Introduction
Subarachnoid hemorrhage (SAH) due to rupture of
intracranial aneurysms (IAs) is a devastating type of stroke
(1). Despite progress in diagnosis
and treatment, the incidence and 30-day mortality rates of SAH have
remained stable for more than three decades (2).
However, the primary cause of IA rupture is not
clearly understood. Previous studies have shown that chronic
vascular inflammation, hemodynamic stress, and other factors lead
to arterial wall remodeling, smooth muscle cell proliferation, and
aneurysm rupture (3). In addition
to known risk factors, genetic predisposition and family history
play a significant role in aneurysm formation and progression
(4). Population-based studies have
reported that first-degree relatives of patients with IAs have up
to eight times greater risk of SAH than the general population
(5). Van Hoe et al reported
that individuals with ≥2 affected first-degree relatives had a
greater prevalence of IA (average 13.1% vs. 3% in the general
population) (6). Whole-exome
sequencing (WES) is the most effective and rational approach for
determining rare genetic variants and identifying the genetic basis
of diseases through the investigation of family forms. Rare
variants with relatively large individual effects need to be
detected, and recent studies have identified associations in family
cases with only a small number of genes (7-14).
The objective of the present study was to identify
the genetic risk factor of IAs/SAH in an affected Kazakh family
using WES while comparing them to 145 ethnicity-matched healthy
individuals.
Materials and methods
Clinical phenotyping
The present study was approved (Approval no.
1/16.02.2015) by the Human Research Ethics Committee of the
National Center for Neurosurgery (Nur Sultan, Kazakhstan).
In accordance with the Familial Intracranial
Aneurysm (FIA) study protocol (15), the following inclusion and exclusion
criteria were applied: Eligible families: i) Families with at least
two living affected siblings; ii) families with at least two
affected siblings, one of whom is living and the other whose
genotype can be reconstructed through the collection of
closely-related living family members (i.e., spouses and children);
iii) Families with ≥3 affected family members (such as cousin,
uncle, aunt), two of whom are alive and have living connecting
relatives; and iv) families with ≥3 affected family members, with
one living affected and at least one other affected relative whose
genotype can be reconstructed through the collection of closely
related living family members.
The exclusion criteria included a family history of
polycystic kidney disease, Ehlers-Danlos syndrome, Marfan syndrome,
fibromuscular dysplasia, Moya-Moya syndrome, or failure to obtain
informed consent from the patient or family members. Patients with
a history of neurological or connective tissue diseases were
excluded.
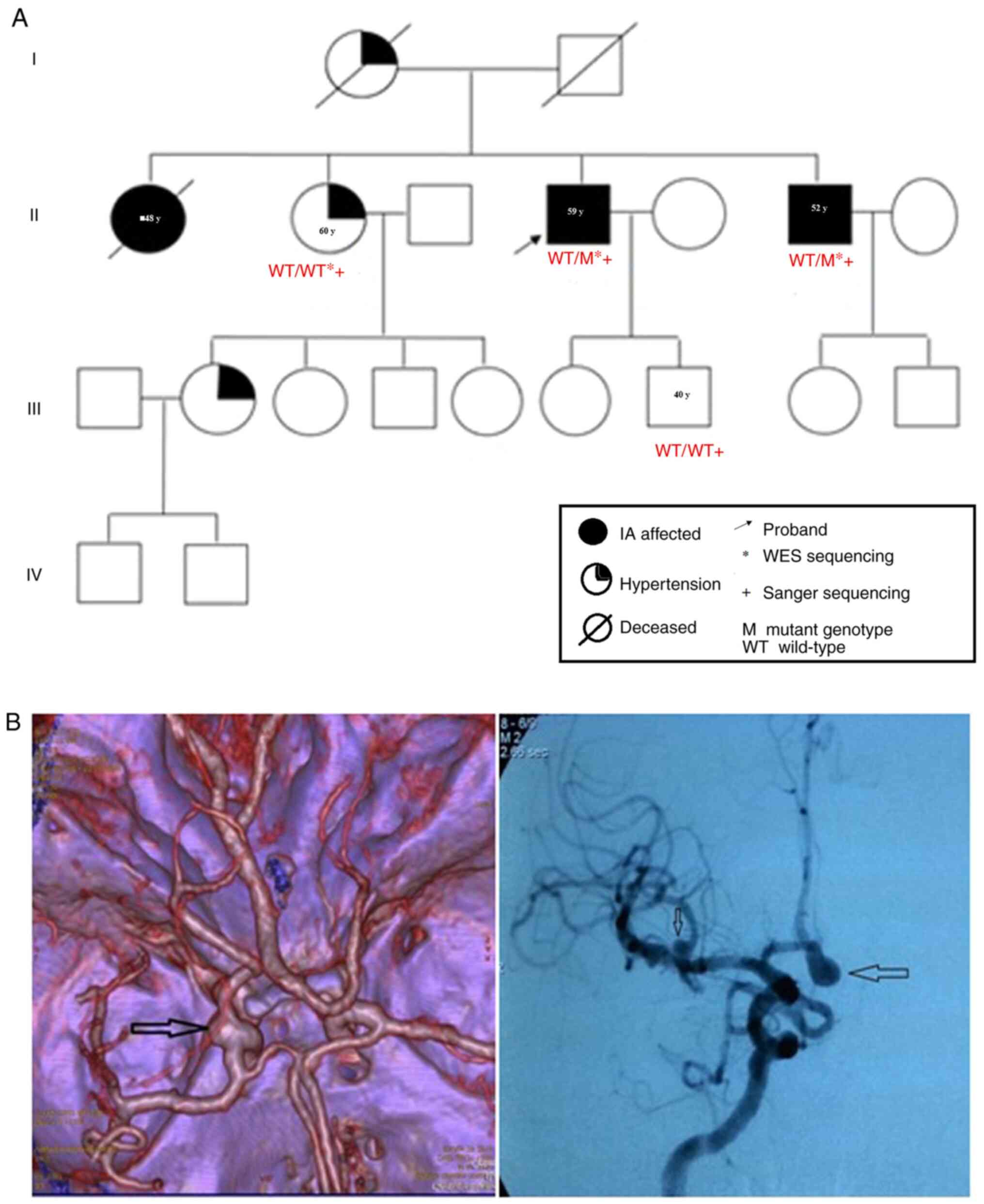
