Introduction
Gastrointestinal cancers encompass a variety of
malignant diseases, with only colorectal cancer (CRC) ranked among
the most common tumors (1). The
integrated conventional staging system (T, tumor size; N, lymph
node status; M, distant metastasis) and molecular classifications
of CRC may aid in reliable personalized treatments and may
contribute to the prediction of the prognosis of cancer (2-4)
Moreover, the success of CRC screening and detection must depend
significantly on molecular parameters and not exclusively on
clinical stage (5,6).
Based on findings concerning peripheral blood
possibly reflecting changes that occur in tissues (7,8),
numerous molecular parameters from blood specimens have been
reported for CRC detection (9-11).
As this type of test does not directly include samples of colonic
cell origin, the molecular parameters of numerous genes should be
composed to achieve adequate sensitivity and specificity (9,11).
Compared with the results of other groups that have used blood
specimens for screening, a previous study by the authors involving
the extraction of expression profiles from stool specimens revealed
a good association of these profiles with CRC progression and
recurrence (12-14).
The direct detection of changes in gene expression in colonic
tissues may contribute to a further understanding of CRC
progression and may allow the development of biomarkers and drug
targets for this malignant disease (15). Human stool has been studied for CRC
screening for several years (16-18),
and several lines of evidence have indicated that cells shed from
the colonic tract may reflect localized diseases (19-21).
Thus, either DNA or RNA extracted from stool specimens can be used
to detect colorectal neoplasia accurately (22-24).
Genes that are actively expressed in human stool specimens have
emerged as specific molecular signatures of CRC (25,26).
These genetic molecules may aid in the understanding of the process
of the development of CRC (27,28).
Moreover, the signatures concurrently reflect CRC biology, and
inform prognoses and treatment responses in a non-invasive manner
(29). The gene expression status
of colonic cells that pass into stool specimens has been considered
to faithfully represent CRC manifestations (30-33).
Herein, the novel, to the best of our knowledge,
advantage of using cells shed from the colonic tract for
cost-effective CRC screening is presented. Previously, genes
expressed in human stool specimens that were used to identify
patients with differentially staged CRC distinguishing them from
healthy donor control samples were acquired using whole-genome
microarrays. Expressed genes were further filtered via custom-made
microarrays. Specific gene sets were tested with a small number of
testing samples using reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) analyses, trained with other samples set
by a series of leave-one-out cross-validation (LOOCV) and
discriminant analyses (34,35), and then confirmed with the third
sample set for the probabilities of group membership (healthy donor
or CRC). The corresponding proteins of target genes were assessed
based on CRC tissue arrays. Furthermore, a logistic regression
model was used to predict diseases for a specific molecular panel
(36,37).
Materials and methods
Study participants and ethical
approval
Human stool specimens were collected from 29 healthy
donor controls (age range, 23-78 years; 8 males and 21 females) and
58 patients with well-diagnosed CRC (age range, 29-87 years; 33
males and 25 females) at the Sijhih Cathay General Hospital for
predictive model training. All the participating subjects provided
their written informed consent. The research was conducted with the
obtained approval (approval no. CGH-P101014) of the Institutional
Review Board of the Cathay General Hospital and according to the
Principles of the Declaration of Helsinki.
All 87 cases (29 healthy donors and 58 CRC cases)
were randomly divided into three independent sets that were used
for different purposes as follows: Set I (n=11, five healthy donor
and six CRC cases); set II (n=56, 20 healthy donor and 36 CRC
cases); and set III (n=20, four healthy donor and 16 CRC cases).
The initial tumor stage of patients with CRC was classified using
the 8th Edition of the American Joint Committee on Cancer (AJCC)
staging system (38), and the
healthy donor controls were requested to undergo a colonoscopy
examination. All patients enrolled in the present study were
managed according to standard guidelines, with regular follow-up.
To characterize the targets of interest in CRC samples, the
proteins encoded by these genes were immunostained in colon cancer
tissue arrays (COC1021; total 102 available cores, including two
non-CRC colonic tissues, one congenital megacolon, two colon
adenomas, four papillary adenocarcinomas, seven mucinous
adenocarcinomas, and 86 colon adenocarcinomas; Pantomics, Inc.).
According to the provided TNM classification from Pantomics, Inc.,
22 tissue cores with adenocarcinoma were diagnosed as AJCC stage I,
39 were AJCC stage II, and 36 were AJCC stage III (Fig. 1) (39). To validate a predictive model, other
stool specimens and one cDNA array of colonic tissues were used.
Briefly, stool specimens of two female patients (59 and 71 years of
age) during pre- and post-surgical treatments at the Sijhih Cathay
General Hospital (New Taipei City, Taiwan) and 119 individuals (age
range, 26-76 years; 67 males and 52 females) following a
colonoscopy at Cathay Healthcare Management (Taipei City, Taiwan)
were collected. In addition, one cDNA array (HCRT104; OriGene
Technologies, Inc.) of 48 colon tissues covering non-CRC status
(n=8) and four CRC stages (n=40) was purchased to examine the
predictive model.
