Introduction
Breast cancer (BC) is the most common malignancy in
females and the second leading cause of cancer-related death after
lung cancer worldwide (1). BC has a
significant impact on a women's health (2), and its incidence rates have been
steadily increasing in recent years in Arab-speaking communities,
with a significant number of cases being diagnosed in the first
instance at advanced stages of the disease (3). BC incidence varies widely globally
(4), and its incidence amongst
Saudi Arabian women has progressively increased (3). BC is a complex and multifactorial
disease, and genetic, hormonal and environmental factors contribute
to its pathogenesis (5). The
interplay between the genetic background and the environment in BC
development has been proposed, but with mixed results on the
importance of each (6-8).
Female sex, age and ethnicity are the strongest risk factors
associated with increased incidence (9). In addition, obesity (10), age at first delivery of a child
(11), early menarche/late
menopause (12), ionizing radiation
exposure (13), breastfeeding
(14), past/current estrogen
treatment (15), breast tissue
density (16), smoking and alcohol
consumption (17) and steroid
hormone receptors (18) are all
known risk factors of BC.
BC aggregates in families, and an estimated 5-10% of
BC cases are hereditary in origin caused by a myriad of
susceptibility genes, transmitted from parent to child (19). These include rare variants in
BRCA1, BRCA2, PALB2, ATM and
CHEK2 genes, which reportedly confer a moderate-high
lifetime risk of the disease. Other variants of >70 loci, which
were identified through genome-wide association study, and
large-scale replication studies (20), were also reported to confer
heightened risk of disease, though to varying extents.
While hereditary BC is linked to a well-established
set of susceptibility genes, the exact contribution of these genes
to disease pathogenesis remains largely unknown (21). BRCA1 (chromosome 17) and
BRCA2 (chromosome 13), described as regulators of DNA
repair, transcription and cell cycle progression in response to DNA
damage, were confirmed genetic loci associated with genetic
susceptibility to BC (22,23). In this regard, it was shown that
pathogenic BRCA1/BRCA2 mutations account for almost
30% of BC cases in high-risk families (24). Polymorphisms in other genes are also
involved in BC, but to variable extents (25), suggesting that genetic variations
may explain the heterogeneous nature of BC, and thus
inter-individual differences with regard to tumor behavior
(20).
AKT1, PIK3CA, PTEN and
TP53 have been identified as recurrently mutated genes, and
somatic mutations in these genes are found at a high frequency in
BC patients. According to the Catalogue of Somatic Mutations in
Cancer (COSMIC) database (26),
high prevalence rates of PIK3CA (26.4%), TP53
(24.7%), PTEN (3.8%), and AKT1 (2.8%) were reported
for BC.
In addition to the aforementioned mutated genes, two
kinds of genomic instability have been often reported in BC and
seldom in proliferative breast disease: Microsatellite instability
(MSI) and loss of heterozygosity (27). Clinical testing for MSI involves
immunohistochemistry and PCR testing for four proteins of the
mismatch repair pathway: MSH2, MSH6, MLH1 and PMS2. MSI has been
documented in BC, but at a lower frequency compared with other
types of cancer. Previous studies reported that 0.9% of primary
Triple Negative BC (28) and 1.53%
of BC of all subtypes (29) have
MSI.
In view of its heterogeneous etiology, the
manifestations of BC vary widely among individual patients, with
each patient having a unique profile, hence highlighting the
potential value of precision medicine and individualized therapies
for effective management (30-32).
Next-Generation sequencing (NGS) was recently employed to improve
identification of novel gene mutations responsible for disease
manifestation amongst BC patients (33-36).
Ion Torrent™ technology allows the parallel
sequencing of several genes, thus overcoming the problems inherent
with conventional sequencing. In this study, Ion semiconductor
sequencing technology with the Ion S5 System and AmpliSeq™ Cancer
Hotspot Panel v2 was used to analyze ~2,800 COSMIC mutations from
50 oncogenes and tumor suppressor genes in a cohort of 32 BC cases
from Saudi Arabia. Therefore, the present study aims to investigate
the efficiency of AmpliSeq™ Cancer Hotspot Panel v2 on the
detection of mutations in the genomic DNA extracted from the whole
blood of the Saudi BC patients.
Patients and methods
Study subjects
Ethical approval for the present study was obtained
from the Ethics Committee of King Fahad Medical City (KFMC; Riyadh,
Saudi Arabia; IRB approval no. FWA00018774), and the study was
performed in accordance with the guidelines described in the
Helsinki Declaration (37). A total
of 32 Saudi Arabian patients with BC (mean age 48.5±8.2 years;
median age 45 years [age range, 31-85 years; interquartile range
(IQR) 42.5-55.5)], and with histologically confirmed invasive BC,
were recruited from Medical Oncology Department, KFMC. None of the
patients had a history of other cancer types and were not subjected
to chemo-, radio or hormone therapy. In addition, 32 healthy Saudi
women with no familial history of any cancer types served as the
controls (mean age 49.1±11.0 years; median age, 47.5 years (age
range, 35-71 years; IQR, 42.5-55), were recruited into this
retrospective case-controlled study from the blood bank (Table I). Demographic and clinical data of
BC patients and control women were collected from the hospital
records. All participants provided signed informed consent prior to
inclusion in this study.
 | Table IDemographics and clinical
characteristics of the cohorts. |
Table I
Demographics and clinical
characteristics of the cohorts.
|
Characteristics | Healthy controls,
n=32 | Patients BC,
n=32 | P-value |
|---|
| Mean age,
yearsc | 49.09±11.02 | 48.80±8.28 | 0.904d |
| BMI,
kg/m2b | 27.60±5.66 | 32.89±7.96 | 0.004b,d |
| Oral contraceptives
use | | | 0.5e |
|
Yes | 19 | 20 | |
|
No | 13 | 12 | |
| Breastfeeding | | | 0.011a,e |
|
Yes | 19 | 9 | |
|
No | 13 | 23 | |
| Tumor size | | | - |
|
<2
cm | - | 4 | |
|
≥2 cm | - | 28 | |
| Tumor stage | | | - |
|
I | - | 3 | |
|
II | - | 11 | |
|
III | - | 13 | |
|
IV | - | 5 | |
| Histological
classification | | | - |
|
IDC | - | 29 | |
|
ILC | - | 2 | |
|
DCIS | - | 1 | |
| Tumor location | | | |
|
Left | - | 28 | |
|
Right | - | 4 | |
| ER status | | | - |
|
ER+ | - | 23 | |
|
ER- | - | 9 | |
| PR status | | | - |
|
PR+ | - | 21 | |
|
PR- | - | 11 | |
| HER2 status | | | - |
|
HER2+ | - | 13 | |
|
HER2- | - | 19 | |
DNA extraction and quantification
Peripheral blood (2 ml) was collected in EDTA tubes
from each participant. Genomic DNA was extracted using a
QIAamp® DNA Mini Kit according to manufacturer's
instructions (Qiagen GmbH) then quantified using a Qubit™ dsDNA BR
Assay Kit on a Qubit 2.0 Fluorometer (Thermo Fisher Scientific,
Inc.), following the manufacturer's instructions.