Eventually, a large family of Kazakh ethnicity
(Fig. 1A) with two IA- and
SAH-affected individuals among the 20 living members were
recruited. IA cases and SAH-affected individuals were examined
independently by three experienced neurologists from the Department
of Vascular and Functional Neurosurgery of the National Center for
Neurosurgery (Nur Sultan, Kazakhstan) using computed tomography or
magnetic resonance angiography (Fig.
1B). Peripheral blood samples were obtained from two
IA-affected individuals (II:4 and II:6) and two available family
members. The clinical characteristics of these four participants
are shown in Table I. Written
informed consent was obtained from all participants.
 | Table IPrimer sequences and
characteristics. |
Table I
Primer sequences and
characteristics.
| Primer name | Sequence (5'–3') | Ta
(˚C) | Amplicon length
(bp) |
|---|
| ALCAM_F |
tctctcctgctgaatacagt | 58 | 560 |
| ALCAM_R |
attcagaagagacactcataga | 58 | |
A total of 145 healthy individuals were selected
from the control group in our previous study of sporadic IA cases
(16). The control group comprised
healthy individuals aged 18-80 years old with no personal or family
history of IA, SAH, or other neurological disorders (such as
arteriovenous malformations of the brain, cavernous angiomas, brain
tumors, craniocerebral trauma, and connective tissue diseases
including Marfan syndrome and Ehlers-Danlos syndrome). All the
individuals included in the present study were unrelated. In all
cases, IA was diagnosed using computed tomography angiography (CTA)
or selective cerebral angiography (SCA). The participants were only
of Kazakh nationality.
WES analysis
Initially, only three family members [the proband
(II:4) and two siblings (II:2, II:6)] were available for WES
(Fig. 1A). From the collected
pedigree with IAs, genomic DNA was isolated from two affected
siblings (II:4, II:6) and one unaffected individual (II:2) using
the Promega Wizard Genomic DNA Purification Kit (cat. no. A1125;
Promega Corporation) according to the manufacturer's protocol. DNA
quantification was performed using PicoGreen method (cat. no.
P7589; Invitrogen; Thermo Fisher Scientific, Inc.) and VICTOR3™
multilabel plate reader. Genomic DNA libraries were prepared
according to the standard protocol provided by Illumina, Inc.
Agilent Technologies 2100 Bioanalyzer using a DNA 1000 chip was
used to verify the library size. The protein-coding regions of
human genomic DNA were captured using an Agilent SureSelect V6-Post
kit (Agilent Technologies, Inc.) and sequenced on a Novaseq 6000
platform (Illumina, Inc.) with 150 bp paired-end reads.
Single-nucleotide variants (SNVs) and insertion/deletions (indels)
were detected using the GATK best practice guidelines (17). ANNOVAR (annotation of genetic
variants) tools were used to annotate the variants for location and
the corresponding gene and transcript length (18).