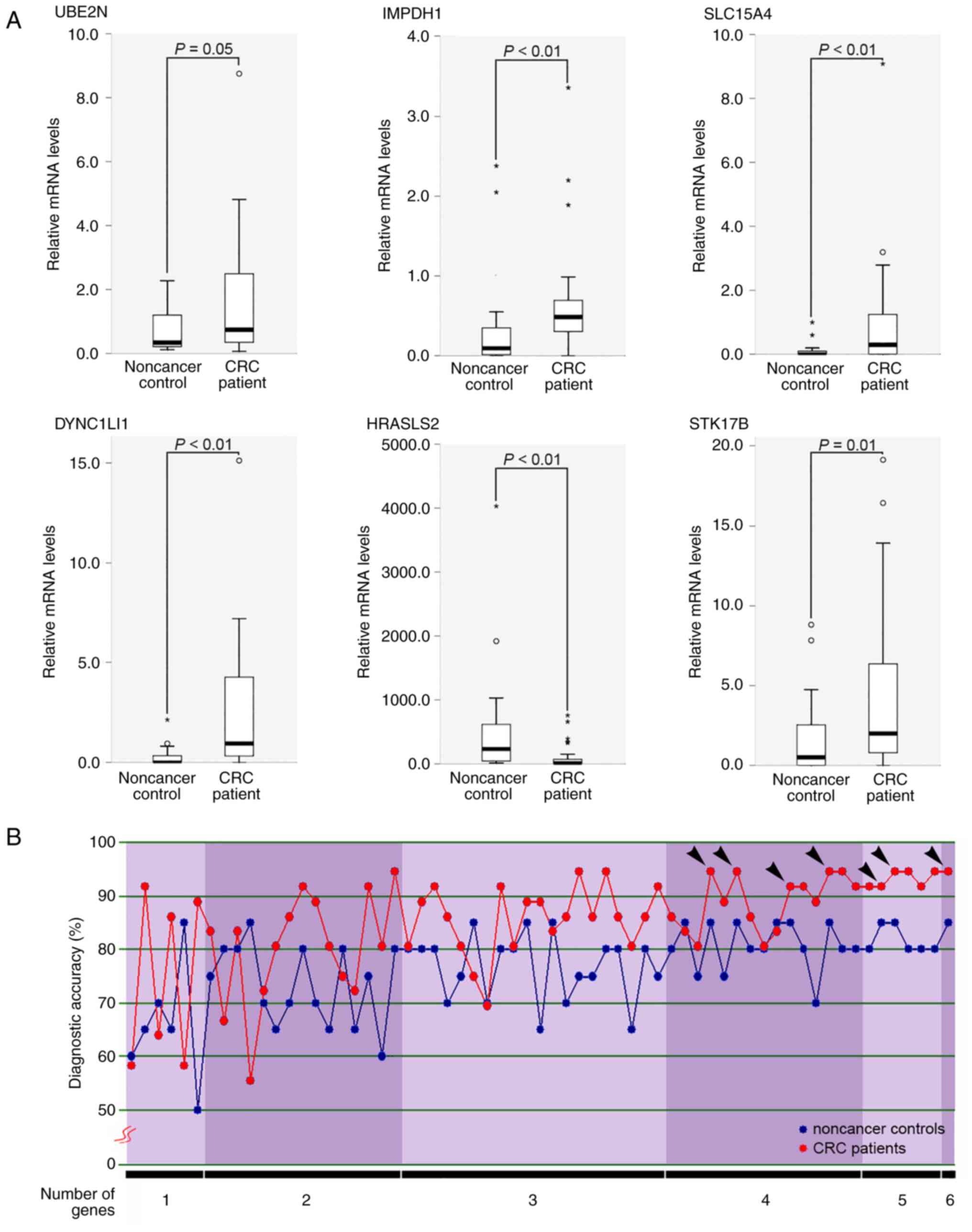 | Figure 1Development of gene panels in stool
specimens for patients with CRC. (A) Statistical comparison of gene
expressions between healthy donor control and CRC patients. Six
genes were differentially expressed in healthy donor controls
(n=20) and CRC patients (n=36). Differences in the relative levels
of the target mRNAs among the samples were determined using the
Mann-Whitney U test. P≤0.05 (B) Diagnostic accuracy of 63 different
molecular panels. The 63 different molecular panels were comprised
of one to six genes. Each panel was used to predict the healthy
donor or CRC disease status using LOOCV analysis. The six genes
used were UBE2N, IMPDH1, SLC15A4, DYNC1LI1, HRASLS2 and STK17B.
Black arrowheads indicate the panels with higher sensitivity (≥90%)
and specificity (≥85%). CRC, colorectal cancer; LOOCV,
leave-one-out cross-validation; UBE2N, ubiquitin-conjugating enzyme
E2 N; IMPDH1, inosine monophosphate dehydrogenase 1; SLC15A4,
phospholipase A and acyltransferase 2 solute carrier family 15
member 4; DYNC1LI1, dynein cytoplasmic 1 light intermediate chain
1; HRASLS2, phospholipase A and acyltransferase 2; STK17B,
serine/threonine kinase 17b. |
LOOCV, discriminant analyses and
predictive model for CRC risk
The stool specimens from set I were initially
applied to screen out genes with a differential expression in
previous microarray hybridizations performed by the authors
(14,40). A LOOCV analysis was then performed
for the additional screened genes on a given set (set II) as a
learning system. Briefly, each stool specimen was excluded in turn
and classified using a defined model based on the non-excluded
samples (41,42). Using the cases in set II as the
well-defined group, significant LOOCV-estimated molecular panels
were used to test the probabilities of group membership (healthy
donor or CRC) in the testing sets (sets I and III) via discriminant
analysis. As a result, a predictive model of an optimal molecular
panel for CRC risk was produced from all 87 well-known cases (sets
I, II, and III) via the analysis of the logistic regression model
(43,44), and a receiver operating
characteristic (ROC) curve was generated to assess model
discrimination (45).
Relative gene expression
quantification
The method of purifying total RNA from stool mud
[0.5 g stool in 1 ml guanidinium thiocyanate buffer; 10 mM Tris (pH
7.4), 200 mM NaCl, 1 mM EDTA (pH 8.0), 4 M guanidinium thiocyanate,
and 1% β-mercaptoethanol] was largely described in previous reports
by the authors (40,46). An appropriate supernatant was then
extracted using a MagCore Nucleic Acid Extract kit in a MagCore
Nucleic Acid Extractor (RBC Bioscience Corp.). The eluted fecal
total RNA was then quantified using a NanoDrop ND 1000
spectrophotometer (Thermo Fisher Scientific, Inc.) and
reverse-transcribed to generate single-stranded cDNAs using random
primers and a PowerScript Reverse Transcriptase Kit (cat. no.
RR037B; Takara Bio USA, Inc.), according to the manufacturer's
instructions.
The genes of interest were quantified by a program
(10 min at 95˚C, proceeding with 60 cycles at 95˚C for 10 sec and
at 60˚C for 20 sec) in the presence of a TaqMan probe and TaqMan
Master Mix using a Roche LightCycler nano system (Roche Diagnostics
GmbH) according to the manufacturer's instructions. Briefly, all
genes were quantified relative to the level of
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each
quantification. All primers and probes used in the present study
are listed in Table I. LightCycler
Software (version 4.05; Roche Diagnostics GmbH) was used to analyze
the PCR kinetics. Expression levels were quantified using the
2-ΔΔCq method (47) and
normalized to the expression level of GAPDH (48). Each run also included an appropriate
and predetermined diluted human reference cDNA (Takara Bio USA,
Inc.), which was used as a standard to estimate relative expression
levels.
 | Table IRT-qPCR primers and probe numbers for
gene expression quantification. |
Table I
RT-qPCR primers and probe numbers for
gene expression quantification.