Library preparation
Manual library preparations were performed using an
Ion AmpliSeq™ Cancer Hotspot Panel v.2, Ion Xpress barcoded
adapters, and an Ion AmpliSeq Library Kit 2.0 (Thermo Fisher
Scientific, Inc.). The panel consisted of 207 amplicons, covering
~20,000 bases surveying hotspot regions, including up to 2,855
COSMIC mutations in 50 oncogenes and tumor suppressor genes, all
with known cancer associations. The genes included in this panel
were: ABL1, AKT1, ALK, APC, ATM,
BRAF, CDH1, CDKN2A, CSF1R,
CTNNB1, EGFR, ERBB2, ERBB4,
EZH2, FBXW7, FGFR1, FGFR2,
FGFR3, FLT3, GNA11, GNAS, GNAQ,
HNF1A, HRAS, IDH1, IDH2, JAK2,
JAK3, KDR, KIT, KRAS, MET,
MLH1, MPL, NOTCH1, NPM1, NRAS,
PDGFRA, PIK3CA, PTEN, PTPN11,
RB1, RET, SMAD4, SMARCB1, SMO,
SRC, STK11, TP53 and VHL.
Multiplex PCR was performed using 10 ng genomic DNA
with a premixed primer pool and Ion AmpliSeq HiFi master mix (Ion
AmpliSeq Library Kit 2.0). The amplicons were treated with 2
µl FuPa reagent to partially digest the primer sequences and
phosphorylate the amplicons. Amplicons were ligated to adapters
with the diluted barcodes of Ion Xpress Barcode Adapters kit
(Thermo Fisher Scientific, Inc.). The adapter-ligated amplicons
(library) were purified using the Agencourt AMPure XP reagent
(Beckman Coulter, Inc.). Quantification of the final libraries was
performed using an Ion Library TaqMan™ Quantitation Kit (Thermo
Fisher Scientific, Inc.) on the Applied Biosystems® 7500
Real-Time PCR System, following the manufacturer's protocols.
Template preparation and chip
loading
After library dilution to ~100 pM, the clonal
amplification of barcoded DNA library (AmpliSeq libraries) onto ion
spheres was performed on an Ion Chef™ Instrument using Ion 520™
& Ion 530™ Kit-Chef, according to the manufacturer's
instructions (Thermo Fisher Scientific, Inc.). Template-positive
spheres from barcoded libraries were multiplexed and loaded onto
Ion 530™ Chips following the manufacturer's protocol, and
sequencing was run on the Ion Gene Studio S5 system (Thermo Fisher
Scientific, Inc.).
Statistical analysis
Samples were evaluated for genomic alterations,
including single nucleotide variants (SNVs), and insertions and
deletions, using Ion Reporter Software v.5.6 (Thermo Fisher
Scientific, Inc.), after alignment to the hg19 human reference
genome. Of note, higher quality requirements for variant analysis
and selection, high-quality SNVs and insertion/deletion variants
were strictly followed in this study and were defined as: i)
FILTER=PASS, ii) QUAL ≥100, iii) depth coverage ≥20X, and iv)
variant fraction ≥20%. The sequencing data analysis using such
approach yielded high-quality variants that did not necessary
require additional confirmatory testing (such as through Sanger
sequencing validation) as recommended by Arteche-López group
(38). Furthermore, the ‘bam’ files
of each clinically actionable variant were carefully reviewed in
order to provide additional confidence to the accuracy and
reliability of the NGS calls.
Qualitative and quantitative data were analyzed
using SPSS v.21 (IBM Corp.). Qualitative data are presented as the
frequency and percentage of total, and these data were compared
using a χ2 goodness-of-fit test, while continuous
variables are presented as the mean ± SD, and were compared using
an unpaired two-tailed Student's t-test. P<0.05 was considered
to indicate a statistically significant difference.
Results
Study subjects
The clinical and demographic characteristics of the
study subjects are summarized in Table
I. Patients were clinically characterized in terms of tumor
size, location, stage, histological classification and
presence/absence of tumor markers, including estrogen receptor
(ER), progesterone receptor (PR), and human epidermal growth factor
receptor 2 (HER2). In addition, age, body mass index (BMI), use of
oral contraception and breastfeeding were compared between the two
groups. No statistically significant differences were noted between
patients and controls regarding mean age (P=0.90), and oral
contraceptive use (P=0.50). However, a significant difference was
noted between BC patients and healthy subjects for mean BMI, which
was higher in patients compared with the control group (P=0.004),
and breastfeeding (P=0.011), which was higher in the healthy
control group.
Invasive ductal carcinoma of no specific type (n=29,
90.6%) was the predominant histological type of primary tumor,
followed by invasive lobular carcinoma (n=2, 6.3%), and ductal
carcinoma in situ (n=1, 3.1%). The majority of cases were
stage III (n=13, 40.6%) and stage II (n=11, 34.4%), whereas stage I
(n=3, 9.3%) and stage IV (n=5, 15.6%) were less common. In
addition, 4 patients (12.5%) had ER+, PR+,
HER2- tumors; 21 patients (65.6%) had ER+,
and/or PR+, HER2+ tumors, 4 patients (12.5%)
had ER-, PR-, HER2+ tumors, and 3
patients (9.4%) had triple-negative (ER-,
PR-, HER2-) tumors.
Somatic mutations
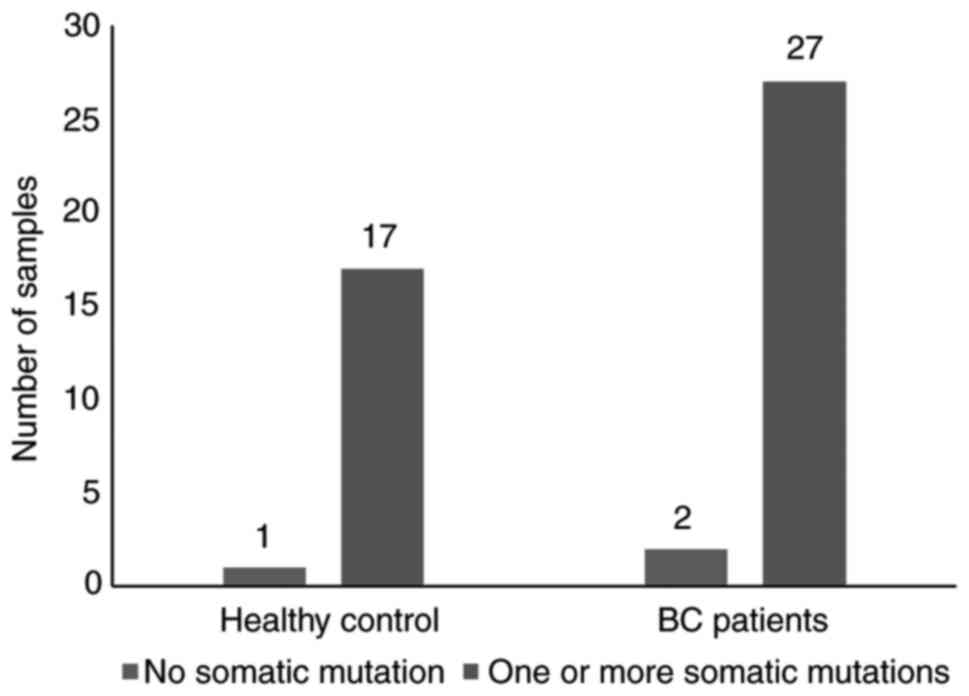
There were only three unclassified samples, two from
patients (sample #2 and sample #13) and 1 from a control individual
(sample #4) that did not possess any identified mutations (Fig. 1). The observed mutations were
detected with varied frequencies across BC patients (Table II) and healthy controls (Table III); 26 mutations, including 17
(65%) missense point mutations (SNV), and 9 (35%) frameshift
(insertion/deletion) mutations in 11 genes (out of 50);
TP53, PIK3CA, KDR, KIT, ATM,
HRAS, ERBB2, FGFR3, GNAQ, APC
and JAK3 (Fig. 2) across the
cohort in the 61 samples (95%).
 | Table IIMutational status of breast cancer
patient samples analyzed using the Ion AmpliSeq™ Cancer Hotspot
Panel v2. |
Table II
Mutational status of breast cancer
patient samples analyzed using the Ion AmpliSeq™ Cancer Hotspot
Panel v2.
