetti A and
Gaetano C: The dark that matters: Long Non-coding RNAs as master
regulators of cellular metabolism in Non-communicable diseases.
Front Physiol. 10(369)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lorenzen JM and Thum T: Long noncoding
RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol.
12:360–373. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ignarski M, Islam R and Müller RU: Long
Non-coding RNAs in kidney disease. Int J Mol Sci.
20(3276)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tian H, Wu M, Zhou P, Huang C, Ye C and
Wang L: The long non-coding RNA MALAT1 is increased in renal
ischemia-reperfusion injury and inhibits hypoxia-induced
inflammation. Ren Fail. 40:527–533. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hu S, Han R, Shi J, Zhu X, Qin W, Zeng C,
Bao H and Liu Z: The long noncoding RNA LOC105374325 causes
podocyte injury in individuals with focal segmental
glomerulosclerosis. J Biol Chem. 293:20227–20239. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Han R, Hu S, Qin W, Shi J, Zeng C, Bao H
and Liu Z: Upregulated long noncoding RNA LOC105375913 induces
tubulointerstitial fibrosis in focal segmental glomerulosclerosis.
Sci Rep. 9(716)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jin LW, Pan M, Ye HY, Zheng Y, Chen Y,
Huang WW, Xu XY and Zheng SB: Down-regulation of the long
non-coding RNA XIST ameliorates podocyte apoptosis in membranous
nephropathy via the miR-217-TLR4 pathway. Exp Physiol. 104:220–230.
2019.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
Liao Z, Ye Z, Xue Z, Wu L, Ouyang Y, Yao
C, Cui C, Xu N, Ma J, Hou G, et al: Identification of renal long
Non-coding RNA RP11-2B6.2 as a positive regulator of type I
interferon signaling pathway in lupus nephritis. Front Immunol.
10(975)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Santer L, López B, Ravassa S, Baer C,
Riedel I, Chatterjee S, Moreno MU, González A, Querejeta R, Pinet
F, et al: Circulating long noncoding RNA LIPCAR predicts heart
failure outcomes in patients without chronic kidney disease.
Hypertension. 73:820–828. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Huang YS, Hsieh HY, Shih HM, Sytwu HK and
Wu CC: Urinary Xist is a potential biomarker for membranous
nephropathy. Biochem Biophys Res Commun. 452:415–421.
2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Forbes JM and Thorburn DR: Mitochondrial
dysfunction in diabetic kidney disease. Nat Rev Nephrol.
14:291–312. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Young TL, Matsuda T and Cepko CL: The
noncoding RNA taurine upregulated gene 1 is required for
differentiation of the murine retina. Curr Biol. 15:501–512.
2005.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Li SY, Park J, Qiu C, Han SH, Palmer MB,
Arany Z and Susztak K: Increasing the level of peroxisome
proliferator-activated receptor γ coactivator-1α in podocytes
results in collapsing glomerulopathy. JCI Insight.
2(e92930)2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li SY and Susztak K: The long noncoding
RNA Tug1 connects metabolic changes with kidney disease in
podocytes. J Clin Invest. 126:4072–4075. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
LeBleu VS, O'Connell JT, Gonzalez Herrera
KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A,
Domingos Chinen LT, Rocha RM, et al: PGC-1α mediates mitochondrial
biogenesis and oxidative phosphorylation in cancer cells to promote
metastasis. Nat Cell Biol. 16:992–1003, 1-15. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
25
|
Uldry M, Yang W, St-Pierre J, Lin J, Seale
P and Spiegelman BM: Complementary action of the PGC-1 coactivators
in mitochondrial biogenesis and brown fat differentiation. Cell
Metab. 3:333–341. 2006.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Finck BN and Kelly DP: PGC-1 coactivators:
Inducible regulators of energy metabolism in health and disease. J
Clin Invest. 116:615–622. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Kang HM, Ahn SH, Choi P, Ko YA, Han SH,
Chinga F, Park AS, Tao J, Sharma K, Pullman J, et al: Defective
fatty acid oxidation in renal tubular epithelial cells has a key
role in kidney fibrosis development. Nat Med. 21:37–46.
2015.PubMed/NCBI View
Article : Google Scholar
|
|
28
|
Long J, Badal SS, Ye Z, Wang Y, Ayanga BA,
Galvan DL, Green NH, Chang BH, Overbeek PA and Danesh FR: Long
noncoding RNA Tug1 regulates mitochondrial bioenergetics in
diabetic nephropathy. J Clin Invest. 126:4205–4218. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
29
|
Association WM: World medical association
declaration of Helsinki: Ethical principles for medical research
involving human subjects. JAMA. 310:2191–2194. 2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Yu C, Li L, Xie F, Guo S, Liu F, Dong N
and Wang Y: LncRNA TUG1 sponges miR-204-5p to promote osteoblast
differentiation through upregulating Runx2 in aortic valve
calcification. Cardiovasc Res. 114:168–179. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ben-Ari Fuchs S, Lieder I, Stelzer G,
Mazor Y, Buzhor E, Kaplan S, Bogoch Y, Plaschkes I, Shitrit A,
Rappaport N, et al: GeneAnalytics: An integrative gene set analysis
tool for next generation sequencing, RNAseq and microarray data.