| Gene name | Accession no. | Sequence (from 5'
to 3') | UPL no. |
|---|
| GAPDH | NM_002046 | Fw:
CTCTGCTCCTCCTGTTCGAC | #60 |
| | | Rv:
ACGACCAAATCCGTTGACTC | |
| UBE2N | NM_003348.3 | Fw:
AAGCCCAAGCCATAGAAACA | #2 |
| | | Rv:
ATGCAAACAAAGAGGAGGAAGT | |
| AKIRIN1 | NM_024595.1 | Fw:
ACTCCTCAGCACTCACAGCA | #80 |
| | | Rv:
CCAACTTGTCGGAGGGTAAA | |
| IMPDH1 | NM_000883.3 | Fw:
GTCCATGGCCTGCACTCT | #22 |
| | | Rv:
GTGGACACTGGGGTGCAT | |
| SLC15A4 | NM_145648.3 | Fw:
GAGCAGTCACACAGACTTTGGT | #71 |
| | | Rv:
CAGGAGGGTAGCTCCTTGAA | |
| DYNC1LI1 | NM_016141.2 | Fw:
CTGGTGTGAGTGGTGGTAGC | #10 |
| | | Rv:
TCTGCATGAACATCTAAGACAGG | |
| HRASLS2 | NM_017878.1 | Fw:
ATCTGCGCTATGGCGTCT | #74 |
| | | Rv:
CAGCAGGATCCCCACAAG | |
| APOA1 | NM_000039.1 | Fw:
CCTTGGGAAAACAGCTAAACC | #39 |
| | | Rv:
CCAGAACTCCTGGGTCACA | |
| STK17B | NM_004226.2 | Fw:
GGAAATCATGGGAACACCAG | #50 |
| | | Rv:
TTGCTGTGGTAATGGGATCAT | |
Immunohistochemistry
COC1021 tissue arrays (Pantomics, Inc.), which have
defined clinical diagnosis and clinicopathological information,
were used for immunohistochemical staining (49). Firstly, tissue arrays with 4 µm core
thickness were deparaffinated and hydrated using routine protocols.
Antigen retrieval was performed by steaming in Tris-EDTA buffer (pH
9.0) for 20 min, and arrays were then blocked in 1.5% (v/v) Normal
Horse Serum Blocking Solution (Vector Laboratories, Inc.) for 2 h
at room temperature. The specific target protein was immunodetected
using an adequate primary antibody concentration
[ubiquitin-conjugating enzyme E2 N (UBE2N): anti-Ube2N, cat. no.
ab117090, dilution 1:20, Abcam; inosine monophosphate dehydrogenase
1 (IMPDH1): anti-IMPDH1, cat. no. ab84957, 1:20, Abcam; dynein
cytoplasmic 1 light intermediate chain 1 (DYNC1LI1): anti-DLC-A,
cat. no. ab154251, 1:20, Abcam; phospholipase A and acyltransferase
2 (HRASLS2): anti-HRASLS2, cat. no. bs-6013R, 1:1,000, Thermo
Fisher Scientific, Inc.] in the blocking solution mentioned above
at 4˚C overnight. Endogenous peroxidases in tissue sections were
removed by incubation with 0.3% H2O2 for 15
min, and these peroxidase-free arrays were further incubated with a
biotinylated secondary antibody (either goat anti-rabbit, cat. no.
BA-1000-1.5, 1:200, Vector Laboratories, Inc. or rabbit anti-goat
immunoglobulin G, cat. no. BA-5000-1.5, 1:200, Vector Laboratories,
Inc.) at room temperature for 60 min. The VECTASTAIN ABC system and
DAB Substrate kit (both from Vector Laboratories, Inc.) were used
to develop the secondary antibodies captured on arrays, according
to the manufacturer's instructions. Following hematoxylin (GHS3; 50
ml, Merck KGaA) counterstaining at room temperature for 5 min and
slide mounting with Malinol (Muto Pure Chemicals, Co., Ltd.),
images were digitalized using a high-resolution scanner (Mirax
Scan, Carl Zeiss AG) at the Taiwan Mouse Clinic (Academia Sinica,
Taipei City, Taiwan). Two independent pathologists reviewed and
evaluated the imaging results. QuPath (Version 0.3.0; https://qupath.github.io) was employed to produce the
cell densities of immunoreactive cells per mm2 for all
target proteins (50).
Statistical analyses
Differences in the relative levels of the target
mRNAs among the various samples were determined using the unpaired
non-parametric two-sample Mann-Whitney U test. The equation for the
probability of CRC was acquired via a logistic regression model. In
addition, one-way analysis of variance (ANOVA) was performed to
compare the densities of positive signaling cells for the
immunohistochemical staining of target proteins. All ANOVA analyses
were followed by a Bonferroni post hoc test. These deduced
probabilities and identifications based on 119 individuals after
colonoscopy were compared using the chi-squared test. To assess the
effectiveness of the predictive model of CRC risk, a ROC curve was
plotted between the sensitivity and (1-specificity), and the total
area under the ROC curve (AUC) was calculated. All statistical
analyses were performed with IBM SPSS Statistics for Windows
(Version 22.0; IBM Corp.). P<0.05 was considered to indicate a
statistically significant difference.
Results
Development of gene panels for
patients with CRC
All genes were selected according to different
clinical statuses from two microarray hybridizations, whole-genome
oligonucleotides and custom-made microarrays with ~4,000 genes from
previous studies by the authors (14,40,51).
Three independent sets of stool specimens (sets I, II and III) were
then used to validate gene expression by using a series of RT-qPCR
assays in order to select the optimal stool-based panels to predict
CRC probability. From sample set I, eight differentially expressed
genes [UBE2N, akirin 1 (AKIRIN1), IMPDH1, solute carrier family 15
member 4 (SLC15A4), DYNC1LI1, HRASLS2, apolipoprotein A1 (APOA1)
and serine/threonine kinase 17b (STK17B)] (P<0.2; Mann-Whitney U
test) were in accordance with the corresponding expression trends
obtained from the custom-made microarray hybridization (Table SI). However, six genes only (UBE2N,
IMPDH1, SLC15A4, DYNC1LI1, HRASLS2 and STK17B) were differentially
expressed in sample set II, which was used as the training set with
statistical significance (P≤0.05; Mann-Whitney U test) (Fig. 1A). These six genes comprised 63
different molecular panels. Moreover, using the same samples of set
II, each panel was used to predict the healthy donor or CRC disease
status using LOOCV analysis. As demonstrated in Fig. 1B, the diagnostic accuracy of each
panel for healthy donor controls or patients with CRC was ≥50%, and
seven different panels (UBE2N, IMPDH1, DYNC1LI1 and HRASLS2 genes;
UBE2N, IMPDH1, HRASLS2 and STK17B genes; UBE2N, DYNC1LI1, HRASLS2
and STK17B genes; IMPDH1, SLC15A4, HRASLS2 and STK17B genes; UBE2N,
IMPDH1, SLC15A4, DYNC1LI1 and STK17B genes; UBE2N, IMPDH1, SLC15A4,
HRASLS2 and STK17B genes; UBE2N, IMPDH1, SLC15A4, DYNC1LI1, HRASLS2
and STK17B genes) yielded higher sensitivity (≥90%) and specificity
(≥85%). A four-gene panel composed of only three upregulated genes
(UBE2N, IMPDH1 and DYNC1LI1) and one downregulated (HRASLS2) in
patients with CRC exhibited the highest accuracy for healthy donor
controls (88.9%, eight of nine) and patients with CRC (95.4%, 21 of
22) in predicting the testing sets (sets I and III) and using
sample set II as the training set, as assessed by discriminant
analyses.
