Introduction
Hepatocellular carcinoma (HCC) is the fourth leading
cause of cancer-related death worldwide (1). Its incidence varies widely according
to geographic location, and the distribution of HCC also differs
among racial and ethnic groups as well as among regions within the
same country (2). HCC has a high
incidence in regions such as sub-Saharan Africa, the Republic of
China, and Hong Kong. The high incidence rate of HCC in developing
countries, such as Asian countries, is due to the high frequency of
hepatitis virus infection (3). HCC
has a lower incidence in regions of North and South America,
Europe, Australia, and the Middle East. However, the incidence of
HCC in the United States has increased over the past two decades
(4). In regions with historically
low incidence rates, the incidence of liver cancer has increased
due to the increased prevalence of obesity and diabetes and the
high prevalence of hepatitis C viral infections due to abuse of
injected drugs (2). The prognosis
of patients with advanced and unresectable HCC is poor. Sorafenib
has long been used as the standard treatment agent for HCC;
however, it only slightly prolongs survival (5,6).
Therefore, more effective treatments for advanced HCC are
needed.
Lenvatinib is a receptor tyrosine kinase inhibitor
active against vascular endothelial growth factors (VEGFR1-3),
fibroblast growth factors (FGFR1-4), platelet-derived growth factor
receptor (PDGFR)α, RET, and KIT (7). It has been used to treat renal and
thyroid cancers (7,8). Lenvatinib has demonstrated
noninferiority to sorafenib in clinical trials for HCC (9) and is an unresectable treatment agent
for HCC approved in >50 countries, including the United States,
Japan, and Europe. The antitumor effects of Lenvatinib have been
explored in various HCC models, and its main mechanism of action
involves inhibition of angiogenesis and the tumor FGF signaling
pathway (10). In addition to
inhibiting VEGFR and FGFR, Lenvatinib, a multi-kinase inhibitor,
inhibits various tyrosine kinases (6). Lenvatinib is anticipated to exert its
antitumor effects via the regulation of the cell cycle, and
angiogenesis in HCC cells.
MicroRNAs (miRNAs/miRs) directly control several
cellular events, including cell cycle progression, by targeting
cell cycle regulators (11,12). Additionally, miRNAs indirectly
control cell cycle progression by targeting signal transduction
pathways in anticancer therapy. We previously reported the role of
a miRNA signature in the antitumor effects of cisplatin in HCC via
modulation of the cell cycle (13).
However, the detailed mechanism of the antiproliferative effects of
Lenvatinib in HCC cells via the cell cycle and cell cycle-related
molecules remains unclear. In this study, we revealed the antitumor
effects of Lenvatinib and its mechanism of action in HCC cell lines
and mice xenografted tumors in vitro and in vivo. We
examined the following: i) The antitumor effects of Lenvatinib on
HCC cell lines in vitro and in vivo; ii) its effects
on cell cycle and cell cycle-related molecules; and iii) its
effects on the miRNA signatures in HCC cells and exosomes.
Materials and methods
Drugs, chemicals, and reagents
Lenvatinib was purchased from AdooQ Bioscience LLC,
and a solution of Lenvatinib was prepared by dilution with DMSO and
stored at -20˚C. RPMI-1640 was obtained from Gibco (Thermo Fisher
Scientific, Inc.). Trypan blue was purchased from MilliporeSigma.
DMEM, minimum essential medium (MEM), and FBS were obtained from
Wako Pure Chemical Industries, Ltd. Penicillin-streptomycin was
obtained from Invitrogen (Thermo Fisher Scientific, Inc.). A cell
cycle phase determination kit was obtained from Cayman Chemical
Company, a protease inhibitor cocktail from iNtRON Biotechnology,
and a Proteome Profiler Human Angiogenesis Antibody Array Kit from
R&D Systems, Inc.
Cell lines and culture
Four HCC cell lines were used in this study. Huh-7
and PLC/PRF/5 cells were obtained from the Japanese Cancer Research
Bank. Hep3B cells were obtained from American Type Culture
Collection. The Li-7 cells were obtained from the Central Institute
for Experimental Animals. HuH-7 cells were cultured in low-glucose
DMEM supplemented with 10% FBS and 100 units/ml penicillin-100
µg/ml streptomycin. Hep3B cells were cultured in MEM supplemented
with 10% FBS, 1% non-essential amino acid solution (NEAA), and
penicillin-streptomycin. Li-7 cells were cultured in RPMI-1640
supplemented with 10% FBS and penicillin-streptomycin. PLC/PRF/5
cells were cultured in DMEM supplemented with 10% FBS and
penicillin-streptomycin. All the cell lines were cultured in a
humidified incubator supplied with 5% CO2 at 37˚C.
Cell proliferation assay
Cell proliferation assays were performed using the
CCK-8 kit (Dojindo Molecular Technologies, Inc.) according to the
manufacturer's instructions. Cells (1x103) were seeded
in 96-well plates. After 24 h, the cells were treated with
Lenvatinib (0, 0.2, 0.4 or 1.0 µM), adjusted for the concentration
that can be used in vitro, and cultured for another 144 h.
At specific time points, the medium was replaced with 100 µl medium
containing the CCK-8 reagent, and after 3 h, the absorbance was
measured at a wavelength of 450 nm using an automatic microplate
reader (Multiskan FC; Thermo Fisher Scientific, Inc.).
Cell cycle analysis
Cell cycle analysis was performed as previously
described (13). HuH-7 cells were
seeded into 100-mm culture dishes at 1x106 per dish and
cultured for 24 h. Cells were treated with 1 µM Lenvatinib adjusted
to close to the maximum blood concentration at the time of
treatment according to the manufacturer's instructions for 48 h.
Cell cycle analysis was performed using flow cytometry on a
Cytomics FC 500 flow cytometer (Beckman Coulter, Inc.), according
to the manufacturer's protocol. Data were analyzed using Kaluza
software 2.1.3 (Beckman Coulter, Inc.).
Western blotting
Western blotting was performed as previously
described (13). HuH-7 cells were
treated with 1 µM Lenvatinib or DMSO control, cultured for 48 h,
and then lysed with PRO-PREP complete protease inhibitor mixture
(iNtRON Biotechnology). Supernatants were obtained by
centrifugation at 13,000 x g, 4˚C for 5 min containing soluble
cellular proteins were collected and stored at -80˚C until
required. Protein concentrations were measured using a NanoDrop
2000 spectrofluorometer (Thermo Fisher Scientific, Inc.),
resuspended in sample buffer to a concentration of 0.1 µg/ml,
loaded on a 10% SDS-gel, and resolved using SDS-PAGE. After
blocking with 5% skimmed milk in 0.05% Tween-20/TBS buffer, the
membranes were incubated with the primary antibodies and then with
horseradish peroxidase (HRP)-conjugated secondary antibody.