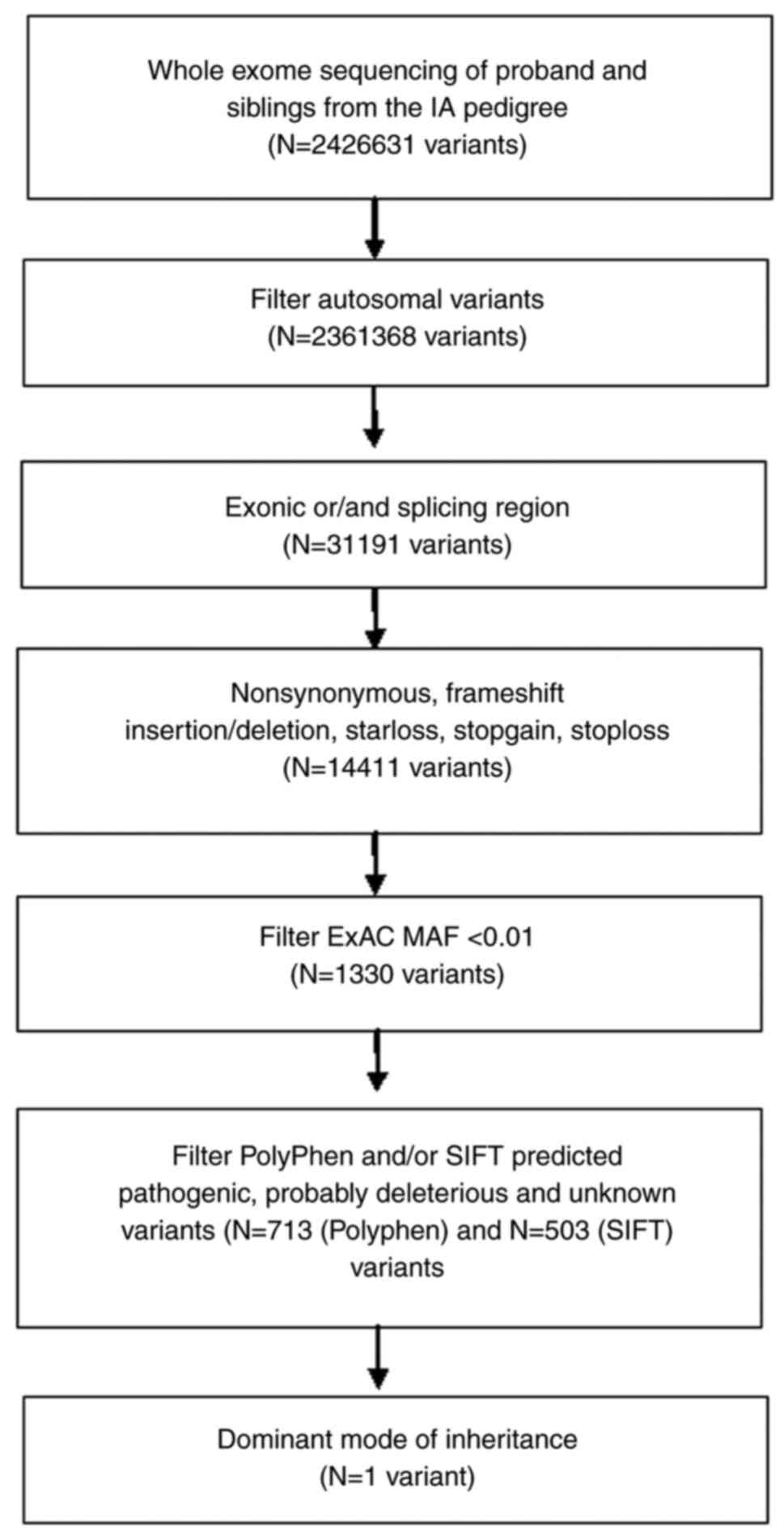
A flowchart detailing variant filtering is
illustrated in Fig. 2.
SNVs located in the intron region and synonymous
variants that did not affect the splicing site were excluded.
Variants with a minor allele frequency (MAF) >0.01 in the 1000
Genomes database (http://www.1000genomes.org) and exome aggregation
consortium (ExAC; http://exac.broadinstitute.org) were discarded.
Programs were used to evaluate the potential pathogenicity of the
SNVs, including SIFT (http://sift.jcvi.org), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2),
LRT (http://www.genetics.wustl.edu/jflab/lrt_query.html)
and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar). All variants
were compared against publicly available databases, such as the
1000 Genomes project (http://internationalgenome.org/), the exome variant
server, NHLBI GO exome sequencing project (ESP; http://evs.gs.washington.edu/EVS/), ExAC
(http://exac.broadinstitute.org/), and
the genome aggregation database (gnomAD; http://gnomad.broadinstitute.org/).
Variant confirmation
To confirm the variants identified by WES, Sanger
sequencing was performed on the DNA obtained from peripheral blood
samples of four members of the participating family. Additionally,
the DNA from the son of the proband (the only one available in the
next generation) was sequenced by the same method. Moreover, the
prevalence of the variant in 145 ethnicity-matched healthy
individuals from our previous study on sporadic cases of IA, was
analyzed (16).