Relative protein expression levels of
the four-gene panel in CRC tissues
Subsequently, it was investigated whether the
corresponding proteins of four genes were differentially expressed
in tumor samples from patients with CRC. An array method based on
established immunohistochemistry protocols was used to detect the
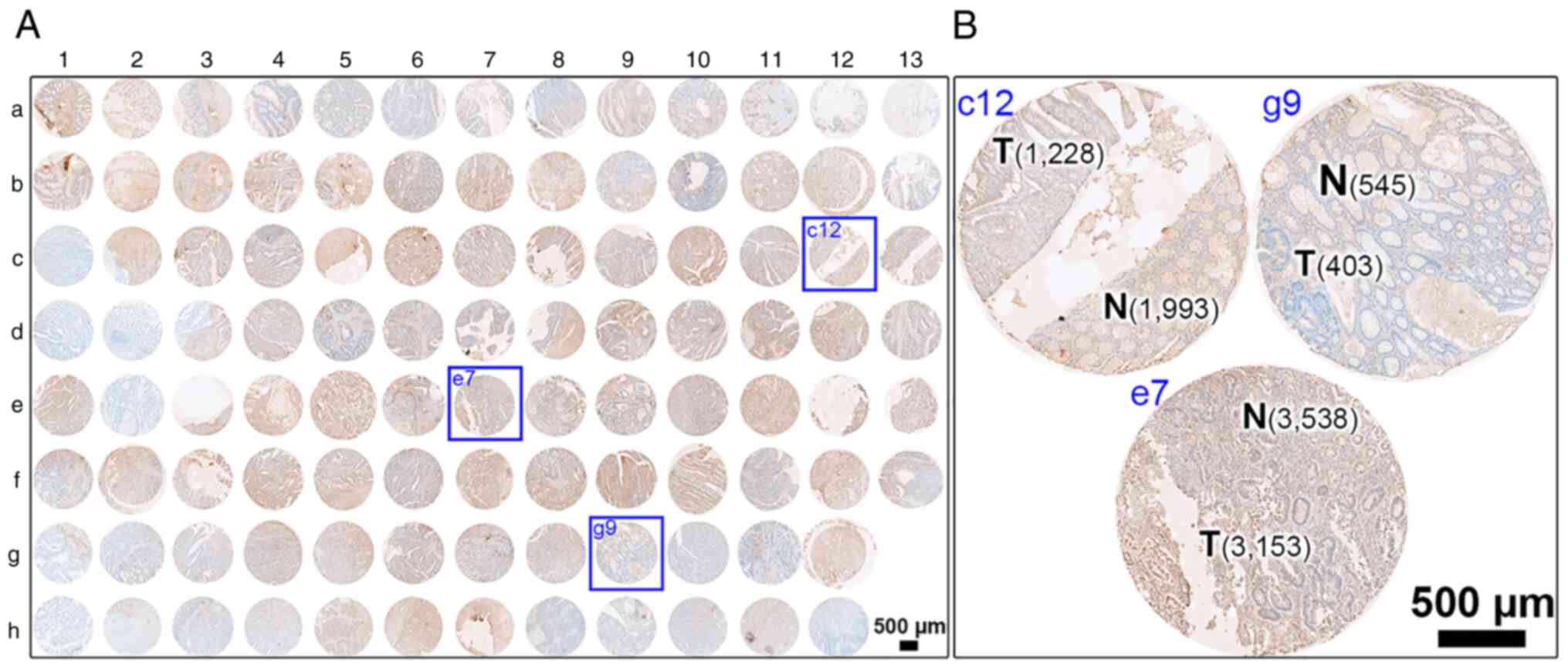
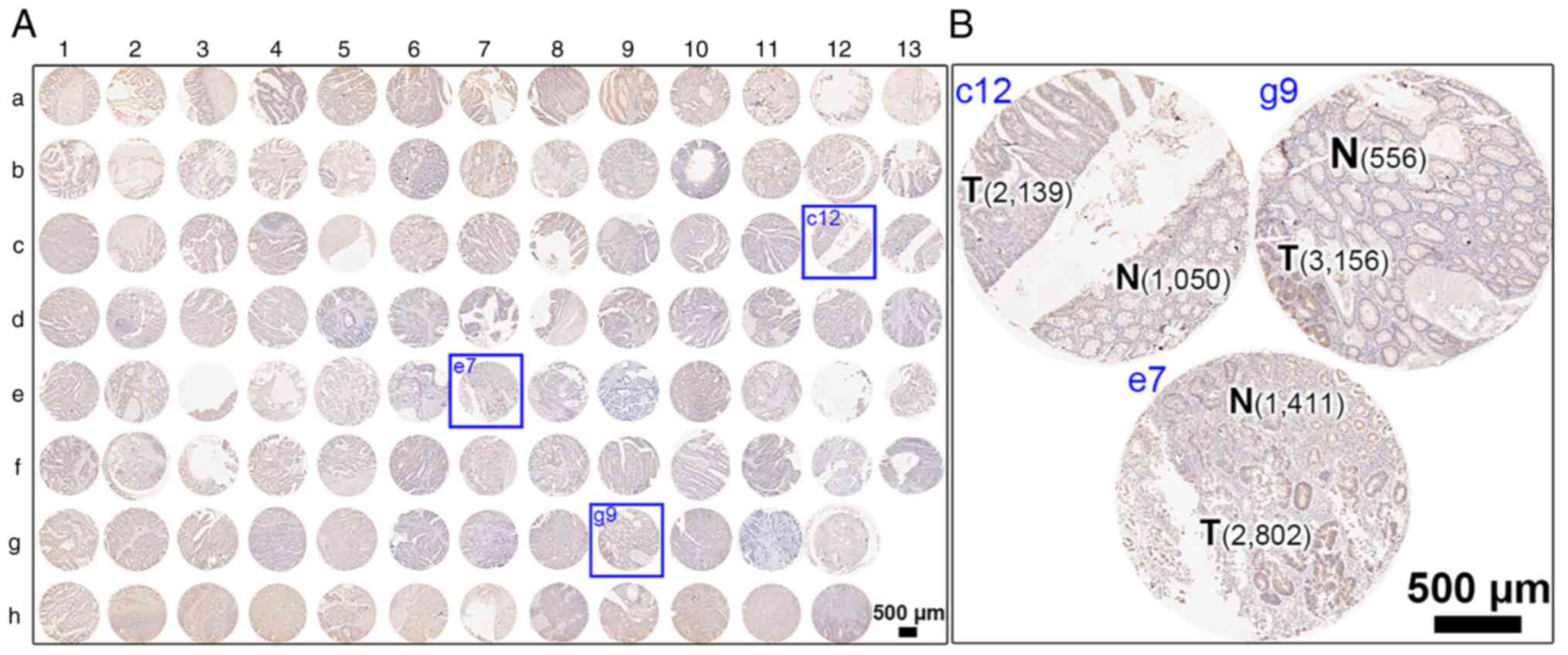
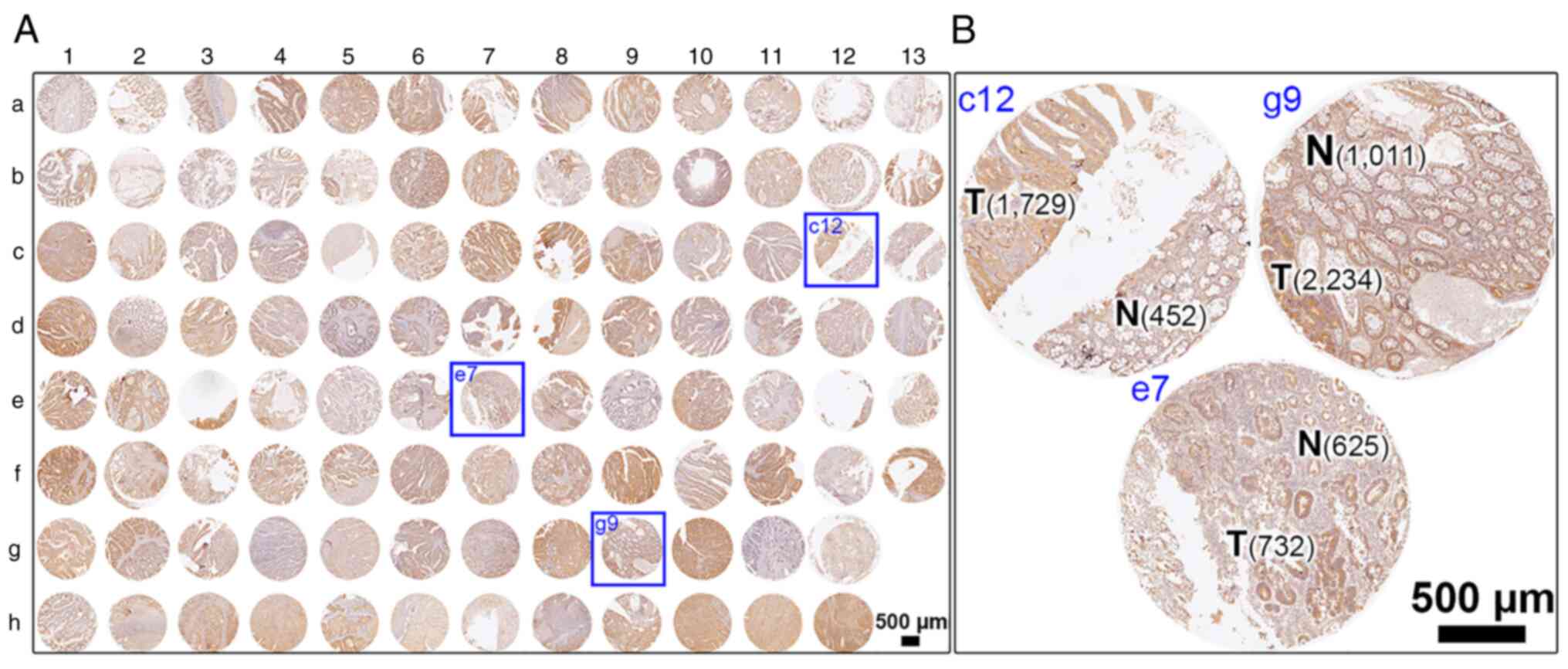
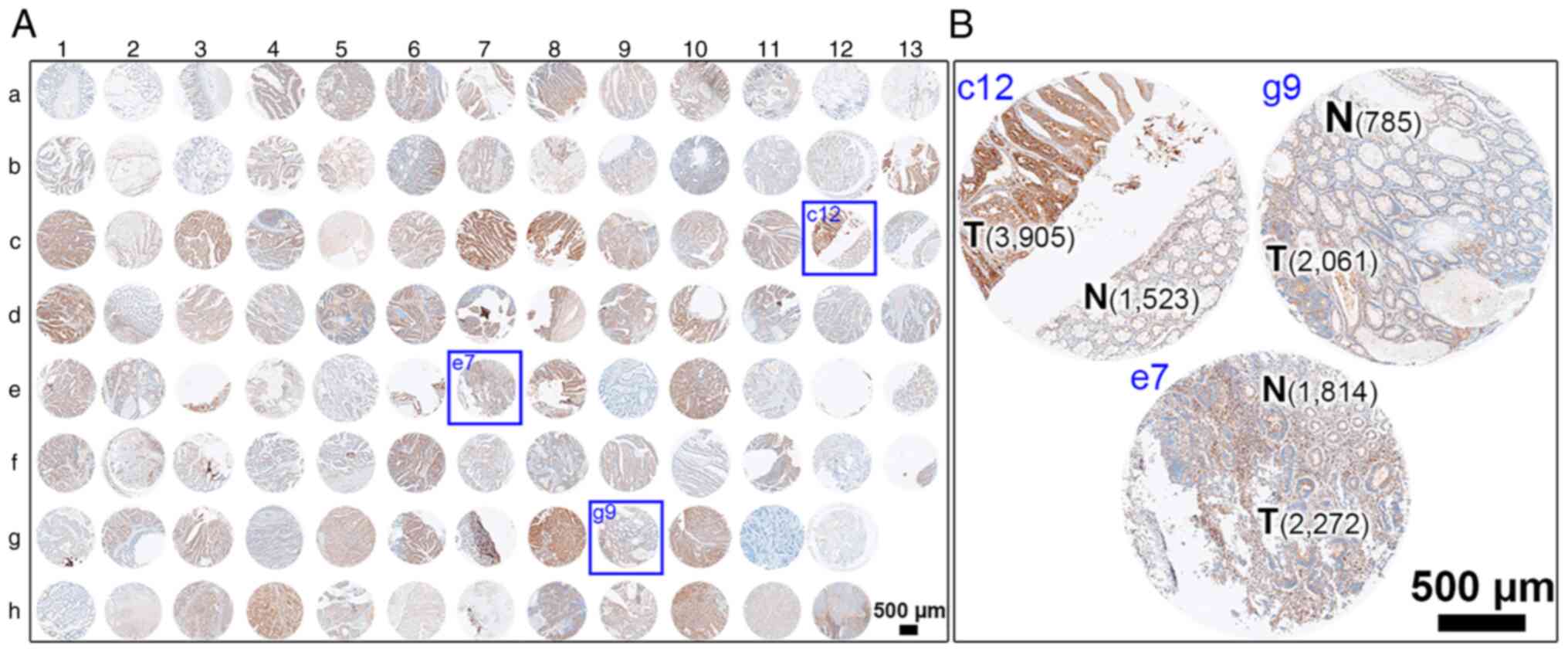
corresponding protein expression in human colonic tissues (Fig. 2 for overview of COC1021 tissue array
with 102 cores (x-axis, nos. 1 to 13; y-axis, a to h); Fig. 3 for immunohistochemical staining of
HRASLS2; Fig. 4 for
immunohistochemical staining of UBE2N; Fig. 5 for immunohistochemical staining of
IMPDH1; Fig. 6 for
immunohistochemical staining of DYNC1LI1). Moreover, CRC tissues at
different stages with CRC and non-CRC fractions in the same donor
of the COC1021 tissue array revealed that the signal density
(positive cells/mm2) of UBE2N (Fig. 4B: 2,139 cells/mm2 in c12,
2,802 cells/mm2 in e7, and 3,156 cells/mm2 in
g9), IMPDH1 (Fig. 5B: 1,729
cells/mm2 in c12, 732 cells/mm2 in e7, and
2,234 cells/mm2 in g9), or DYNC1LI1 (Fig. 6B: 3,905 cells/mm2 in c12,
2,272 cells/mm2 in e7, and 2,061 cells/mm2 in