Antibodies used for western blot analysis were obtained from the
following sources: β-actin (monoclonal; cat. no. 8H10D10; Cell
Signaling Technology, Inc.), cyclin D1 (cat. no. MA5-14512; Thermo
Fisher Scientific, Inc.), cyclin E (cat. no. MS-870-P1; Thermo
Fisher Scientific, Inc.), Cdk2 (cat. no. sc-163, #A2810; Santa Cruz
Biotechnology, Inc.), Cdk4 (cat. no. sc-749, #G0516; Santa Cruz
Biotechnology, Inc.), and Cdk6 (cat. no. sc-177, #G1610; Santa Cruz
Biotechnology, Inc.). The secondary antibodies included anti-mouse
and anti-rabbit IgG and HRP-conjugated antibodies (cat. no. #7074,
Cell Signaling Technology, Inc.). Primary and secondary antibodies
were diluted 1,000-fold in blocking solution. All experiments were
performed in triplicate. The membranes were exposed to X-ray film
using a chemiluminescence detection system (Perkin-Elmer). The
immunoreactive band density obtained from the array was analyzed
using ImageJ version 1.52a (National Institutes of Health).
Xenograft model
Six-week-old BALB/c-nu/nu mice (n=16) were obtained
from Japan SLC and were housed under barrier conditions. A standard
sterilized laboratory diet and water were provided ad
libitum. All the animals were treated in accordance with the
guidelines of the Kagawa University Committee on Experimental
Animals. The Kagawa University Animal Care Committee approved all
animal protocols, including any animal ethical concerns (approval
no. 18675). Mice were housed at least 1 week before experiments in
temperature-controlled rooms at 20-22˚C with free access to food
and water supply and a light/dark cycle of 14/10 h. Inhalation of
3% sevoflurane in a controlled chamber was used for anesthesia
during the implantation of HCC cell lines. An inhalation anesthesia
machine for small animals (RC2, VetEquip, Inc.) was used to
continuously monitor the concentrations of sevoflurane and oxygen
in the anesthesia box and deliver sevoflurane at a rate of 5 l/min.
To establish the model, the mice were subcutaneously inoculated
with HuH-7 cells (3x106/animal) into their flanks. After
~2 weeks, when the tumors reached a maximal diameter of >6 mm,
the 16 mice were randomly assigned to one of two groups: Mice were
orally administered PBS only (vehicle control; n=8) or 0.2 mg/day
Lenvatinib (n=8). Bodyweight and tumor volume was monitored every 3
days. Tumor volumes were calculated using the formula V=length x
width2/2, as reported previously (14). The maximum size of the implanted
tumours was <1,500 mm3. We monitored the mice's
weight, and no significant changes were observed during in
vivo experiments. The animals were sacrificed on day 8 after
the start of treatment using 100% CO2 for 5 min and were
observed for 20 min. The flow rate of CO2 was 50% of the
chamber volume per min at the end of the experiment.
Microarray analysis of miRNAs
miRNA array analysis was performed as previously
described (11). Briefly, total RNA
was isolated from HuH-7 cells treated with 1 µM Lenvatinib for 96 h
using an miRNeasy Mini Kit (Qiagen GmbH) according to the
manufacturer's instructions. Exosomal RNA was extracted from the
culture medium using the exoRNeasy Serum/Plasma Maxi Kit (Qiagen
GmbH). RNA concentration and purity were confirmed using absorbance
measurements with a NanoDrop 2000 spectrophotometer (Thermo Fisher
Scientific, Inc.). miRNA expression analysis was performed using
the miRCURYHy3/HI Power Labeling Kit and a human miRNA Oligo chip
(v. 21.0; Toray Industries). The arrays were scanned using a
3D-Gene Scanner 3000 (Toray Industries), and the fluorescence
images were analyzed using 3D-Gene extraction version 1.2 software
(Toray Industries). Quantile normalization was performed on the raw
data, which exceeded the background level. Differentially expressed
miRNAs were identified using the Mann-Whitney U test. Hierarchical
clustering was performed using the farthest end method with the
absolute non-central Pearson correlation coefficient as the metric.
A heatmap was created based on the relative expression intensity of
each miRNA. The log2 value was centered on the median
value of each row. All the microarray data in this study were
submitted to NCBI Gene Expression Omnibus, accession no. GSE201775.
(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE201775).
Statistical analysis
GraphPad Prism version 8.0 (GraphPad Software, Inc.)
was used for all statistical analyses. Data excluding miRNAs were
analyzed using a two-way ANOVA followed by a Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference. A Mann-Whitney U test was used to identify miRNAs with
different expression levels.
Results
Lenvatinib treatment suppresses human
HCC cell growth in vitro
Four human HCC cell lines, HuH-7, Hep3B, PLC/PRF/5,
and Li-7 cells, were treated with various concentrations of
Lenvatinib (0, 0.2, 0.4 or 1.0 µM) for 6 days. Lenvatinib
suppressed the proliferation of HuH-7 and Hep3B cells after 144 h
of treatment (Fig. 1). Notably,
PLC/PRF/5 and Li-7 cells were less susceptible to Lenvatinib than
other HCC cell lines. We examined the differences in sensitivity to
Lenvatinib between HuH-7 (Lenvatinib-sensitive) and PLC/PRF/5
(Lenvatinib resistant) cells in subsequent experiments.