Specifically, polymerase chain reaction (PCR)
primers (forward and reverse) were designed, and their sequences
are listed in Table I. Amplified
PCR was performed with a final volume of 20 µl, containing 50 ng/µl
of the genomic DNA, 10 pmol of forward and reverse primers (ALCAM_F
and ALCAM_R), 0.4 µl of Phusion™ High-Fidelity DNA Polymerase
(Thermo Fisher Scientific, Inc.), 2.5 mM of dNTPs (Fermentas;
Thermo Fisher Scientific, Inc.), and 4 µl of 5X PCR buffer. The
thermal cycling conditions included initial denaturation for 30 sec
at 98˚C, followed by 25 cycles of denaturation at 98˚C for 10 sec,
annealing at 58˚C for 10 sec, elongation at 72˚C for 20 sec, and a
final elongation for 10 min at 72˚C. PCR products confirmed by gel
electrophoresis were purified using Exo-Sap enzymes (cat. nos.
EN0581 and EF0651; Thermo Fisher Scientific, Inc.). DNA sequencing
was performed using the BigDye Terminator Cycle Sequencing v.3.1
kit (cat. no. 4337455; Applied Biosystems; Thermo Fisher
Scientific, Inc.). DNA sequencing analysis was performed using an
ABI 3730XL Genetic Analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Using co-segregation analysis, SNVs with
phenotypes confirmed in the affected family members and absent in
the non-family member controls were identified as susceptibility
genes for IAs.
The Conserved Domains Database of NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi)
and UniProt (https://www.uniprot.org/uniprot/Q13740#family_and_domains)
were searched to analyze conservation of the protein in
evolution.
Results
Clinical data
The proband (II:4, Fig.
1A) was a 59-year-old Kazakh man diagnosed with SAH. CTA
revealed multiple aneurysms, with one ruptured aneurysm in the
anterior communicating artery and two unruptured IAs in the middle
cerebral artery (II:4; Fig. 1B). No
other cerebrovascular disease was found in the proband, except for
aneurysms. Previously, his brother (II:6) was affected by SAH at 50
years of age. His brother was 52 years old at the time of the
study. The patient had a giant aneurysm in the middle cerebral
artery (II:6; Fig. 1B). The
diagnosis was independently confirmed by two neurologists.
Furthermore, one of the older sisters (II:1) of the
proband died at the age of 48 years after an episode suggestive of
a hemorrhagic stroke. The other sister (II:2) had hypertension.
The niece (IV:1, 22 years old) of the proband was
diagnosed with early-onset hypertension. The father of the proband
had no familial history of IA and died at 70 years of age, whereas
the mother of the proband was diagnosed with ischemic heart disease
and hypertension and died at 57 years of age. Therefore, blood
samples from the parents of the proband were unavailable for
mutation analysis. Two IA-affected (II:4 and II:6) and one
unaffected (II:2) individual, from whom sufficient DNA samples were
available, were selected for WES analysis (Table II).
 | Table IICharacteristics of the family
members. |
Table II
Characteristics of the family
members.
| Patient | Age, years | Sex | IA
localization | Number of IAs | IA diameter
(mm) | Rupture | Hypertension | Smoking |
|---|
| II:2 | 60 | Female | - | - | - | - | Yes | No |
| II:4 | 59 | Male | ACA, MCA | 3 | <5 | Yes | Yes | No |
| II:6 | 52 | Male | MCA | 1 | >5 | Yes | Yes | No |
| III:7 | 40 | Male | - | - | - | - | No | No |
WES analysis
WES analysis was conducted using DNA samples
isolated from the blood of three selected individuals. A total of
13.6-17.5 billion bases were generated for each individual, with an
average coverage depth of 97.9X. In addition, 96.35% (95.85-96.65%)
of the target exon regions were covered by at least 200X (Table III). A flowchart detailing variant
filtering is illustrated in Fig. 2.
After alignment and a series of quality control procedures, a total
of 2,426,633 SNVs were identified (from three individuals). Novel
heterozygous variants in the coding region, as predicted by
conceptual translation, which affect protein-coding sequences, were
investigated. After filtering against references from the public
ExAC database <0.01, 1,330 SNVs were retained. Furthermore,
benign or tolerated variants, as predicted by PolyPhen-2, were
removed. This resulted in 713 variants (Table SI). A dominant mode of inheritance
was then assumed. This inheritance mode assumes that one risk
allele is sufficient to be affected by the disease. In our case,
the criteria were to have a wild-type genotype in the unaffected
individual and a heterozygous or homozygous mutant genotype in
individuals with IA.
 | Table IIIExome sequencing coverage of three
individuals from the pedigree. |
Table III
Exome sequencing coverage of three
individuals from the pedigree.