| No. | Tumor histologic
type | Stage | Hormone receptor
status | Genes | Mutations
detected | Effect | Amino acid
change |
|---|
| 1 | IDC | III |
HER2+ | ER+ | PR+ | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | KDR | c.1416A>T | Missense | p.Gln472His |
| | | | | | | GNAQ | c.625C>A | Missense | p.Gln209Lys |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 3 | IDC | IV |
HER2- | ER- | PR- | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | HRAS | c.84_85insT, | Frameshift | p.Val29fs, |
| | | | | | | HRAS | c.80_81insC | Frameshift | p.Val29fs |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| | | | | | | JAK3 | c.394C>A | Missense | p.Pro132Thr |
| 4 | IDC | III |
HER2- | ER+ | PR+ | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | KDR | c.1416A>T | Missense | p.Gln472His |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 5 | IDC | II |
HER2+ | ER+ | PR+ | KIT | c.1621A>C | Missense | p.Met541Leu |
| | | | | | | TP53 |
c.21+I9:K95C>G | Missense | p.Pro72Arg |
| 6 | IDC | III |
HER2+ | ER+ | PR+ | KDR | c.1416A>T | Missense | p.Gln472His |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 7 | IDC | III |
HER2+ | ER+ | PR- | APC | c.3949G>C | Missense | p.Glu1317Gln |
| | | | | | | ATM | c.2572T>C | Missense | p.Phe858Leu |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 8 | IDC | II |
HER2+ | ER+ | PR+ | KDR | c.1416A>T | Missense | p.Gln472His |
| | | | | | | TP53 | c.21G | Missense | p.Pro72Arg |
| 9 | IDC | II |
HER2- | ER+ | PR+ | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 10 | IDC | II |
HER2+ | ER+ | PR+ | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 11 | IDC | IV |
HER2- | ER- | PR- | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 12 | DCIS | III |
HER2+ | ER- | PR- | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | KDR | c.1416A>T | Missense | p.Gln472His |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 14 | IDC | I |
HER2+ | ER- | PR- | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 15 | IDC | III |
HER2+ | ER+ | PR+ | ATM | c.2525C>G | Missense | p.Thr842Ser |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 16 | IDC | IV |
HER2+ | ER+ | PR+ | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | PR+ | APC | c.3920T>A | Missense | p.Ile1307Lys |
| | | | | | | APC |
c.3920_3921delTA | Frameshift | p.Ile1307fs |
| | | | | | | APC | c.3920delT | Frameshift | p.Ile1307fs |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 17 | IDC | IV |
HER2+ | ER+ | PR+ | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | KIT | c.1621A>C | Missense | p.Met541Leu |
| | | | | | | TP53 | c.215_216insG | Frameshift | p.Val73fs |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 18 | IDC | II |
HER2+ | ER+ | PR+ | KDR | c.1416A>T | Missense | p.Gln472His |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 19 | IDC | III |
HER2- | ER- | PR- | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 20 | IDC | II |
HER2+ | ER+ | PR+ | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 21 | ILC | I |
HER2+ | ER+ | PR+ | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | FGFR3 | c.2389G>C | Missense | p.Ala797Pro |
| | | | | | | ATM | c.7313C>T | Missense | p.Thr2438Ile |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 22 | IDC | III |
HER2+ | ER+ | PR+ | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 23 | IDC | III |
HER2+ | ER+ | PR+ | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 24 | IDC | III |
HER2+ | ER+ | PR+ | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | HRAS | c.84_85insT | Frameshift | p.Val29fs, |
| | | | | | | HRAS | c.80_81insC | Frameshift | p.Val29fs |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 25 | IDC | III |
HER2- | ER- | PR- | KIT | c.1621A>C | Missense | p.Met541Leu |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 26 | ILC | III |
HER2+ | ER+ | PR+ | KIT | c.1621A>C | Missense | p.Met541Leu |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 27 | IDC | II |
HER2+ | ER+ | PR+ | PIK3CA | c.233A>G | Missense | p.Glu78Gly |
| | | | | | | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| 28 | IDC | IV |
HER2+ | ER+ | PR+ | PIK3CA | c.233A>G | Missense | p.Glu78Gly |
| | | | | | | ATM | c.1810C>T | Missense | p.Pro604Ser |
| | | | | | | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 29 | IDC | II |
HER2- | ER+ | PR+ | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | KDR | c.1416A>T | Missense | p.Gln472His |
| 30 | ILC | II |
HER2- | ER+ | PR+ | KDR | c.1416A>T | Missense | p.Gln472His |
| | | | | | | ATM | c.3905G>T | Missense | p.Gly1302Val |
| 31 | IDC | II |
HER2+ | ER- | PR- | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 32 | IDC | II |
HER2+ | ER- | PR- | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | | | | | | KDR | c.1416A>T | Missense | p.Gln472His |
 | Table IIIMutational status of healthy control
samples analyzed using the Ion AmpliSeq™ Cancer Hotspot Panel
v2. |
Table III
Mutational status of healthy control
samples analyzed using the Ion AmpliSeq™ Cancer Hotspot Panel
v2.