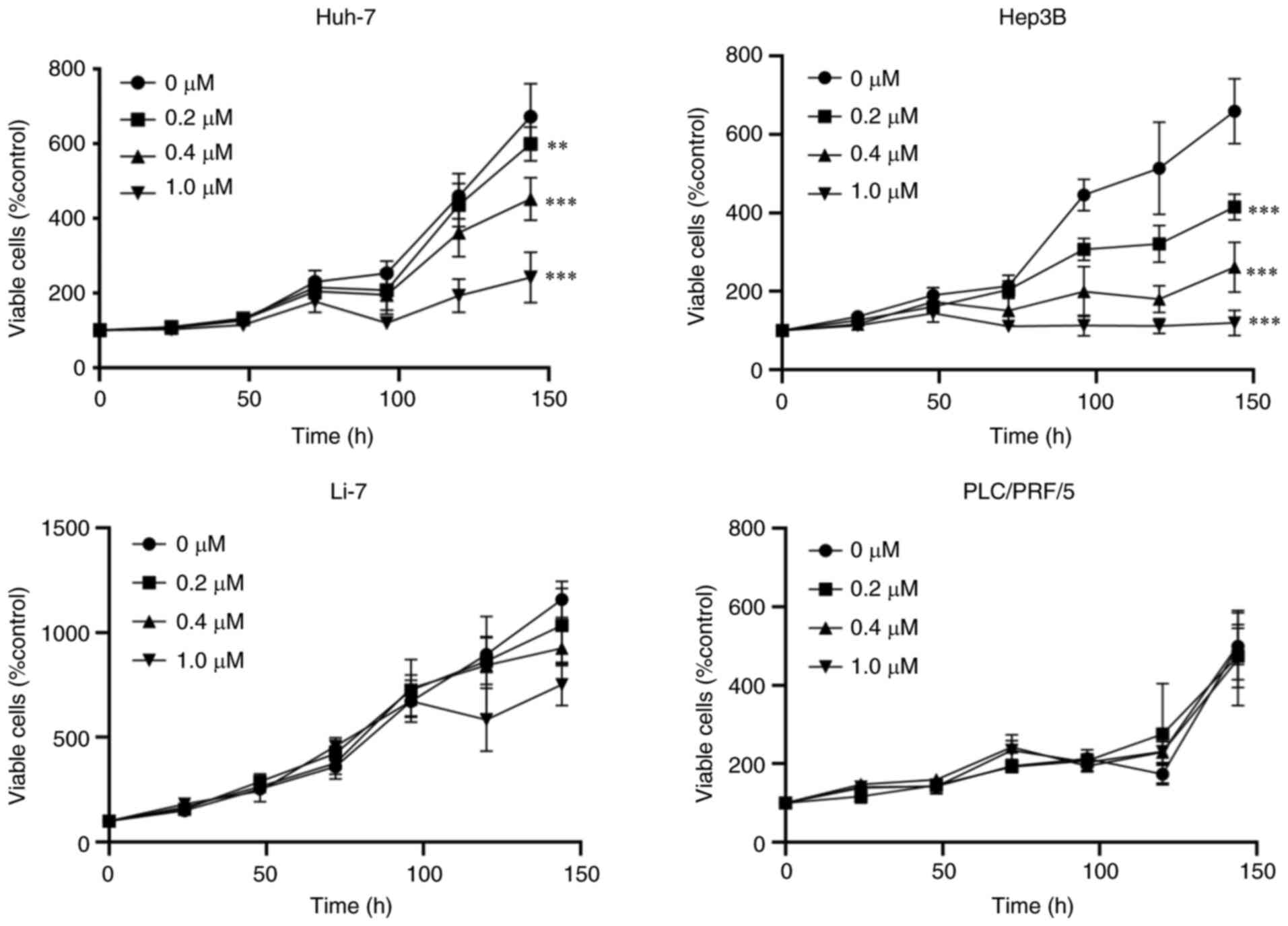 | Figure 1Cell proliferation assays. Lenvatinib
suppressed the proliferation of HCC cells. HuH-7, Hep3B, PLC/PRF/5,
and Li-7 cells were treated with 0, 0.2, 0.4, 1.0 µM Lenvatinib for
0, 24, 48, 72 and 144 h. Cell proliferation was assessed using
CCK-8 assays. Data points are present as the mean ± SD of three
independent repeats. **P<0.01,
***P<0.001 vs. control. |
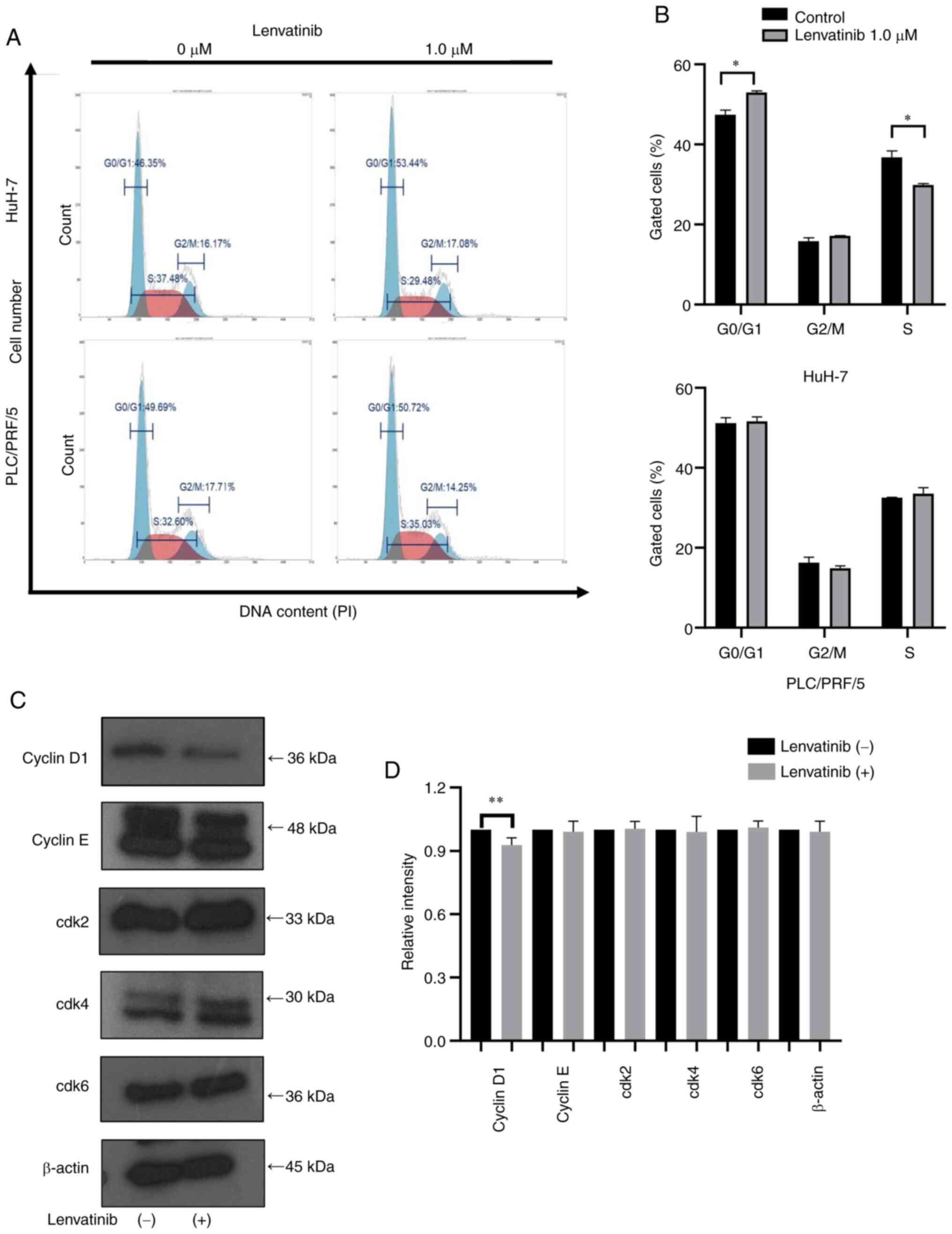
Lenvatinib treatment induces
G0/G1 phase cell cycle arrest and regulates
cell cycle-related molecules
We next investigated the effects of Lenvatinib
treatment on the cell cycle of Huh-7 and PLC/PRF/5 cells using flow
cytometry analysis (Fig. 2A). In
HuH-7 cells incubated with 1 µM Lenvatinib for 48 h, the number of
cells in the S phase decreased and those in the
G0/G1 phase increased (Fig. 2B). In contrast, the G1
and S phase distributions showed no significant changes in
PLC/PRF/5 cells after Lenvatinib treatment (Fig. 2B). These results indicated that
Lenvatinib suppressed the growth of the Lenvatinib-sensitive HuH-7
cells by arresting them in the G0/G1
phase.