| Individual | Q20 (%) | Q30 (%) | Clean reads | Coverage (%)
(1X) | 10X (%) | 20X (%) | 50X (%) | Mean depth of
target regions |
|---|
| II-2 | 96.6 | 91.3 | 92.194.480 | 99.6 | 99.0 | 97.6 | 86.6 | 160.8 |
| II-4 | 97.9 | 94.1 | 117.266.490 | 99.9 | 99.4 | 98.2 | 90.1 | 136.6 |
| II-6 | 97.4 | 93 | 110.392.196 | 99.7 | 99.1 | 97.9 | 89.0 | 149.5 |
Finally, one candidate genetic variant, activated
leukocyte cell adhesion molecule (ALCAM) c1382 G>A
(p.Gly229Val) was selected, by considering known disease genes and
ontology associations with SAH morphogenesis or other known
diseases. Considering its pathogenicity, this variant was
prioritized as a putative candidate, although it was not found in
the ClinVar database. The score predictor of c1382 G>A
(p.Gly229Val) substitution pathogenicity was 0.994 for the
Poly-Phen2 algorithm, and this mutation was predicted as ‘probably
damaging’. The allele A MAF of rs10933819 polymorphism was 0.0010
in the ExAC database.
Validation and co-segregation analysis
of candidate variants
Candidate genetic variants in the four family
members were directly sequenced using Sanger sequencing. Moreover,
the variant was validated in 145 healthy individuals to eliminate
any false-positive findings.
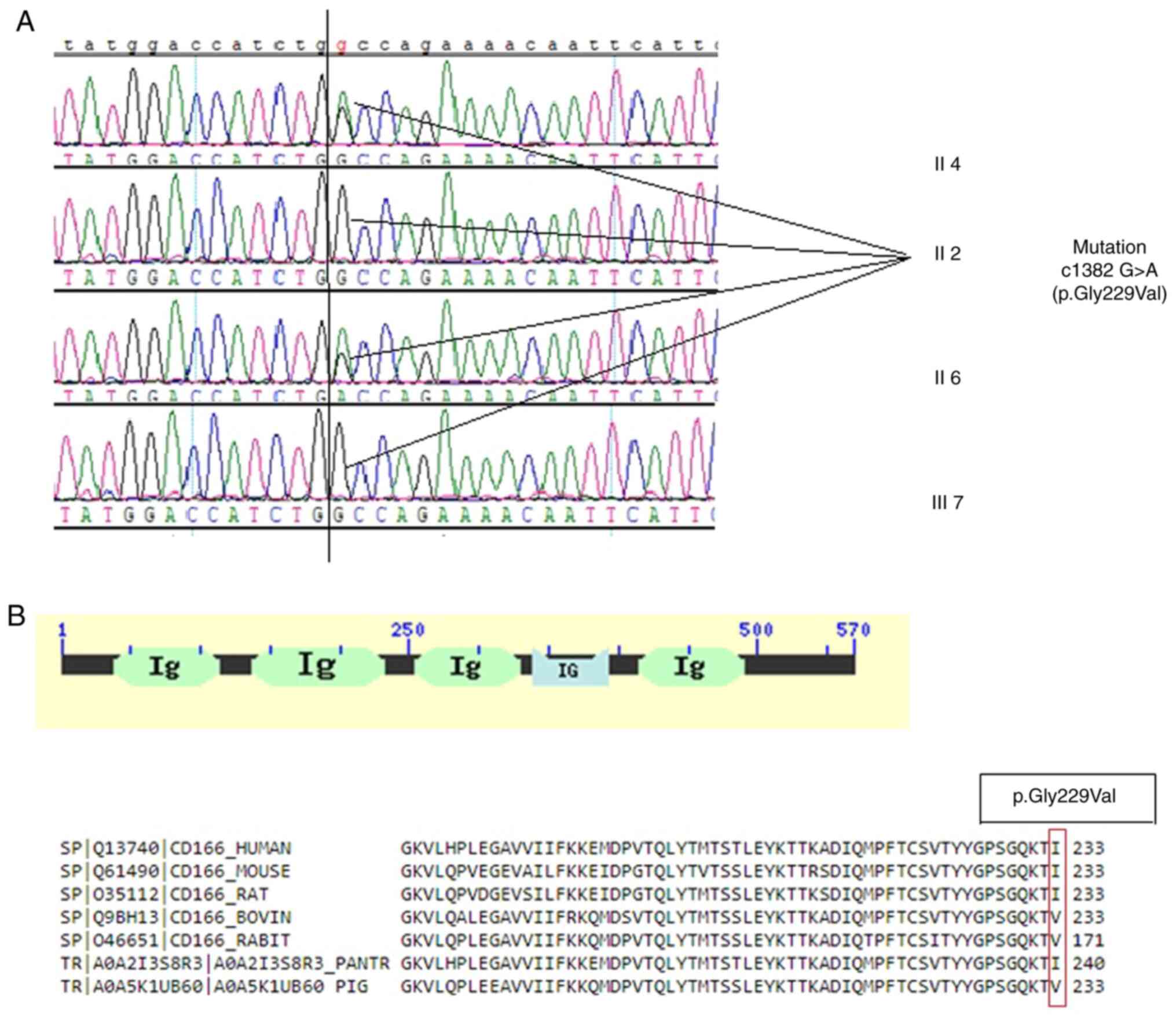
The missense variant c1382 G>A (p.Gly229Val) in
ALCAM was confirmed to be heterozygous in the proband
(pedigree II:4; Fig. 3). A
heterozygous mutation in his brother with IA (pedigree II:6;
Fig. 3A) was also identified,
whereas his elder sister and son had the wild-type gene and a
normal phenotype (pedigrees II:2 and III:7, respectively; Fig. 3A). These results indicated that the
c1382 G>A (p.Gly229Val) gene co-segregated with the disease
phenotype in the family members that were tested and were absent
from the unaffected second-generation members. Furthermore, this
mutant was absent in 145 normal ethnically-matched individuals.