g9) was high in the CRC fractions and low in the adjacent non-CRC
fractions (Fig. 4B: 1,050
cells/mm2 in c12, 1,411 cells/mm2 in e7, and
556 cells/mm2 in g9; Fig.
5B: 452 cells/mm2 in c12, 625 cells/mm2
in e7, and 1,011 cells/mm2 in g9; Fig. 6B: 1,523 cells/mm2 in c12,
1,814 cells/mm2 in e7, and 785 cells/mm2 in
g9). By contrast, HRASLS2 was expressed with a high density of
positive cells in the non-CRC fractions (1,993 cells/mm2
in c12, 3,538 cells/mm2 in e7, and 545
cells/mm2 in g9), but relatively less densely expressed
in the adjacent CRC fractions (1,228 cells/mm2 in c12,
3,153 cells/mm2 in e7, and 403 cells/mm2 in
g9) (Fig. 3B).
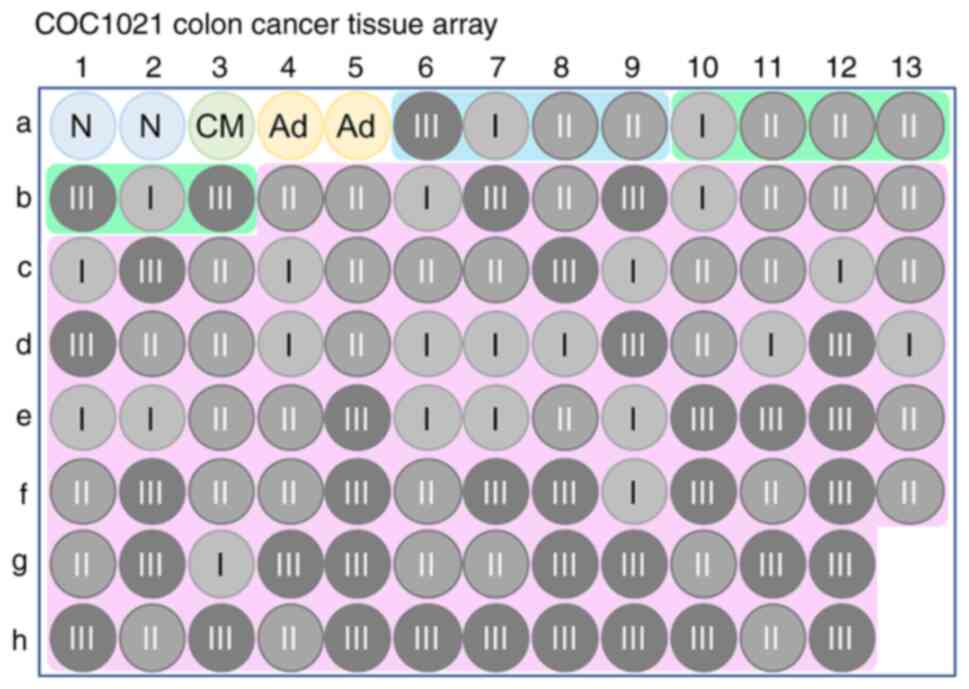 | Figure 2COC1021 colon cancer tissue array. A
total number of 102 available cores was enrolled (102 cores:
x-axis, nos. 1 to 13; y-axis, a to h). N, non-CRC colonic tissue;
CM, congenital megacolon; Ad, colon adenoma; I, AJCC stage I; II,
AJCC stage II; III, AJCC stage III. Blue area, papillary
adenocarcinoma; green area, mucinous adenocarcinoma; pink area,
colon adenocarcinoma. CRC, colorectal cancer; AJCC, American Joint
Committee on Cancer. |
A logistic regression model to predict
CRC disease
Three sets of stool specimens (n=87) were then
pooled to produce a binary logistic regression equation as follows:
ProbCRC=1/1 + e-z, where ProbCRC
is the probability of CRC disease, ‘e’ denotes the exponential
function, and ‘z’ is equal to 5.468 + 1.394 x log(UBE2N) + 0.859 x
log(IMPDH1) + 1.129 x log(DYNC1LI1)-1.471 x log(HRASLS2). The
overall accuracy of this model to predict CRC in the participants
was 94.3%. The sensitivity was 96.6% (56 out of 58), and the
specificity was 89.7% (26 out of 29). The positive predictive value
(PPV) was 95.1% (56 out of 59), and the negative predictive value
(NPV) was 93.9% (26 out of 28). Thus, this logistic regression
model was used for the prediction of increased risk for CRC in
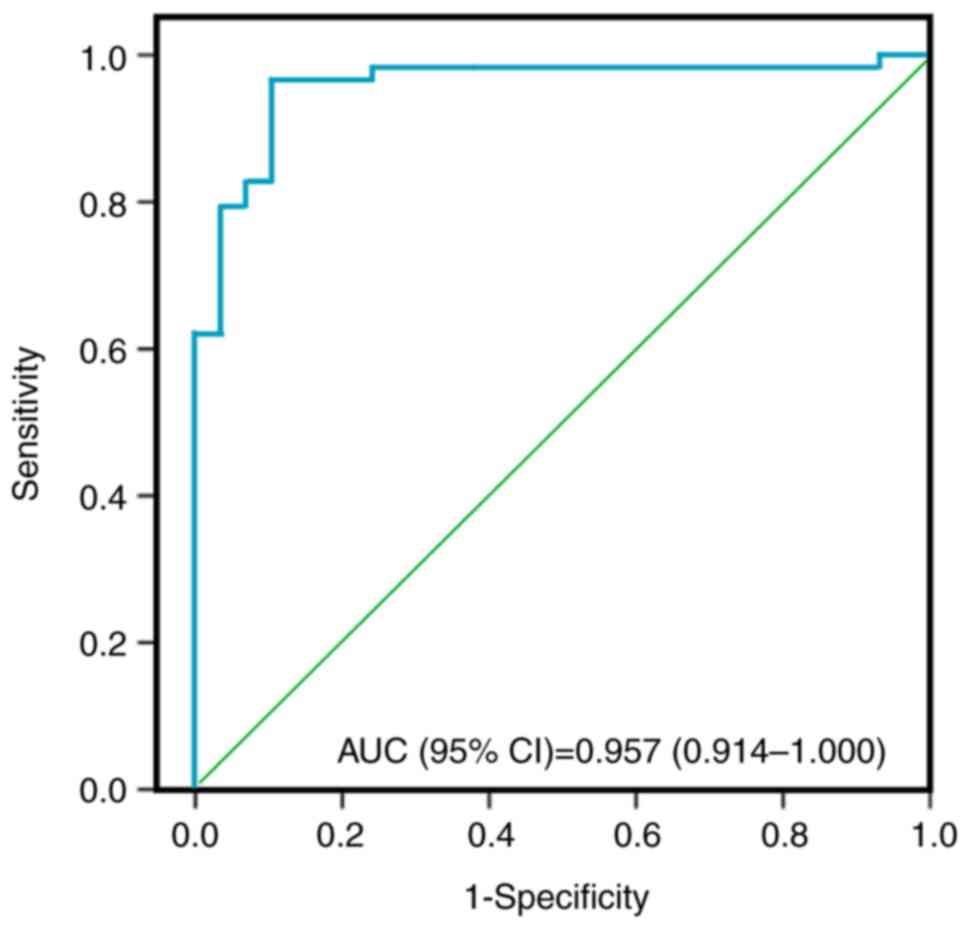
patients, among new study participants. Furthermore, the AUC for
the assessment of the model discrimination was 0.957 [95%
confidence interval (CI), 0.914-1.000; P<0.01] (Fig. 7). The cut-off value for predicting
CRC (ProbCRC) was 0.540, with 96.6% (95% CI, 88.1-99.6%)
sensitivity and 89.7% (95% CI, 72.6-97.8%) specificity (Table II).
 | Table IISensitivity and specificity for
predicting colorectal cancer. |
Table II
Sensitivity and specificity for
predicting colorectal cancer.
| Criterion | Sensitivity
(%) | 95% CI | Specificity
(%) | 95% CI | Accuracy (%) |
|---|
| 0.006 | 98.3 | 90.8-100.0 | 6.9 | 0.8-22.8 | 69.2 |
| 0.264 | 98.3 | 90.8-100.0 | 75.9 | 56.5-89.7 | 90.1 |