Previous studies on Lenvatinib and cell cycle arrest
have reported that Lenvatinib induces cell cycle arrest by
downregulating cyclin D1 expression in thyroid cancer cells
(15). We further examined the key
cell cycle regulators by western blotting. In HuH7 cells, the
expression levels of cyclin D1 decreased, while the expression of
Cdk4 and Cdk6, which are the catalytic binding partners of cyclin
D1, remained unchanged after 48 h of treatment with 1 µM Lenvatinib
treatment (Fig. 2C and D). No changes were observed in cyclin E
and Cdk2, expression, which are related to the
G1/G2 phase transition (Fig. 2C and D). Together, these results indicated that
Lenvatinib inhibited G0/G1 phase transition
and suppressed the growth of Lenvatinib-sensitive cells by reducing
cyclin D1.
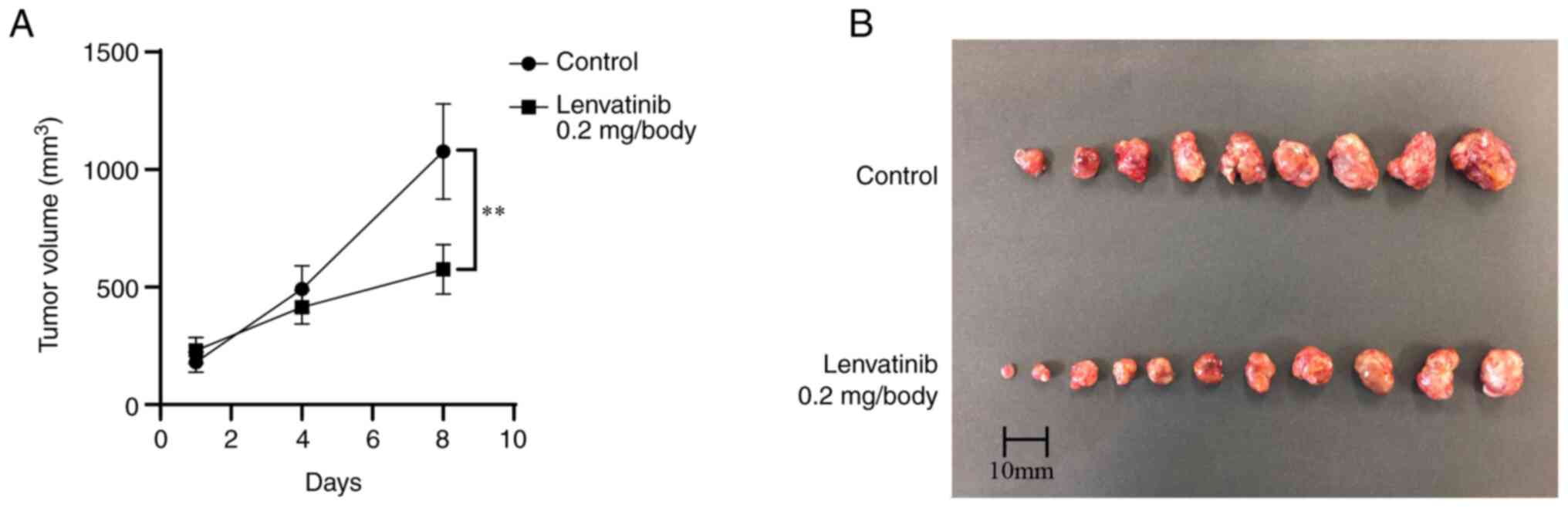
Lenvatinib suppresses tumor growth in
vivo
Next, we investigated the antitumor effects of
Lenvatinib in vivo. The xenografted mice were treated with
Lenvatinib (0.2 mg/day) or PBS orally after subcutaneous
implantation of HuH-7 cells (Fig.
3A). In the Lenvatinib treatment group, tumor growth was
significantly suppressed by 46.6% on day 8 after administration
compared with that in the untreated group (P<0.01, Fig. 3B). Lenvatinib treatment had no
significant effect on the body weight of animals during the course
of treatment.