Collectively, these findings showed complete co-segregation of the
mutation in the pedigree of the IA/SAH-affected family, indicating
its role in the pathogenesis of this disease.
Conservation of the protein in
evolution
The cluster of differentiation 166 (CD166) antigen
(ALCAM) has five conserved Ig-like domains (Fig. 3B): V-type 1, V-type 2, C2-type 1,
C2-type 2, and C2-type 3. The missense mutation, p.Gly229Val, is
located in the Ig-like V-type 2 domain of the CD166 antigen. The
CD166 antigen protein sequences from several species, including
humans, chimps, mice, rats, rabbits, bovines, and pigs, were
compared. Multiple sequence alignment analysis revealed that the
Gly229 residue was strongly conserved (Fig. 3B).
Discussion
Previous research has shown that the sex and
hormonal background of a patient, alcohol abuse, and the presence
of arterial hypertension (AH) are general risk factors for the
development of aneurysmal SAH (19). Environmental and genetic factors
play important roles in the pathogenesis and rupture of IAs
(20).
A practical WES method was used to systematically
explore rare coding variants and identify potentially causative
variants in a family case of IA.
After sequencing two affected individuals and one
unaffected member, bioinformatic filtering revealed that only
ALCAM c1382 G>A was a candidate variant. Sanger
sequencing showed that this variant was fully co-segregated with
definite IA phenotypes in the family.
Very little information is available on the
rs10933819 polymorphism of ALCAM. ALCAM (or CD166) is an
immunoglobulin-like protein belonging to the cell adhesion molecule
family with three C2 and two V extracellular domains. It functions
by mediating homotypic and heterotypic cell-to-cell contacts via
the CD166-CD6 interaction (21).
Notably, the heterotopically interaction affinity with CD6 is much
higher than the homotypic interaction with itself. Hassan et
al (21) reported that the
interaction with CD166 was 100-fold lower than that with CD6.
Together, the heterotypically interacting CD6 and CD166 play a role
in T cell activation. CD6 is a lymphocyte membrane protein that
contains an scavenger receptor cysteine-rich (SRCR) domain that
interacts with CD166. Interestingly, soluble monomeric forms
inhibit T cell activation.
Cell adhesion molecules (CAMs) are essential for
inflammatory processes. They are highly expressed in vascular
endothelial tissues (22).
CD166-CD6 interactions form immunological synapses at T cells and
CD166-presenting cells. ALCAM contributes to the movement of T
cells and monocytes through the endothelium and blood-brain barrier
(18). ALCAM and vascular cell
adhesion molecule (VCAM) levels were high in patients with systemic
lupus erythematosus (SLE) vs. healthy individuals, and there is a
theory that elevated expression of ALCAM may breach T cell
tolerance (23). Willrodt et
al (24) investigated the role
of ALCAM in corneal allograft rejection. The cornea is usually
avascular, but inflammation-induced neovascularization in the
cornea of the receptor increases the risk of allograft rejection.
ALCAM blockade reduced the angiogenic process and T cell
activation, which adds to the evidence regarding the dual role of
ALCAM in vasculogenesis and mediation of dendritic cells.
In 2001, Ohneda et al (25) described the role of ALCAM in
hematopoietic and endothelial cell development. However, they
reported that ALCAM is highly expressed in the YSCL-72 endothelial
cell line (derived from the yolk sac) and not in EOMA (derived from
the adult aorta). Moreover, they experimentally examined the role
of ALCAM in vascular tube formation. They found that ALCAM
facilitates cord-like endothelial cluster formation but has no
effect on sheet-like clusters.
Thus, ALCAM physically interacts with the T
cell-expressed scavenger receptor CD6, endothelial L1CAM, and
galectin-8(24). Notably, the other
lectins, namely galectin-1 and galectin-3, have been reported to be
associated with abdominal aortic aneurysms (26,27).