| No. | Genes | Mutations
detected | Effect | Amino acid
change |
|---|
| 1 | KDR | c.1416A>T | Missense | p.Gln472His |
| | HRAS | c.80_81insC | Frameshift | p.Val29fs |
| | HRAS | c.80_81insC | Frameshift | p.Val29fs |
| | ERBB2 | c.2380G>T | Missense | p.Val794Leu |
| 2 | KDR | c.1416A>T | Missense | p.Gln472His |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 3 | TP53 | c.215C>G | Missense | p.Pro72Arg |
| | |
c.209_215delCTCCCCCinsTCCCCCG | Frameshift |
p.Ala70_Pro72delinsValProArg |
| 5 | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 6 | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | APC | c.3920T>A | Missense | p.Ile1307Lys |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 7 | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 8 | PIK3CA | c.233A>G | Missense | p.Glu78Gly |
| | FGFR3 | c.2389G>C | Missense | p.Ala797Pro |
| | KDR | c.1416A>T | Missense | p.Gln472His |
| 9 | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 10 | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 11 | APC | c.3920T>A | Missense | p.Ile1307Lys |
| | APC | c.3920delT | Frameshift | p.Ile1307fs |
| 12 | ATM | c.1811delC | Frameshift | p.Pro604fs |
| | ATM | c.1810C>T | Missense | p.Pro604Ser |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | KIT | c.1621A>C | Missense | p.Met541Leu |
| 13 | KDR | c.1416A>T | Missense | p.Gln472His |
| | APC | c.3920T>A | Missense | p.Ile1307Lys |
| | APC |
c.3920_3921delTA | Frameshift | p.Ile1307fs |
| 14 | PIK3CA | c.233A>G | Missense | p.Glu78Gly |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 15 | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | GNAQ | c.625C>A | Missense | p.Gln209Lys |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| | TP53 |
c.209_215delCTCCCCCinsTCCCCCG | Frameshift |
p.Ala70_Pro72delinsValProArg |
| 16 | KDR | c.1416A>T | Missense | p.Gln472His |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 17 | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| 18 | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | KDR | c.1416A>T | Missense | p.Gln472His |
| | ATM | c.2572T>C | Missense | p.Phe858Leu |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 19 | KDR | c.1416A>T | Missense | p.Gln472His |
| | JAK3 | c.394C>A | Missense | p.Pro132Thr |
| 20 | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 21 | ATM | c.2572T>C | Missense | p.Phe858Leu |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 22 | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | KDR | c.1416A>T | Missense | p.Gln472His |
| 23 | KIT | c.1621A>C | Missense | p.Met541Leu |
| | KDR | c.1416A>T | Missense | p.Gln472His |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 24 | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | APC | c.3920T>A | Missense | p.Ile1307Lys |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 25 | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 26 | KIT | c.1621A>C | Missense | p.Met541Leu |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 27 | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 28 | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 29 | PIK3CA | c.1173A>G | Missense | p.Ile391Met |
| | APC | c.3920delT | Frameshift | p.Ile1307fs |
| | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 30 | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 31 | TP53 | c.215C>G | Missense | p.Pro72Arg |
| 32 | TP53 | c.215C>G | Missense | p.Pro72Arg |
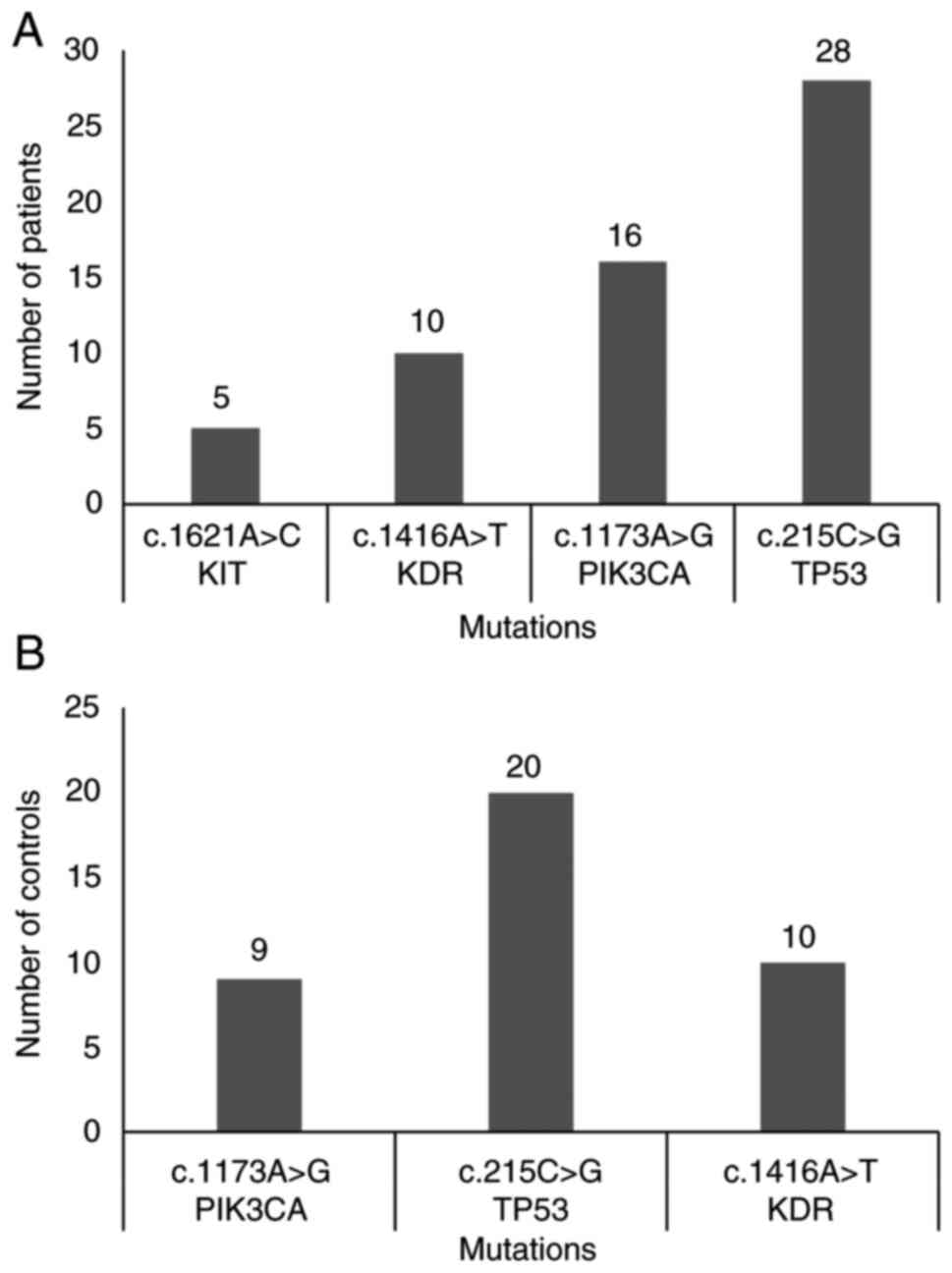
Amongst BC patients, 27 patients (84.8%) were
positive for >1 somatic mutation, compared with 17 control
subjects (53.1%) (Fig. 1). The most
frequently observed concurrent mutations were some combination of
c.215C>G (TP53), c.1173A>G (PIK3CA),
c.1416A>T (KDR) and c.1621A>C (KIT) in patients
(Fig. 3A), and c.215C>G
(TP53), c.1173A>G (PIK3CA) and
c.1416A>T (KDR) in healthy controls (Fig. 3B).
The site of the most frequent mutations within
TP53 and PIK3CA differed between samples. The most
common TP53 mutation detected was a missense mutation
(c.215C>G) in 26 patients (86.70%), with only 1 frameshift
mutation (c.215_216insG) identified in 1 patient (3.33%). The most
common PIK3CA mutation detected was c.1173A>G, located in
exon 9, and was identified in 15 (50%) patients.
Table IV summarizes
the distribution of the most frequent missense mutations in
TP53 and PIK3CA (c.215C>G and c.1173A>G,
respectively), between patients and healthy control samples. A
significant difference (P=0.020) was observed in the frequency of
c.215C>G (Pro72Arg), which was higher in patients (0.87) than in
controls (0.61). Similarly, a significant difference was observed
in the frequency of c.1173A>G (Pro72Arg) (P=0.041) that was
higher in patients (0.53) compared with the healthy control
participants (0.25).
 | Table IVFrequency distributions of the most
frequent mutations TP53 and PIK3CA between the breast cancer
patients and the healthy controls. |
Table IV
Frequency distributions of the most
frequent mutations TP53 and PIK3CA between the breast cancer
patients and the healthy controls.