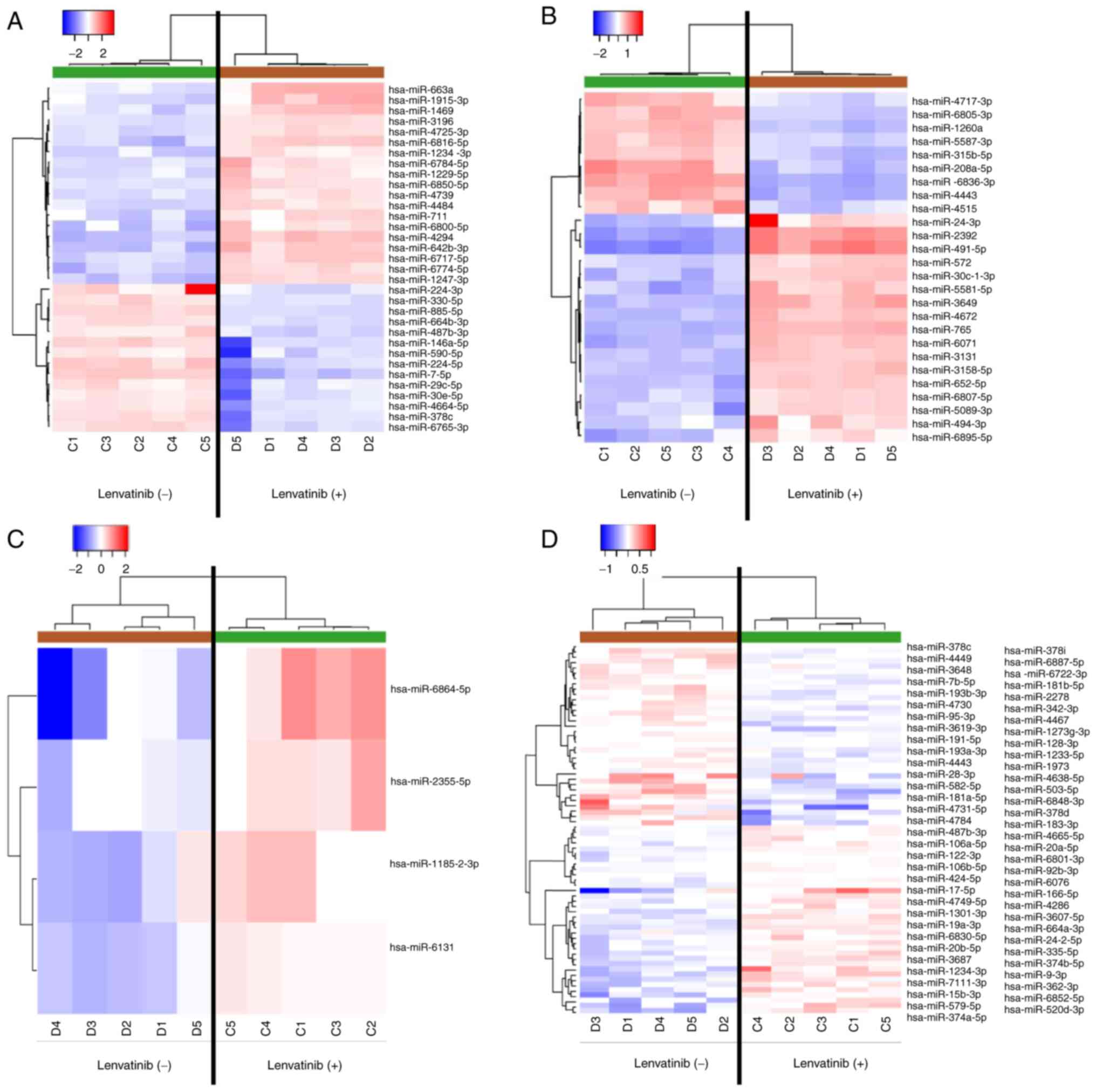
miRNA expression signatures differ
between the Lenvatinib-treated and -untreated HuH-7 cells
Heat maps generated by miRNA microarray analysis
identified several miRNAs with dysregulated expression in HuH-7
cells treated with 1 µM Lenvatinib for 96 h. After normalization
and removal of miRNAs with missing values, 33 significantly
differentially expressed miRNAs were identified in
Lenvatinib-treated HuH-7 cells, including 19 upregulated and 14
downregulated miRNAs (Fig. 4A and
B). We also found 26 significantly
differentially expressed miRNAs in the exosomes of
Lenvatinib-treated cells, including 17 significantly upregulated
and 9 downregulated miRNAs (Fig. 4C
and D, Table I).
 | Table IResults of the miRNA array analysis
in the HuH-7-derived exosomes treated with Lenvatinib. |
Table I
Results of the miRNA array analysis
in the HuH-7-derived exosomes treated with Lenvatinib.
| A, Upregulated |
|---|
| miRNA | FCa | P-value | FDR | Chromosomal
location |
|---|
| hsa-miR-491-5p | 5.456318 | 0.007937 | 0.037576 | 9p21.3 |
| hsa-miR-2392 | 4.177528 | 0.007937 | 0.037576 | 14q32.2 |
| hsa-miR-24-3p | 3.53251 | 0.007937 | 0.037576 | 9q22.32 |
| hsa-miR-3649 | 2.931781 | 0.007937 | 0.037576 | 12p13.33 |
| hsa-miR-765 | 2.916814 | 0.007937 | 0.037576 | 1q23.1 |
| hsa-miR-6071 | 2.684267 | 0.007937 | 0.037576 | 2p11.2 |
| hsa-miR-4672 | 2.58229 | 0.007937 | 0.037576 | 9q34.11 |
|
hsa-miR-5581-5p | 2.44319 | 0.007937 | 0.037576 | 1p34.3 |
| hsa-miR-652-5p | 2.419417 | 0.007937 | 0.037576 | Xq23 |
|
hsa-miR-3158-5p | 2.399236 | 0.007937 | 0.037576 | 10q24.32 |
| hsa-miR-3131 | 2.370459 | 0.007937 | 0.037576 | 2q35 |
|
hsa-miR-30c-1-3p | 2.310656 | 0.007937 | 0.037576 | 1p34.2 |
|
hsa-miR-5089-3p | 2.084861 | 0.007937 | 0.037576 | 17q21.32 |
|
hsa-miR-6807-5p | 2.046529 | 0.007937 | 0.037576 | 19q13.43 |
|
hsa-miR-6895-5p | 2.040822 | 0.007937 | 0.037576 | Xp11.22 |
| hsa-miR-572 | 2.037145 | 0.007937 | 0.037576 | 4p15.33 |
| hsa-miR-494-3p | 2.013227 | 0.007937 | 0.037576 | 14q32.31 |
| B,
Downregulated |
| miRNA | FCa | P-value | FDR | Chromosomal
location |
|
hsa-miR-6836-3p | 0.270333 | 0.007937 | 0.037576 | 7p22.3 |
| hsa-miR-4443 | 0.316821 | 0.007937 | 0.037576 | 3p21.31 |
|
hsa-miR-208a-5p | 0.325378 | 0.007937 | 0.037576 | 14q11.2 |
| hsa-miR-1260a | 0.391065 | 0.007937 | 0.037576 | 14q24.3 |
|
hsa-miR-6805-3p | 0.415766 | 0.007937 | 0.037576 | 19q13.42 |
|
hsa-miR-3150b-5p | 0.435225 | 0.007937 | 0.037576 | 8q22.1 |
| hsa-miR-4515 | 0.444861 | 0.007937 | 0.037576 | 15q25.2 |
|
hsa-miR-5587-3p | 0.475811 | 0.007937 | 0.037576 | 16p13.3 |
|
hsa-miR-4717-3p | 0.478848 | 0.007937 | 0.037576 | 16p13.3 |
Discussion
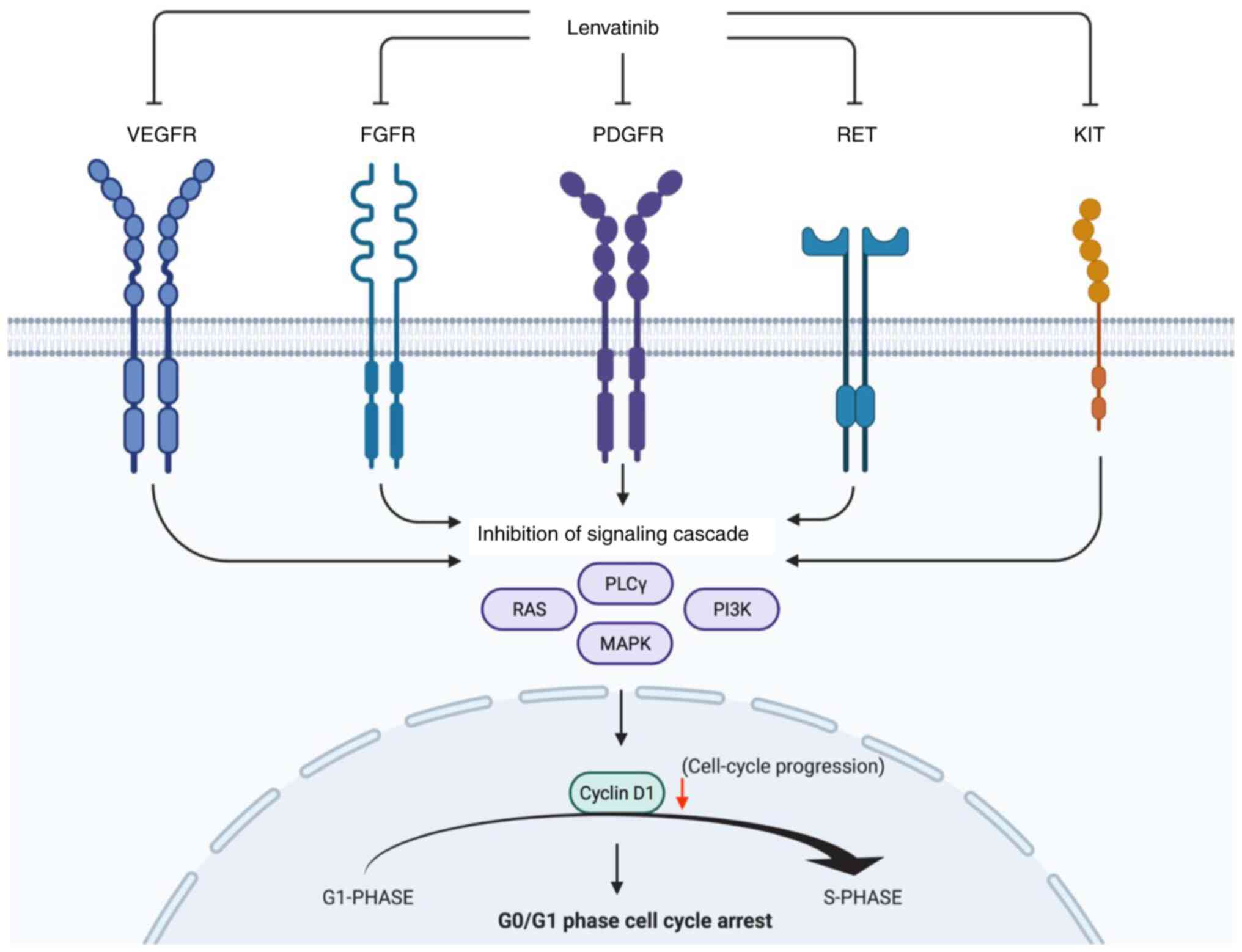
In the present study, we focused on the antitumor
effects of Lenvatinib in HCC. The findings of this study are
significant as we investigated the antitumor effect of Lenvatinib
on HCC cell growth both in vitro and in vivo.