However, secreted ALCAM (sALCAM) performs
differently. Galectin-8 has been reported to play a role in blood
and lymph vessel angiogenesis (28). Noticeably, galectin-8 interacts with
podoplanin in lymphatic angiogenesis and with ALCAM in vascular
endothelial cells. Galectin-3 inhibits galectin-8(29). Cheng et al reported higher
galectin-3 expression and lower galectin-8 expression in
endothelial cells than in the epithelial cells (29).
In summary, a rare variant, c1382 G>A
(p.Gly229Val), in ALCAM, which has not been previously
reported in the Kazakh population was identified. The present study
has the limitation that ALCAM c1382 G>A was fully
co-segregated with definite IA phenotypes in only one family.
Therefore, further studies are required to evaluate the functional
impact of ALCAM c1382 G>A, which may warrant further
replication and biological investigations related to IA.
Supplementary Material
PolyPhen predicted variants
shortlisted after filtration.
Acknowledgements
The authors would like to thank Macrogen, Inc.
(Seoul, South Korea) for their support with next-generation
sequencing.
Funding
Funding: This research was funded by the Ministry of Education
and Science of the Republic of Kazakhstan (grant no.
AP08955996).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the SRA database (https://www.ncbi.nlm.nih.gov/sra) repository
under accession no. PRJNA819841 (BioProject) (SRR18475941,
SRR18475942 and SRR18475943).
Authors' contributions
AA, KM, YM and EZ conceived the study. GK, AA, EZ,
KM and YR wrote the first draft of the manuscript. YM, BD and SA
performed the sample collection, processed the clinical data and
test results. IA conducted the design and synthesis of primers. KK
performed the laboratory experiments. TU, KK and AA validated the
data. GK, SA and YR analyzed data. TU and KM prepared the figures.
EZ and KM confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
1/16.02.2015) by the Human Research Ethics Committee of the
National Center for Neurosurgery (Nur Sultan, Kazakhstan). Written
informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Macdonald RL and Schweizer TA: Spontaneous
subarachnoid haemorrhage. Lancet. 389:655–666. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lee VH, Ouyang B, John S, Conners JJ, Garg
R, Bleck TP, Temes RE, Cutting S and Prabhakaran S: Risk
stratification for the in-hospital mortality in subarachnoid
hemorrhage: The HAIR score. Neurocrit Care. 21:14–19.
2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Jung KH: New pathophysiological
considerations on cerebral aneurysms. Neurointervention. 13:73–83.
2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Xu Z, Rui YN, Hagan JP and Kim DH:
Intracranial aneurysms: Pathology, genetics, and molecular
mechanisms. NeuroMolecular Med. 21:325–343. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Slot EMH, Rinkel GJE, Algra A and Ruigrok
YM: Patient and aneurysm characteristics in familial intracranial
aneurysms. A systematic review and meta-analysis. PLoS One.
14(e0213372)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Van Hoe W, van Loon J, Demeestere J,
Lemmens R, Peluso J and De Vleeschouwer S: Screening for
intracranial aneurysms in individuals with a positive first-degree
family history: A systematic review. World Neurosurg.
151:235–248.e5. 2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bourcier R, Le Scouarnec S, Bonnaud S,
Karakachoff M, Bourcereau E, Heurtebise-Chrétien S, Menguy C, Dina
C, Simonet F, Moles A, et al: Rare coding variants in ANGPTl6 are
associated with familial forms of intracranial aneurysm. Am J Hum
Genet. 102:133–141. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhou S, Ambalavanan A, Rochefort D, Xie P,
Bourassa CV, Hince P, Dionne-Laporte A, Spiegelman D, Gan-Or Z,
Mirarchi C, et al: RNF213 is associated with intracranial aneurysms
in the French-Canadian population. Am J Hum Genet. 99:1072–1085.
2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sauvigny T, Alawi M, Krause L, Renner S,
Spohn M, Busch A, Kolbe V, Altmüller J, Löscher BS, Franke A, et
al: Exome sequencing in 38 patients with intracranial aneurysms and
subarachnoid hemorrhage. J Neurol. 267:2533–2545. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Santiago-Sim T, Fang X, Hennessy ML,
Nalbach SV, DePalma SR, Lee MS, Greenway SC, McDonough B,
Hergenroeder GW, Patek KJ, et al: THSD1 (thrombospondin Type 1
domain containing Protein 1) mutation in the pathogenesis of
intracranial aneurysm and subarachnoid hemorrhage. Stroke.