| Gene | Mutation | Amino acid
change | Chromosome | Exon | NCBI 1000 Genomes
Browser ID | Variant frequency
in the patients | Variant frequency
in the healthy controls |
P-valueb |
|---|
| TP53 | c.215C>G | p.Pro72Arg | 17 | 3 | rs1042522 | 0.87 | 0.61 | 0.020a |
| PIK3CA | c.1173A>G | p.Ile391Met | 3 | 9 | rs2230461 | 0.53 | 0.25 | 0.041a |
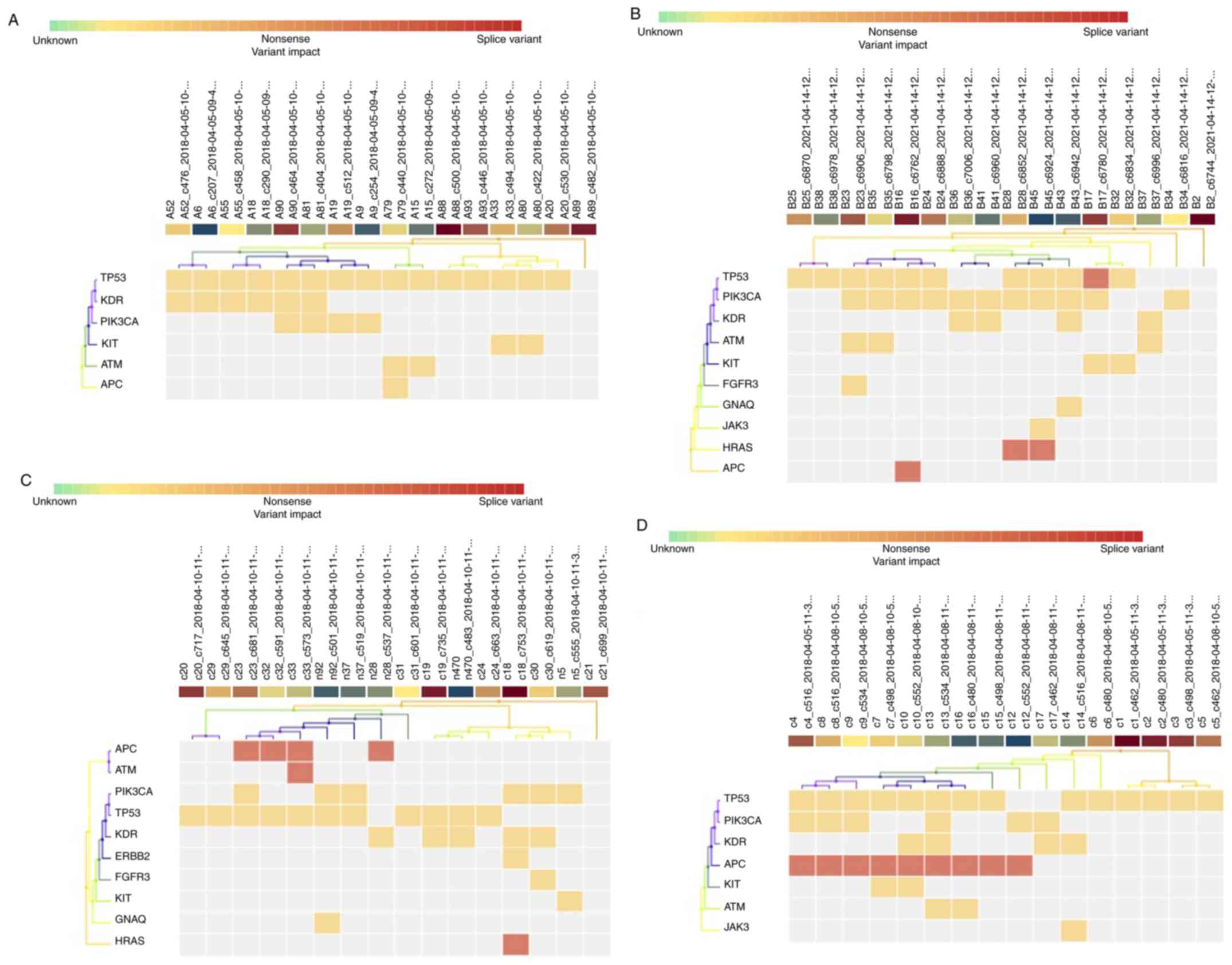
A unique mutational status was identified for every
BC patient except for patients #8, #10, #18, and #20 (Table II).
Discussion
BC is the most common type of cancer amongst Arabian
women, and the major cause of disease-associated mortality in women
worldwide (39). In the Middle
East, Arabian women face a significantly higher risk of mortality,
as the cancer is often diagnosed at a later stage in disease
progression (40-45).
Current efforts to manage BC include on improving
prevention, diagnosis and an increased armamentarium of effective
treatment choices for patients with BC (46). Due to the heterogeneity of BC and
the interactions between genetic and environmental factors, each
patient's tumor possibly exhibits an unique gene mutation profile
(47). By profiling an individual's
cancer genome, it becomes possible to differentiate the oncogenic
mechanisms that regulate cancer and, therefore, the genetic
biomarkers that may be specifically associated with the disease
state (5).
In this study, massively parallel sequencing was
performed to identify frequent mutations in 32 BC Saudi Arabian
patients and 32 healthy controls using Ion Torrent sequencing
technology.
In this study, 32 BC patients were clinically
characterized in terms of tumor size, location, stage, histological
classification, and the presence or absence of tumor markers such
as ER, PR, and HER2. In addition, four common risk factors,
including age, BMI, oral contraceptives use and breastfeeding, were
also evaluated and compared between healthy controls and BC
patients. The statistical analysis showed no significant difference
in the mean age between BC patients and healthy controls as the two
study groups were already age matched.
BMI was significantly higher in BC patients compared
with the healthy control group. This finding is in agreement with
several reports, highlighting BMI as one of the most important risk
factors for the BC (48,49). Chronic low-levels of inflammation
that are usually observed in obese people can lead to BC through
the increased likelihood of DNA damage (50). Additionally, fat cells can produce
an excess amount of estrogen and adipokines, which can stimulate
cell growth, as observed in BC (49,51).
Similarly, a significant difference was observed for breastfeeding,
which was higher in control group compared to the BC patients
group, supporting the previously published literature linking
breastfeeding to reduced BC risk (52).
PIK3CA and TP53 are the most
frequently mutated genes, and harbor most of the mutations in this
cohort. The p53 tumor suppressor gene, located on chromosome 17p13,
is 20 kb long, encompassing 11 exons encoding a 53 kDa
phosphoprotein (53). In the
present study the detected mutations in this gene were 1 missense
mutations (c.215C>G) in 26 patients (86.70%) and 1 frameshift
mutation (c.215_216insG) in 1 patient (3.33%). These results are in
agreement with the current release of the International Agency for
Research on Cancer TP53 database (http://www-p53.iarc.fr/), which also shows that all
TP53 mutations were missense mutations in the coding region
(54). The detected mutations were
located within exons 3 and 5, encoding the Proline-rich domain,
which plays a role in p53-mediated apoptosis and in the
DNA-binding and oligomerization domain. These regions are required
for interactions with FBX042, HIPK1 and AXIN1, the DNA major
groove, and a domain containing a nuclear export signal (46,55,56).
Dysfunction of p53 can cause defective DNA replication and
malignant transformation, common in dysplasia's of BC (53). The p53 gene exhibits several genetic
alterations in patients with BC (57). This highlights the need to
administer effective treatments such as cell-cycle inhibitors in
the form of target therapies and combinatorial target therapies
against the wide range of TP53 mutations.
In the current study, the high TP53 mutation
rate in the cohort could be explained by the high number of
ER+ cases, given that 67% of the observed TP53
mutations occurred in the ER+ tumors. ER status is
tightly associated with the molecular subtypes, and a significantly
higher TP53 mutation rate was demonstrated in the basal-like
subtype, mainly ER-, and HER2-enriched (both ER- and
ER+) tumors compared with the primarily ER+
luminal type (56,58). A recent study by Bai et al
(59) conducted in 2021 using NGS
to detect TP53 mutations in the cell free DNA in Chinese
metastatic BC (MBC) patients indicated that TP53 mutations could be
used as a prognostic marker for worse outcomes in MBC and for the
response of adjuvant endocrine therapy. TP53-mutated MBC
patients had a significantly worse outcome than TP53
wild-type patients, especially those in the
HR+/HER2- and triple-negative BC (TNBC)
cohorts. TP53 mutations were also associated with endocrine
resistance (59).
In the present study TP53 mutations were not
associated with HER2- tumors, which is comparable to the
previously published research. TP53 mutation status was an
independent predictive factor of survival especially in
HR+/HER2- and TNBC cohorts, but not in the
HER2+ cohort (59,60).