| 0.540 | 96.6 | 88.1-99.6 | 89.7 | 72.6-97.8 | 94.5 |
| 0.714 | 82.8 | 70.6-91.4 | 93.1 | 77.2-99.2 | 86.8 |
| 0.950 | 62.1 | 48.4-74.5 | 96.6 | 82.2-99.9 | 74.7 |
Model validity of patients with CRC
and healthy individuals
First, pre- and post-surgical stool specimens were
obtained from two patients with CRC (pCCG1: AJCC stage III; pCCG2:
AJCC stage II). In addition, stool specimens were obtained from
another patient with CRC (a 49-year-old female) as a positive CRC
control and a healthy donor individual (a 72-year-old male) with
negative colonoscopy results as a negative control. As demonstrated
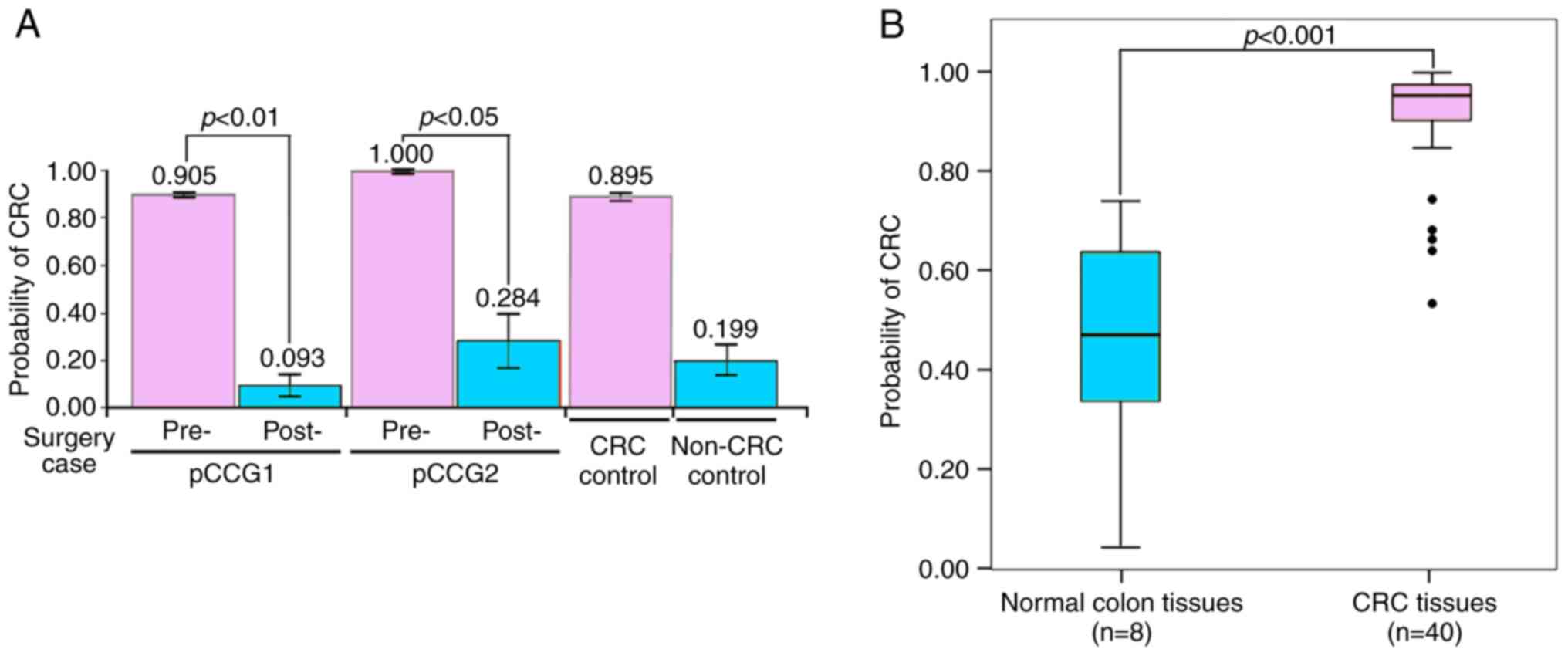
in Fig. 8A, the highest positive
predicted values for CRC disease were detected for the two
pre-surgical samples (pCCG1, 0.905; pCCG2, 1.000) as compared with
the healthy donor control (0.199), as assessed using the logistic
regression model of the four-gene panel. By contrast, markedly
lower predicted values were detected in post-surgical samples
(0.093 for pCCG1; 0.284 for pCCG2), which were prepared from the
feces of two patients whose tumors were completely removed. It is
notable that the timepoint of collection of the postsurgical
samples was at least 1 month after surgery, with patient pCCG1
receiving further chemotherapy.