47:3005–3013. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lorenzo-Betancor O, Blackburn PR, Edwards
E, Vázquez-do-Campo R, Klee EW, Labbé C, Hodges K, Glover P,
Sigafoos AN, Soto AI, et al: PCNT point mutations and familial
intracranial aneurysms. Neurology. 91:e2170–e2181. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen S, Li M, Xin W, Liu S, Zheng L, Li Y,
Li M, Zhan M and Yang X: Intracranial aneurysm's association with
genetic variants, transcription abnormality, and methylation
changes in ADAMTS genes. PeerJ. 8(e8596)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ding X, Zhao S, Zhang Q, Yan Z, Wang Y, Wu
Y, Li X, Liu J, Niu Y, Zhang Y, et al: Exome sequencing reveals a
novel variant in NFX1 causing intracranial aneurysm in a Chinese
family. J Neurointerv Surg. 12:221–226. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Liu J, Liao X, Zhou J, Li B, Xu L, Liu S,
Li Y, Yuan D, Hu C, Jiang W and Yan J: A rare variant of ANK3 is
associated with intracranial aneurysm. Front Neurol.
12(672570)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Broderick JP, Sauerbeck LR, Foroud T,
Huston J, III Pankratz N, Meissner I and Brown RD Jr: The familial
intracranial aneurysm (FIA) study protocol. BMC Med Genet.
6(17)2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zholdybayeva EV, Medetov YZ, Aitkulova AM,
Makhambetov YT, Akshulakov SK, Kaliyev AB, Talzhanov YA,
Kulmambetova GN, Iskakova AN and Ramankulov YM: Genetic risk
factors for intracranial aneurysm in the Kazakh population. J Mol
Neurosci. 66:135–145. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wang K, Li M and Hakonarson H: ANNOVAR:
Functional annotation of genetic variants from high-throughput
sequencing data. Nucleic Acids Res. 38(e164)2010.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Caranci F, Briganti F, Cirillo L, Leonardi
M and Muto M: Epidemiology and genetics of intracranial aneurysms.
Eur J Radiol. 82:1598–1605. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Tromp G, Weinsheimer S, Ronkainen A and
Kuivaniemi H: Molecular basis and genetic predisposition to
intracranial aneurysm. Ann Med. 46:597–606. 2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Hassan NJ, Barclay AN and Brown MH:
Frontline: Optimal T cell activation requires the engagement of CD6
and CD166. Eur J Immunol. 34:930–940. 2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Cayrol R, Wosik K, Berard JL,
Dodelet-Devillers A, Ifergan I, Kebir H, Haqqani AS, Kreymborg K,
Krug S, Moumdjian R, et al: Activated leukocyte cell adhesion
molecule promotes leukocyte trafficking into the central nervous
system. Nat Immunol. 9:137–145. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Parodis I, Gokaraju S, Zickert A, Vanarsa
K, Zhang T, Habazi D, Botto J, Serdoura Alves C, Giannopoulos P,
Larsson A, et al: ALCAM and VCAM-1 as urine biomarkers of activity
and long-term renal outcome in systemic lupus erythematosus.
Rheumatology (Oxford). 59:2237–2249. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Willrodt AH, Salabarria AC, Schineis P,
Ignatova D, Hunter MC, Vranova M, Golding-Ochsenbein AM, Sigmund E,
Romagna A, Strassberger V, et al: ALCAM mediates DC migration
through afferent lymphatics and promotes allospecific immune
reactions. Front Immunol. 10(759)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ohneda O, Ohneda K, Arai F, Lee J,
Miyamoto T, Fukushima Y, Dowbenko D, Lasky LA and Suda T: ALCAM
(CD166): Its role in hematopoietic and endothelial development.
Blood. 98:2134–2142. 2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lu HY, Shih CM, Sung SH, Wu ATH, Cheng TM,
Lin YC and Shih CC: Galectin-3 as a biomarker for stratifying
abdominal aortic aneurysm size in a Taiwanese population. Front
Cardiovasc Med. 8(663152)2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Roldán-Montero R, Perez-Saez JM,
Cerro-Pardo I, Martinez-Lopez D, Nuñez E, Maller S, Gutierrez-Muñoz
C, Mendez-Barbero N, Escola-Gil JC, Michel JB, et al: Galectin-1
prevents pathological vascular remodeling in atherosclerosis and
abdominal aortic aneurysm. Sci Adv. 8(eabm7322)2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Troncoso MF, Ferragut F, Bacigalupo ML,
Cárdenas Delgado VM, Nugnes LG, Gentilini L, Laderach D,
Wolfenstein-Todel C, Compagno D, Rabinovich GA and Elola MT:
Galectin-8: A matricellular lectin with key roles in angiogenesis.
Glycobiology. 24:907–914. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cheng YL, Wu YW, Kuo CF, Lu SL, Liu FT,
Anderson R, Lin CF, Liu YL, Wang WY, Chen YD, et al: Galectin-3
inhibits galectin-8/parkin-mediated ubiquitination of group A
Streptococcus. mBio. 8:e00899–17. 2017.PubMed/NCBI View Article : Google Scholar
|