In the present study, the somatic TP53 mutation
c.215C>G p.(Pro72Arg) was the most frequently detected mutation
in all BC patients, particularly those with Stage III BC. A
previous study showed that TP53 mutation NM_000546.5:c.824G>A
p.(Cys275Tyr) was the most common mutation detected in 82 patients
with Stage I-III BC who underwent NGS using tissue and blood
samples, and they showed that TP53 pathogenic somatic mutations
were associated with an 8-fold risk of recurrence in the univariate
Cox regression analysis (61). The
same study showed that the coexistence of TP53 and PIK3CA mutations
was a common finding in BC patients (61). PIK3CA mutation (c.1173A>G)
and p.(Ile391Met) located in exon 9, was identified in 15 (50%)
patients. This exon encodes the helical domain, and mutations,
represented by single amino acid substitutions, in this domain are
associated with increased lipid kinase activity, and thus induce
oncogenic transformation (62,63).
The PI3K pathway has been identified as a major
player in cancer development and progression (62-66).
PI3K is a heterodimeric enzyme composed of a p110α catalytic
subunit encoded by the PIK3CA gene and a p85 regulatory subunit
encoded by the PIK3R1 gene (67).
In the present study, 47% of the BC patients were carriers of
PIK3CA mutations. This result corroborates the findings of
the previous work, reporting that PIK3CA mutations occur in
20-40% of BC and ≈30% of tumors of the prostate, cervix and
endometrium (68,69). Several studies have suggested that
PIK3CA mutations are more frequent in ER+ and
HER2+ BC cases (68,69).
Accordingly, the low mutation rate can be explained by a bias in
the subtype distribution of the cohort. In the present study, 2 out
of 5 (40%) of the PIK3CA mutations occurred in
ER+ primary tumors. The small size of the cohort may
have influenced this distribution. Martínez-Sáez et al
(70) showed that 28% of
PIK3CA mutations identified in circulating tumor DNA (ctDNA)
in 48 patients with advanced HR+/HER2- BC
were not part of the therascreen® PIK3CA test
(QIAGEN GmbH), a FDA approved kit used to select patients who
possessed PIK3CA mutations in tumor tissue specimens and/or
in circulating tumor DNA (ctDNA) isolated from plasma specimens
(71). Therascreen PIK3CA detects
11 PIK3CA hotspot mutations, mostly found in exons 9 and
20(71).
It is important to remember that only a subset of
genes was examined in the present study, and that for a deeper
understanding of the mutational profiles of BC patients,
considerably more extensive sequencing is required. The hotspot
panel was not specifically developed for BC, and it includes areas
and genes that are more commonly mutated in other cancer types
(72).
Recently, Hempel et al (73) utilized a wide NGS panel to
investigate 41 MBC samples and found that PIK3CA mutations
appear in 34% of the patients. A recent study reported that 22% of
the total population and 28% of patients with HR+ BC
have a PIK3CA mutation (74). Further research using a large NGS
panel targeting 1,021 genes in 193 MBC samples by Tang et al
(75) detected 36 (18.7%) mutations
in the kinase domain and 26 (13.5%) substitutions in the helical
domain, with 10 (5.2%) additional alterations distributed in the
remaining PIK3CA sequence.
PIK3CA mutations in the ctDNA of patients
with BC have also been reported (76). Board et al (77) was able to detect PIK3CA
mutations in the vast majority (80%) of ctDNA samples from
PIK3CA-mutated MBC, but not in early BC (78).
In conclusion, the results of this investigation
showed that Ion Torrent DNA Sequencing technology using AmpliSeq
Cancer Hotspot Panel v2 was found to be a suitable method to
perform molecular characterization of the genotype of BC patients
and healthy controls using peripheral blood samples. The present
study revealed specific mutational profiles for every BC patient;
for this reason, it may be a possible to improve diagnosis and
prognosis, and recommend personalized treatments for each BC
patient based on the mutational profile. The primary benefits of
NGS are that it allows for the identification of multiple mutations
at the same time, eliminating the need for sequential individual
tests. Therefore, this technique could be routinely implemented in
cancer diagnosis, as it can precisely identify fusions, SNPs, copy
number variants, and insertion/deletions. Additionally,
confirmation of NGS variants must be carefully investigated and
validated through Sanger sequencing to avoid false positive
outcomes. The results of the present study add to the existing body
of knowledge and practice in the diagnosis and treatment of BC
patients.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by Naif Arab University for
Security Sciences, Kingdom of Saudi Arabia.
Availability of data and materials
The datasets used and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
SAM conceived the study, curated the data, analyzed
the data, performed the experiments and wrote and reviewed the
manuscript. NAAS, BA and SRB contributed to the design of the
study, performed the experiments and drafted the manuscript. ABA
and AMA performed the experiments and contributed to data analysis
and interpretation. MA, AB and WYA conceived the study, performed
data interpretation, and wrote and critically reviewed the
manuscript. SAM, MA and AB confirm the authenticity of all the raw
data. All the authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Ethical approval for this study was obtained from
the Ethics Committee of King Fahad Medical City (KFMC; Riyadh,
Saudi Arabia; IRB approval no. FWA00018774), and the study was
performed in accordance with the guidelines described in the
Helsinki Declaration. All participants provided signed informed
consent prior to inclusion in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Stewart BW and Wild CP (eds): World Cancer
Report 2014. International Agency for Research on Cancer, WHO,
2014.
|
|
2
|
Testa U, Castelli G and Pelosi E: Breast
cancer: A molecularly heterogenous disease needing subtype-specific
treatments. Med Sci (Basel). 8(18)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Almutlaq BA, Almuazzi RF, Almuhayfir AA,
Alfouzan AM, Alshammari BT, AlAnzi HS and Ahmed HG: Breast cancer
in Saudi Arabia and its possible risk factors. J Cancer Policy.
12:83–89. 2017.
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang J, Späth SS, Marjani SL, Zhang W and
Pan X: Characterization of cancer genomic heterogeneity by
next-generation sequencing advances precision medicine in cancer
treatment. Precis Clin Med. 1:29–48. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Fasching PA, Ekici AB, Adamietz BR,
Wachter DL, Hein A, Bayer CM, Häberle L, Loehberg CR, Jud SM,
Heusinger K, et al: Breast cancer risk-genes, environment and
clinics. Geburtshilfe Frauenheilkd. 71:1056–1066. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rudolph A, Chang-Claude J and Schmidt MK:
Gene-environment interaction and risk of breast cancer. Br J
Cancer. 114:125–133. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hiatt RA, Haslam SZ and Osuch J: Breast
Cancer and the Environment Research Centers. The breast cancer and
the environment research centers: Transdisciplinary research on the
role of the environment in breast cancer etiology. Environ Health
Perspect. 117:1814–1822. 2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Winters S, Martin C, Murphy D and Shokar
NK: Breast cancer epidemiology, prevention, and screening. Prog Mol
Biol Transl Sci. 151:1–32. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Parkin DM and Boyd L: 8. Cancers
attributable to overweight and obesity in the UK in 2010. Br J
Cancer. 105 (Suppl 2):S34–S37. 2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Nelson HD, Zakher B, Cantor A, Fu R,
Griffin J, O'Meara ES, Buist DS, Kerlikowske K, van Ravesteyn NT,
Trentham-Dietz A, et al: Risk factors for breast cancer for women
aged 40 to 49 years: A systematic review and meta-analysis. Ann
Intern Med. 156:635–648. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Collaborative Group on Hormonal Factors in
Breast Cancer. Menarche, menopause, and breast cancer risk:
Individual participant meta-analysis, including 118 964 women with
breast cancer from 117 epidemiological studies. Lancet Oncol.
13:1141–1151. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Fenga C: Occupational exposure and risk of
breast cancer. Biomed Rep. 4:282–292. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Elkum N, Al-Tweigeri T, Ajarim D,
Al-Zahrani A, Amer SM and Aboussekhra A: Obesity is a significant
risk factor for breast cancer in Arab women. BMC Cancer.