Lenvatinib induced cell cycle arrest at the
G0/G1 phase by modulating the cell
cycle-regulating protein cyclin D1 in Lenvatinib-sensitive HCC
cells (Fig. 5). These results are
consistent with the findings of previous studies on the effects of
Lenvatinib on various cancer cells (7,8).
Importantly, the antiproliferative effects of Lenvatinib on HCC
cells were validated by modulating miRNAs in HCC cells and
exosomes. To the best of our knowledge, this is the first study to
show that Lenvatinib suppresses HCC cell proliferation by inducing
cell cycle arrest and altering miRNA expression.
We examined the effect of Lenvatinib treatment on
cell proliferation in vitro using four different HCC cell
lines. Lenvatinib has been shown to inhibit HCC cell proliferation
previously (10,16). Our results suggest that Lenvatinib
treatment led to a dose-dependent suppression of proliferation in
HuH-7 and Hep3B cells, but not in PLC/PRF/5 and Li-7 cells. HuH-7
and Hep3B cells, which abundantly express FGF19 and FGFR4, are more
sensitive to Lenvatinib than Li-7 cells, which do not express
FGF19(10). PLC/PRF/5 cells are
also insensitive to pan-FGFR inhibitors (14,10).
Additionally, FGF19 expression is increased in HCC cells compared
with that in normal hepatocytes, which negatively correlates with
E-cadherin expression and is involved in tumor progression
(17,18). PLC/PRF/5 cells do not deactivate
their Akt signaling compared with other HCC cells (Huh-7 and Hep3B)
(19). It has also been reported
that sorafenib, a drug with a molecular target similar to
Lenvatinib, when used as an adjuvant with existing anticancer
drugs, exerts anticancer effects through inactivation of Akt
signaling (20). Therefore, we
conducted an additional study of the Akt pathway in Huh-7 and
PLC/PRF/5 cells. While p-Akt expression decreased in HuH-7 cells
treated with Lenvatinib, no change was observed in PLC/PRF/5 cells.
Therefore, the Akt signaling pathway may be important in the
mechanism of Lenvatinib action, and differences in the molecular
characteristics of the cells may underlie the differences in the
pharmacological activities of Lenvatinib (16). Our in vitro experiments were
conducted using a lower dose of 1 µM Lenvatinib than that used in
previous studies in which 3 and 30 µM doses (10,14,16)
were used. This lower dose of Lenvatinib showed the
antiproliferative effects on HCC cell lines long-term (≥72 h).
Lenvatinib treatment also markedly suppressed the growth of
subcutaneous HuH-7-derived tumors in a xenograft mouse model.
Inhibition of xenograft tumor growth by Lenvatinib has been
observed at doses as low as 1 and 10 mg/kg (21-23).
Our in vivo study was conducted using a dose of Lenvatinib
similar to that used in previous studies (21-23).
No significant differences in body weight, appetite, or reactions
were detected between nude mice treated with Lenvatinib and
controls. These findings suggest that Lenvatinib is an effective
and relatively safe treatment option for HCC.