OMICS. 20:139–151. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4(e05005)2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Levey AS, Eckardt KU, Tsukamoto Y, Levin
A, Coresh J, Rossert J, De Zeeuw D, Hostetter TH, Lameire N and
Eknoyan G: Definition and classification of chronic kidney disease:
A position statement from Kidney Disease: Improving Global Outcomes
(KDIGO). Kidney Int. 67:2089–2100. 2005.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Fukuda A, Wickman LT, Venkatareddy MP,
Wang SQ, Chowdhury MA, Wiggins JE, Shedden KA and Wiggins RC: Urine
podocin:nephrin mRNA ratio (PNR) as a podocyte stress biomarker.
Nephrol Dial Transplant. 27:4079–4087. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Civelek M, Hagopian R, Pan C, Che N, Yang
WP, Kayne PS, Saleem NK, Cederberg H, Kuusisto J, Gargalovic PS, et
al: Genetic regulation of human adipose microRNA expression and its
consequences for metabolic traits. Hum Mol Genet. 22:3023–3037.
2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Saez-Rodriguez J, Rinschen MM, Floege J
and Kramann R: Big science and big data in nephrology. Kidney Int.
95:1326–1337. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kretzler M, Cohen CD, Doran P, Henger A,
Madden S, Gröne EF, Nelson PJ, Schlöndorff D and Gröne HJ:
Repuncturing the renal biopsy: Strategies for molecular diagnosis
in nephrology. J Am Soc Nephrol. 13:1961–1972. 2002.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Sun IO and Lerman LO: Urinary microRNA in
kidney disease: Utility and roles. Am J Physiol Renal Physiol.
316:F785–F793. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Brandenburger T, Salgado Somoza A, Devaux
Y and Lorenzen JM: Noncoding RNAs in acute kidney injury. Kidney
Int. 94:870–881. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Patrakka J, Kestilä M, Wartiovaara J,
Ruotsalainen V, Tissari P, Lenkkeri U, Männikkö M, Visapää I,
Holmberg C, Rapola J, et al: Congenital nephrotic syndrome (NPHS1):
Features resulting from different mutations in Finnish patients.
Kidney Int. 58:972–980. 2000.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Boute N, Gribouval O, Roselli S, Benessy
F, Lee H, Fuchshuber A, Dahan K, Gubler MC, Niaudet P and Antignac
C: NPHS2, encoding the glomerular protein podocin, is mutated in
autosomal recessive steroid-resistant nephrotic syndrome. Nat
Genet. 24:349–354. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
43
|
Sekulic M and Pichler Sekulic S: A
compendium of urinary biomarkers indicative of glomerular
podocytopathy. Patholog Res Int. 2013(782395)2013.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hara M, Yanagihara T and Kihara I: Urinary
podocytes in primary focal segmental glomerulosclerosis. Nephron.
89:342–347. 2001.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Szeto CC, Wang G, Chow KM, Lai FM, Ma TK,
Kwan BC, Luk CC and Li PK: Podocyte mRNA in the urinary sediment of
minimal change nephropathy and focal segmental glomerulosclerosis.
Clin Nephrol. 84:198–205. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Pätäri A, Forsblom C, Havana M, Taipale H,
Groop PH and Holthöfer H: Nephrinuria in diabetic nephropathy of
type 1 diabetes. Diabetes. 52:2969–2974. 2003.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ng DP, Tai BC, Tan E, Leong H, Nurbaya S,
Lim XL, Chia KS, Wong CS, Lim WY and Holthöfer H: Nephrinuria
associates with multiple renal traits in type 2 diabetes. Nephrol
Dial Transplant. 26:2508–2514. 2011.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Chang JH, Paik SY, Mao L, Eisner W,
Flannery PJ, Wang L, Tang Y, Mattocks N, Hadjadj S, Goujon JM, et
al: Diabetic kidney disease in FVB/NJ Akita mice: Temporal pattern
of kidney injury and urinary nephrin excretion. PLoS One.
7(e33942)2012.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Lynch MR, Tran MT and Parikh SM: PGC1α in
the kidney. Am J Physiol Renal Physiol. 314:F1–F8. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Tan J, Qiu K, Li M and Liang Y:
Double-negative feedback loop between long non-coding RNA TUG1 and
miR-145 promotes epithelial to mesenchymal transition and
radioresistance in human bladder cancer cells. FEBS Lett.
589:3175–3181. 2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Cai H, Xue Y, Wang P, Wang Z, Li Z, Hu Y,
Li Z, Shang X and Liu Y: The long noncoding RNA TUG1 regulates
blood-tumor barrier permeability by targeting miR-144. Oncotarget.
6:19759–19779. 2015.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Flores-Pérez A, Marchat LA,
Rodríguez-Cuevas S, Bautista-Piña V, Hidalgo-Miranda A, Ocampo EA,
Martínez M, Palma-Flores C, Fonseca-Sánchez MA, Astudillo-de la
Vega H, et al: Dual targeting of ANGPT1 and TGFBR2 genes by miR-204
controls angiogenesis in breast cancer. Sci Rep.
6(34504)2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Maniyadath B, Chattopadhyay T, Verma S,
Kumari S, Kulkarni P, Banerjee K, Lazarus A, Kokane SS, Shetty T,
Anamika K and Kolthur-Seetharam U: Loss of hepatic oscillatory fed
microRNAs Abrogates Refed transition and causes liver dysfunctions.
Cell Rep. 26:2212–2226. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Houzelle A, Dahlmans D, Nascimento EBM,
Schaart G, Jörgensen JA, Moonen-Kornips E, Kersten S, Wang X and
Hoeks J: MicroRNA-204-5p modulates mitochondrial biogenesis in
C2C12 myotubes and associates with oxidative capacity in humans. J
Cell Physiol. 235:9851–9863. 2020.PubMed/NCBI View Article : Google Scholar
|