14(788)2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gnant M, Mlineritsch B, Schippinger W,
Luschin-Ebengreuth G, Pöstlberger S, Menzel C, Jakesz R, Seifert M,
Hubalek M, Bjelic-Radisic V, et al: Endocrine therapy plus
zoledronic acid in premenopausal breast cancer. N Engl J Med.
360:679–691. 2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
McCormack VA and dos Santos Silva I:
Breast density and parenchymal patterns as markers of breast cancer
risk: A meta-analysis. Cancer Epidemiol Biomarkers Prev.
15:1159–1169. 2006.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Druesne-Pecollo N, Touvier M, Barrandon E,
Chan DS, Norat T, Zelek L, Hercberg S and Latino-Martel P: Excess
body weight and second primary cancer risk after breast cancer: A
systematic review and meta-analysis of prospective studies. Breast
Cancer Res Treat. 135:647–654. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Abderrahman B and Jordan VC: Rethinking
extended adjuvant antiestrogen therapy to increase survivorship in
breast cancer. JAMA Oncol. 4:15–16. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Apostolou P and Fostira F: Hereditary
breast cancer: The era of new susceptibility genes. Biomed Res Int.
2013(747318)2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Michailidou K, Beesley J, Lindstrom S,
Canisius S, Dennis J, Lush MJ, Maranian MJ, Bolla MK, Wang Q, Shah
M, et al: Genome-wide association analysis of more than 120,000
individuals identifies 15 new susceptibility loci for breast
cancer. Nat Genet. 47:373–380. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Luen S, Virassamy B, Savas P, Salgado R
and Loi S: The genomic landscape of breast cancer and its
interaction with host immunity. Breast. 29:241–250. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Mundhofir FE, Wulandari CE, Prajoko YW and
Winarni TI: BRCA1 gene mutation screening for the hereditary breast
and/or ovarian cancer syndrome in breast cancer cases: A first high
resolution DNA melting analysis in Indonesia. Asian Pac J Cancer
Prev. 17:1539–1546. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yoshida K and Miki Y: Role of BRCA1 and
BRCA2 as regulators of DNA repair, transcription, and cell cycle in
response to DNA damage. Cancer Sci. 95:866–871. 2004.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Couch FJ, Nathanson KL and Offit K: Two
decades after BRCA: Setting paradigms in personalized cancer care
and prevention. Science. 343:1466–1470. 2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Byler S, Goldgar S, Heerboth S, Leary M,
Housman G, Moulton K and Sarkar S: Genetic and epigenetic aspects
of breast cancer progression and therapy. Anticancer Res.
34:1071–1077. 2014.PubMed/NCBI
|
|
26
|
Forbes SA, Beare D, Boutselakis H, Bamford
S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al:
COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids
Res. 45:D777–D783. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Marcus L, Lemery SJ, Keegan P and Pazdur
R: FDA approval summary: Pembrolizumab for the treatment of
microsatellite instability-high solid tumors. Clin Cancer Res.
25:3753–3758. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kurata K, Kubo M, Mori H, Kawaji H,
Motoyama Y, Kuroki L, Yamada M, Kaneshiro K, Kai M and Nakamura M:
Abstract P1-06-11: Microsatellite instability in triple negative
breast cancers. Cancer Res. 79 (Suppl 4)(P1-06-11)2019.
|
|
29
|
Bonneville R, Krook MA, Kautto EA, Miya J,
Wing MR, Chen HZ, Reeser JW, Yu L and Roychowdhury S: Landscape of
microsatellite instability across 39 cancer types. JCO Precis
Oncol: Oct 3, 2017 (Epub ahead of print). doi:
10.1200/PO.17.00073.
|
|
30
|
Le Du F, Eckhardt BL, Lim B, Litton JK,
Moulder S, Meric-Bernstam F, Gonzalez-Angulo AM and Ueno NT: Is the
future of personalized therapy in triple-negative breast cancer
based on molecular subtype? Oncotarget. 6:12890–12908.
2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu YR, Jiang YZ, Xu XE, Yu KD, Jin X, Hu
X, Zuo WJ, Hao S, Wu J, Liu GY, et al: Comprehensive transcriptome
analysis identifies novel molecular subtypes and subtype-specific
RNAs of triple-negative breast cancer. Breast Cancer Res.
18(33)2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Sachdev JC, Sandoval AC and Jahanzeb M:
Update on precision medicine in breast cancer. Cancer Treat Res.
178:45–80. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Nagahashi M, Shimada Y, Ichikawa H,
Kameyama H, Takabe K, Okuda S and Wakai T: Next generation
sequencing-based gene panel tests for the management of solid
tumors. Cancer Sci. 110:6–15. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Smith NG, Gyanchandani R, Shah OS, Gurda
GT, Lucas PC, Hartmaier RJ, Brufsky AM, Puhalla S, Bahreini A, Kota
K, et al: Targeted mutation detection in breast cancer using
MammaSeq™. Breast Cancer Res. 21(22)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Desmedt C, Voet T, Sotiriou C and Campbell
PJ: Next-generation sequencing in breast cancer: First take home
messages. Curr Opin Oncol. 24:597–604. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ma R, Gong J and Jiang X: Novel
applications of next-generation sequencing in breast cancer
research. Genes Dis. 4:149–153. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Shrestha B and Dunn L: The declaration of
Helsinki on medical research involving human subjects: A review of
seventh revision. J Nepal Health Res Counc. 17:548–552.
2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Arteche-López A, Ávila-Fernández A, Romero
R, Riveiro-Álvarez R, López-Martínez MA, Giménez-Pardo A,
Vélez-Monsalve C, Gallego-Merlo J, García-Vara I, Almoguera B, et
al: Sanger sequencing is no longer always necessary based on a
single-center validation of 1109 NGS variants in 825 clinical
exomes. Sci Rep. 11(5697)2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Donnelly TT, Khater AH, Al-Bader SB, Al
Kuwari MG, Malik M, Al-Meer N, Singh R and Fung T: Factors that
influence awareness of breast cancer screening among Arab women in
Qatar: Results from a cross sectional survey. Asian Pac J Cancer
Prev. 15:10157–10164. 2014.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Azaiza F and Cohen M: Health beliefs and
rates of breast cancer screening among Arab women. J Womens Health
(Larchmt). 15:520–530. 2006.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Baron-Epel O, Friedman N and Lernau O:
Reducing disparities in mammography-use in a multicultural
population in Israel. Int J Equity Health. 8(19)2009.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Bener A, Ayub H, Kakil R and Ibrahim W:
Patterns of cancer incidence among the population of Qatar: A
worldwide comparative study. Asian Pac J Cancer Prev. 9:19–24.
2008.PubMed/NCBI
|
|
43
|
Najjar H and Easson A: Age at diagnosis of
breast cancer in Arab nations. Int J Surg. 8:448–452.
2010.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Soskolne V, Marie S and Manor O: Beliefs,
recommendations and intentions are important explanatory factors of
mammography screening behavior among Muslim Arab women in Israel.
Health Educ Res. 22:665–676. 2007.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Tarabeia J, Baron-Epel O, Barchana M,
Liphshitz I, Ifrah A, Fishler Y and Green MS: A comparison of
trends in incidence and mortality rates of breast cancer, incidence
to mortality ratio and stage at diagnosis between Arab and Jewish
women in Israel, 1979-2002. Eur J Cancer Prev. 16:36–42.
2007.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Bai X, Zhang E, Ye H, Nandakumar V, Wang
Z, Chen L, Tang C, Li J, Li H, Zhang W, et al: PIK3CA and TP53 gene
mutations in human breast cancer tumors frequently detected by ion
torrent DNA sequencing. PLoS One. 9(e99306)2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Cho SH, Jeon J and Kim SI: Personalized
medicine in breast cancer: A systematic review. J Breast Cancer.