Our in vitro experiments verified that
Lenvatinib induced cell cycle arrest at the
G0/G1 phase, which was accompanied by
downregulation of cyclin D1 in the Lenvatinib-sensitive HuH-7
cells. A few studies have shown a link between the cell cycle and
cell cycle-related proteins in combination therapies of Lenvatinib
with other chemopreventive agents for papillary thyroid cancer
(15,24). One of the antitumor effects of
Lenvatinib is the induction of cell cycle arrest. Kim et al
(15) reported that alternating
sorafenib/Lenvatinib treatment reduced the levels of cyclin D1,
Cdk4, and Cdk6 and arrested the cell cycle of patient-derived
thyroid cancer cells compared with treatment with sorafenib or
Lenvatinib alone. Jing et al (24) showed that the combination therapy of
Lenvatinib with paclitaxel inhibited cell proliferation and tumor
growth in anaplastic thyroid cancer cells by inducing G2/M phase
cell cycle arrest compared with Lenvatinib or paclitaxel
monotherapy (24). These results
indicated that combination therapy may be more effective than
Lenvatinib monotherapy in HCC cells.
We further identified the aberrantly expressed
miRNAs in response to Lenvatinib treatment in HuH-7 cells and
exosomes using miRNA expression arrays. Exosomes are small EVs
secreted by almost all cell types, including cultured cells in
vitro. Exosomes play critical roles in cell-to-cell
communication as they carry mRNAs, miRNAs, and proteins as cargo to
recipient cells from donor cells to regulate their functions
(25). miRNAs account for 2-7% of
all small exosomal RNAs obtained from supernatants of HCC cells
cultured in vitro (26). In
our study, the levels of numerous miRNAs were significantly altered
following Lenvatinib treatment in HuH-7 cells and exosomes.
miR-218-5p, which was significantly upregulated in HuH-7 cells,
specifically targets the 3'-UTR region of Cdk6 and cyclin D1 to
induce cell cycle arrest and inhibit the growth of gastric cancer
cells (27). To clarify the
functional role of miR-218-5p in HCC, we analyzed the effects of
miR-218-5p overexpression and inhibition on HCC cell proliferation
in vitro (data not shown). Neither overexpression nor
silencing of these miRNAs affected proliferation. We also found
that miR-491-5p was significantly upregulated in exosomes. In GC,
miR-491-5p exhibits tumor suppressor functions by suppressing
Wnt3α/β-catenin signaling (28). Lu
et al (29) reported that
miR-491-5p suppressed the proliferation of colorectal cancer cells
via insulin-like growth factor 2(29). In addition, overexpression of
miR-320b, which was significantly upregulated in this study,
induced cell cycle arrest in the G0/G1 phase
and inhibited tumor growth in gliomas (30). Downregulated miR-1470 expression has
been reported to promote cell proliferation by targeting ALX4 in
HCC (31). These studies suggest
that Lenvatinib exerts its antitumor functions through miRNA
regulation. However, this study lacked an analysis of the function
of the regulated miRNAs when overexpressed. Further experiments are
needed to explore the relationship between these miRNAs and cyclin
D1 as well as its upstream signaling. In PLC/PRF/5 cells, which are
less sensitive to Lenvatinib, there were significant changes in
miRNAs in the presence and absence of Lenvatinib; however,
miR-218-5p, which targets cyclin D1, was only significantly altered
in HuH-7 cells as described above, and exosomally upregulated
miR-320b was not significantly altered in PLC/PRF/5 cells. The
differences in sensitivity to Lenvatinib amongst cells may be due
to differences in FGFR expression, as described above, as well as
in the miRNAs involved in cell cycle arrest. Future studies should
explore the potential role of aberrantly expressed miRNAs in the
antitumor effects of Lenvatinib.
In conclusion, the present study revealed that
Lenvatinib directly suppresses HCC cell proliferation and tumor
growth and exerts antitumor effects by inducing cell cycle arrest
in Lenvatinib-sensitive HCC cells.
Acknowledgements
We would like to thank Ms Keiko Fujikawa, Ms Megumi
Okamura, and Ms Fuyuko Kokado (Kagawa University Faculty of
Medicine, Japan) for their technical assistance.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
MN and TM designed the experiments. SF, HI, KT, KO,
TT, KF, JT, AM, HK, and TH performed the experiments, analyzed the
data, and drafted and wrote the final manuscript. TM was involved
in drafting of the final manuscript. All the authors have read and
approved the final version of the manuscript. MN and TM confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
This study was approved by the Committee on Animal
Experimentation of Kagawa University (approval no. 18675; Kagawa,
Japan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Global Burden of Disease Liver Cancer
Collaboration. Akinyemiju T, Abera S, Ahmed M, Alam N, Alemayohu
MA, Allen C, Al-Raddadi R, Alvis-Guzman N, Amoako Y, et al: The
burden of primary liver cancer and underlying etiologies from 1990
to 2015 at the global, regional, and national level: Results from
the global burden of disease study 2015. JAMA Oncol. 3:1683–1691.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
El-Serag HB: Hepatocellular carcinoma:
Recent trends in the United States. Gastroenterology. 127 (Suppl
1):S27–S34. 2004.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Munoz N and Bosch X: Epidemiology of
hepatocellular carcinoma. In: Neoplasms of the liver. Okuda K and
Ishak KG (eds). Springer, Tokyo, p3, 1989.
|
|
4
|
Davila JA, Morgan RO, Shaib Y, McGlynn KA
and El-Serag HB: Hepatitis C infection and the increasing incidence
of hepatocellular carcinoma: A population-based study.
Gastroenterology. 127:1372–1380. 2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Llovet JM, Ricci S, Mazzaferro V, Hilgard
P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A,
et al: Sorafenib in advanced hepatocellular carcinoma. N Engl J
Med. 359:378–390. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sanoff HK, Chang Y, Lund JL, O'Neil BH and
Dusetzina SB: Sorafenib effectiveness in advanced hepatocellular
carcinoma. Oncologist. 21:1113–1120. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Tohyama O, Matsui J, Kodama K, Hata-Sugi
N, Kimura T, Okamoto K, Minoshima Y, Iwata M and Funahashi Y:
Antitumor activity of lenvatinib (e7080): An angiogenesis inhibitor
that targets multiple receptor tyrosine kinases in preclinical
human thyroid cancer models. J Thyroid Res.
2014(638747)2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Motzer RJ, Hutson TE, Glen H, Michaelson
MD, Molina A, Eisen T, Jassem J, Zolnierek J, Maroto JP, Mellado B,
et al: Lenvatinib, everolimus, and the combination in patients with
metastatic renal cell carcinoma: A randomised, phase 2, open-label,
multicentre trial. Lancet Oncol. 16:1473–1482. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kudo M, Finn RS, Qin S, Han KH, Ikeda K,
Piscaglia F, Baron A, Park JW, Han G, Jassem J, et al: Lenvatinib
versus sorafenib in first-line treatment of patients with
unresectable hepatocellularcarcinoma: A randomised phase 3
non-inferiority trial. Lancet. 391:1163–1173. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Matsuki M, Hoshi T, Yamamoto Y,
Ikemori-Kawada M, Minoshima Y, Funahashi Y and Matsui J: Lenvatinib
inhibits angiogenesis and tumor fibroblast growth factor signaling
pathways in human hepatocellular carcinoma models. Cancer Med.