15:265–272. 2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
van den Brandt PA, Spiegelman D, Yaun SS,
Adami HO, Beeson L, Folsom AR, Fraser G, Goldbohm RA, Graham S,
Kushi L, et al: Pooled analysis of prospective cohort studies on
height, weight, and breast cancer risk. Am J Epidemiol.
152:514–527. 2000.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Gallagher EJ and LeRoith D: Obesity and
diabetes: The increased risk of cancer and cancer-related
mortality. Physiol Rev. 95:727–748. 2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Gregor MF and Hotamisligil GS:
Inflammatory mechanisms in obesity. Annu Rev Immunol. 29:415–445.
2011.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Seo BR, Bhardwaj P, Choi S, Gonzalez J,
Andresen Eguiluz RC, Wang K, Mohanan S, Morris PG, Du B, Zhou XK,
et al: Obesity-dependent changes in interstitial ECM mechanics
promote breast tumorigenesis. Sci Transl Med.
7(301ra130)2015.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Anstey EH, Shoemaker ML, Barrera CM,
O'Neil ME, Verma AB and Holman DM: Breastfeeding and breast cancer
risk reduction: Implications for black mothers. Am J Prev Med. 53
(Suppl 1):S40–S46. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Levine AJ: p53, the cellular gatekeeper
for growth and division. Cell. 88:323–331. 1997.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Langerød A, Zhao H, Borgan Ø, Nesland JM,
Bukholm IR, Ikdahl T, Kåresen R, Børresen-Dale AL and Jeffrey SS:
TP53 mutation status and gene expression profiles are powerful
prognostic markers of breast cancer. Breast Cancer Res.
9(R30)2007.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Petitjean A, Mathe E, Kato S, Ishioka C,
Tavtigian SV, Hainaut P and Olivier M: Impact of mutant p53
functional properties on TP53 mutation patterns and tumor
phenotype: Lessons from recent developments in the IARC TP53
database. Hum Mutat. 28:622–629. 2007.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Silwal-Pandit L, Vollan HK, Chin SF, Rueda
OM, McKinney S, Osako T, Quigley DA, Kristensen VN, Aparicio S,
Børresen-Dale AL, et al: TP53 mutation spectrum in breast cancer is
subtype specific and has distinct prognostic relevance. Clin Cancer
Res. 20:3569–3580. 2014.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Ko LJ and Prives C: p53: Puzzle and
paradigm. Genes Dev. 10:1054–1072. 1996.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Cancer Genome Atlas Network. Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Bai H, Yu J, Jia S, Liu X, Liang X and Li
H: Prognostic value of the TP53 mutation location in metastatic
breast cancer as detected by next-generation sequencing. Cancer
Manag Res. 13:3303–3316. 2021.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Ren J, Wang B and Li J: Integrating
proteomic and phosphoproteomic data for pathway analysis in breast
cancer. BMC Syst Biol. 12 (Suppl 8)(S130)2018.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Andrikopoulou A, Terpos E, Chatzinikolaou
S, Apostolidou K, Ntanasis-Stathopoulos I, Gavriatopoulou M,
Dimopoulos MA and Zagouri F: TP53 mutations determined by targeted
NGS in breast cancer: A case-control study. Oncotarget.
12:2206–2214. 2021.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Bader AG, Kang S and Vogt PK:
Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc
Natl Acad Sci USA. 103:1475–1479. 2006.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Kang S, Bader AG and Vogt PK:
Phosphatidylinositol 3-kinase mutations identified in human cancer
are oncogenic. Proc Natl Acad Sci USA. 102:802–807. 2005.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Fresno Vara JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and González-Barón M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Noorolyai S, Shajari N, Baghbani E,
Sadreddini S and Baradaran B: The relation between PI3K/AKT
signalling pathway and cancer. Gene. 698:120–128. 2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Janku F, Yap TA and Meric-Bernstam F:
Targeting the PI3K pathway in cancer: Are we making headway? Nat
Rev Clin Oncol. 15:273–291. 2018.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Liu P, Cheng H, Roberts TM and Zhao JJ:
Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev
Drug Discov. 8:627–644. 2009.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Cizkova M, Susini A, Vacher S,
Cizeron-Clairac G, Andrieu C, Driouch K, Fourme E, Lidereau R and
Bièche I: PIK3CA mutation impact on survival in breast cancer
patients and in ERα, PR and ERBB2-based subgroups. Breast Cancer
Res. 14(R28)2012.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Saal LH, Holm K, Maurer M, Memeo L, Su T,
Wang X, Yu JS, Malmström PO, Mansukhani M, Enoksson J, et al:
PIK3CA mutations correlate with hormone receptors, node metastasis,
and ERBB2, and are mutually exclusive with PTEN loss in human
breast carcinoma. Cancer Res. 65:2554–2559. 2005.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Martínez-Sáez O, Chic N, Pascual T, Adamo
B, Vidal M, González-Farré B, Sanfeliu E, Schettini F, Conte B,
Brasó-Maristany F, et al: Frequency and spectrum of PIK3CA somatic
mutations in breast cancer. Breast Cancer Res.
22(45)2020.PubMed/NCBI View Article : Google Scholar
|
|
71
|
QIAGEN Manchester, Ltd.
therascreen® PIK3CA RGQ PCR Kit Instructions for Use
(Handbook), 2019 (cited 2022 16 january); Available from:
https://www.accessdata.fda.gov/cdrh_docs/pdf19/P190001C.pdf.
|
|
72
|
Bellevicine C, Sgariglia R, Nacchio M, De
Luca C, Pisapia P, Pepe F and Troncone G: Molecular testing of
thyroid fine-needle aspiration: Local issues and solutions. An
interventional cytopathologist perspective. J Mol Pathol.
2:233–240. 2021.
|
|
73
|
Hempel D, Ebner F, Garg A, Trepotec Z,
Both A, Stein W, Gaumann A, Güttler L, Janni W, DeGregorio A, et
al: Real world data analysis of next generation sequencing and
protein expression in metastatic breast cancer patients. Sci Rep.
10(10459)2020.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Mosele F, Stefanovska B, Lusque A, Tran
Dien A, Garberis I, Droin N, Le Tourneau C, Sablin MP, Lacroix L,
Enrico D, et al: Outcome and molecular landscape of patients with
PIK3CA-mutated metastatic breast cancer. Ann Oncol. 31:377–386.
2020.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Tang Y, Li J, Xie N, Yang X, Liu L, Wu H,
Tian C, He Y, Wang X, He Q, et al: PIK3CA gene mutations in the
helical domain correlate with high tumor mutation burden and poor
prognosis in metastatic breast carcinomas with late-line therapies.
Aging (Albany NY). 12:1577–1590. 2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Anderson EJ, Mollon LE, Dean JL, Warholak
TL, Aizer A, Platt EA, Tang DH and Davis LE: A systematic review of
the prevalence and diagnostic workup of PIK3CA mutations in
HR+/HER2-metastatic breast cancer. Int J Breast Cancer.
2020(3759179)2020.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Board RE, Wardley AM, Dixon JM, Armstrong
AC, Howell S, Renshaw L, Donald E, Greystoke A, Ranson M, Hughes A
and Dive C: Detection of PIK3CA mutations in circulating free DNA
in patients with breast cancer. Breast Cancer Res Treat.
120:461–467. 2010.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Fusco N, Malapelle U, Fassan M, Marchiò C,
Buglioni S, Zupo S, Criscitiello C, Vigneri P, Dei Tos AP, Maiorano
E and Viale G: PIK3CA mutations as a molecular target for hormone
receptor-positive, HER2-negative metastatic breast cancer. Front
Oncol. 11(644737)2021.PubMed/NCBI View Article : Google Scholar
|