7:2641–2653. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Morishita A, Iwama H, Fujihara S, Sakamoto
T, Fujita K, Tani J, Miyoshi H, Yoneyama H, Himoto T and Masaki T:
MicroRNA profiles in various hepatocellular carcinoma cell lines.
Oncol Lett. 12:1687–1692. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Liang LH and He XH: Macro-management of
microRNAs in cell cycle progression of tumor cells and its
implications in anti-cancer therapy. Acta Pharmacol Sin.
32:1311–1320. 2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Miyata M, Morishita A, Sakamoto T, Katsura
A, Kato K, Nishioka T, Toyota Y, Fujita K, Maeda E, Nomura T, et
al: MicroRNA profiles in cisplatin-induced apoptosis of
hepatocellular carcinoma cells. Int J Oncol. 47:535–542.
2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Futami T, Okada H, Kihara R, Kawase T,
Nakayama A, Suzuki T, Kameda M, Shindoh N, Terasaka T, Hirano M and
Kuromitsu S: ASP5878, a novel inhibitor of FGFR1, 2, 3, and 4,
inhibits the growth of FGF19-expressing hepatocellular carcinoma.
Mol Cancer Ther. 16:68–75. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kim SY, Kim SM, Chang HJ, Kim BW, Lee YS,
Park CS, Park KC and Chang HS: SoLAT (sorafenib lenvatinib
alternating treatment): A new treatment protocol with alternating
sorafenib and lenvatinib for refractory thyroid cancer. BMC Cancer.
18(956)2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ogasawara S, Mihara Y, Kondo R, Kusano H,
Akiba J and Yano H: Antiproliferative effect of lenvatinib on human
liver cancer cell lines in vitro and in vivo. Anticancer Res.
39:5973–5982. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhao H, Lv F, Liang G, Huang X, Wu G,
Zhang W, Yu L, Shi L and Teng Y: FGF19 promotes
epithelial-mesenchymal transition in hepatocellular carcinoma cells
by modulating the GSK3β/β-catenin signaling cascade via FGFR4
activation. Oncotarget. 7:13575–13586. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Latasa MU, Salis F, Urtasun R,
Garcia-Irigoyen O, Elizalde M, Uriarte I, Santamaria M, Feo F,
Pascale RM, Prieto J, et al: Regulation of amphiregulin gene
expression by β-catenin signaling in human hepatocellular carcinoma
cells: A novel crosstalk between FGF19 and the EGFR system. PLoS
One. 7(e52711)2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen KF, Yeh PY, Yeh KH, Lu YS, Huang SY
and Cheng AL: Down-regulation of phospho-Akt is a major molecular
determinant of bortezomib-induced apoptosis in hepatocellular
carcinoma cells. Cancer Res. 68:6698–6707. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chen KF, Yu HC, Liu TH, Lee SS, Chen PJ
and Cheng AL: Synergistic interactions between sorafenib and
bortezomib in hepatocellular carcinoma involve PP2A-dependent Akt
inactivation. J Hepatol. 52:88–95. 2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Matsui J, Funahashi Y, Uenaka T, Watanabe
T, Tsuruoka A and Asada M: Multi-kinase inhibitor E7080 suppresses
lymph node and lung metastases of human mammary breast tumor
MDA-MB-231 via inhibition of vascular endothelial growth
factor-receptor (VEGF-R) 2 and VEGF-R3 kinase. Clin Cancer Res.
14:5459–5465. 2008.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Matsui J, Yamamoto Y, Funahashi Y,
Tsuruoka A, Watanabe T, Wakabayashi T, Uenaka T and Asada M: E7080,
a novel inhibitor that targets multiple kinases, has potent
antitumor activities against stem cell factor producing human small
cell lung cancer H146, based on angiogenesis inhibition. Int J
Cancer. 122:664–671. 2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Ikuta K, Yano S, Trung VT, Hanibuchi M,
Goto H, Li Q, Wang W, Yamada T, Ogino H, Kakiuchi S, et al: E7080,
a multi-tyrosine kinase inhibitor, suppresses the progression of
malignant pleural mesothelioma with different proangiogenic
cytokine production profiles. Clin Cancer Res. 15:7229–7237.
2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Jing C, Gao Z, Wang R, Yang Z, Shi B and
Hou P: Lenvatinib enhances the antitumor effects of paclitaxel in
anaplastic thyroid cancer. Am J Cancer Res. 7:903–912.
2017.PubMed/NCBI
|
|
25
|
Han Q, Zhao H, Jiang Y, Yin C and Zhang J:
HCC-derived exosomes: critical player and target for cancer immune
escape. Cells. 8(558)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yu LX, Zhang BL, Yang Y, Wang MC, Lei GL,
Gao Y, Liu H, Xiao CH, Xu JJ, Qin H, et al: Exosomal microRNAs as
potential biomarkers for cancer cell migration and prognosis in
hepatocellular carcinoma patient-derived cell models. Oncol Rep.
41:257–269. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Deng M, Zeng C, Lu X, He X, Zhang R, Qiu
Q, Zheng G, Jia X, Liu H and He Z: miR-218 suppresses gastric
cancer cell cycle progression through the CDK6/Cyclin D1/E2F1 axis
in a feedback loop. Cancer Lett. 403:175–185. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sun R, Liu Z, Tong D, Yang Y, Guo B, Wang
X, Zhao L and Huang C: miR-491-5p, mediated by Foxi1, functions as
a tumor suppressor by targeting Wnt3a/β-catenin signaling in the
development of gastric cancer. Cell Death Dis.
8(e2714)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lu L, Cai M, Peng M, Wang F and Zhai X:
miR-491-5p functions as a tumor suppressor by targeting IGF2 in
colorectal cancer. Cancer Manag Res. 11:1805–1816. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lv QL, Du H, Liu YL, Huang YT, Wang GH,
Zhang X, Chen SH and Zhou HH: Low expression of microRNA-320b
correlates with tumorigenesis and unfavorable prognosis in glioma.
Oncol Rep. 38:959–966. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lu Y, Yang L, Qin A, Qiao Z, Huang B,
Jiang X and Wu J: miR-1470 regulates cell proliferation and
apoptosis by targeting ALX4 in hepatocellular carcinoma. Biochem
Biophys Res Commun. 522:716–723. 2020.PubMed/NCBI View Article : Google Scholar
|