This specific four-gene panel was then used to test
the 119 stool specimens that were obtained before colonoscopy
examination. Briefly, 112 cases presented with various hemorrhoids,
51 with polyps, eight with colitis, four with CRC, and only one was
entirely healthy. Among the 43 cases (36.1%, 43 out of 119) with a
higher ProbCRC (>0.540), four were proven to have CRC (mean
ProbCRC=0.968), five had colitis (mean ProbCRC=0.991) and 16 had
polyps (mean ProbCRC=0.834) following a colonoscopy examination
(Table III). Conversely, up to
62.5% (5 out of 8) of the cases with colitis or proctitis were
predicted to be positive, with only 31.4% (16 out of 51) of the
cases with polyps exhibiting increased predictive rates.
 | Table IIIPrediction of CRC by the four-gene
panel for the 119 patients with CRC. |
Table III
Prediction of CRC by the four-gene
panel for the 119 patients with CRC.
| Features | Case no. | Probability of CRC
(%) >0.540 (positive no.) | P-value |
|---|
| Age, years | | | |
|
<49 | 57 | 36.8% (21) | 0.878 |
|
≥49 | 62 | 35.5% (22) | |
| Sex | | | |
|
Male | 65 | 41.5% (27) | 0.178 |
|
Female | 54 | 29.6% (16) | |
| CRC | | | |
|
Negative | 115 | 33.9% (39) | 0.016 |
|
Positive | 4 | 100.0% (4) | |
|
Colitis/proctitis | | | |
|
Negative | 107 | 31.8% (34) | 0.077 |
|
Positive | 8 | 62.5% (5) | |
| Polyp | | | |
|
Negative | 64 | 35.9% (23) | 0.607 |
|
Positive | 51 | 31.4% (16) | |
| Hemorrhoid | | | |
|
Negative | 3 | 33.3% (1) | 0.265 |
|
Internal | 94 | 37.2% (35) | |
|
External | 2 | 50.0% (1) | |
|
Mixed | 16 | 12.5% (2) | |
Finally, the ProbCRC of each case was calculated in
the HCRT104 cDNA array composed of 48 colonic tissues. The mean
ProbCRC of 40 patients with CRC was 0.913 (range: 0.535-0.997),
which was significantly higher than that of eight non-CRC colon
tissues (0.459; range: 0.042-0.740) (Fig. 8B). In particular, the mean ProbCRC
of 10 patients with CRC with distant metastasis was up to 0.973
(range, 0.924-0.997) (Table SII).
Using this predictive model, 39 patients with CRC were identified
as positive cases (ProbCRC >0.540), and the PPV was up to 97.5%
(39 of 40). In addition, five non-CRC cases were correctly
diagnosed as negative cases (ProbCRC ≤0.540), with the
corresponding NPV being 62.5% (5 of 8) (Table SIII).
Discussion
The early detection of CRC would permit timely
surgical intervention in patients and this in turn would halt tumor
progression, thus enabling effective therapy (52,53).
Sigmoidoscopy and colonoscopy are simultaneously used at present
for the early detection of CRC. However, these are invasive
techniques (54-56).
By contrast, clinical examinations are limited due to their risk
and inconvenience. Consequently, a non-invasive fecal occult blood
test is currently and widely applied for CRC screening, even though
it has poor specificity and sensitivity (57).
CRC is believed to develop slowly via the
progressive accumulation of genetic mutations (58,59).
Genes involved in human diseases, tumorigenesis, or even those with
unrecognized functions are potential markers that may aid in the
diagnosis of CRC. A variety of molecular approaches are emerging to
improve the effectiveness and user-friendliness of non-invasive CRC
screening (60,61). In the present study, it was
demonstrated that limited genetic markers rarely discussed in CRC
were able to screen or detect CRC. In other words, it is not
necessary to examine additional genes in the field of clinical
practice for CRC when the sample type used for examination is stool
instead of blood (61,62). For example, assays that aim to
determine early alterations in gene expression in stool specimens
due to malignancy, appear to be a promising alternative to the
detection methods currently used (23,63).
In fact, stool specimens have been used as a representative of CRC
manifestations for a number of years (30,64,65).
Either CRC tissues or human stool specimens can serve as an
appropriate object to explore CRC development (27,28).
Genetic information from these two clinical materials is
significantly interrelated (56,66).
This is also reflected in the results of the present study, where
the differentially expressed genes detected in the stool specimens
of patients with CRC were also detected at the mRNA and protein
levels, in CRC tissues. These findings suggest that a high gene
expression in stool specimens may indicate the presence of
colorectal neoplasia in the colonic tract and may provide an
instrument for understanding CRC development (67-69).
Moreover, this non-invasive approach can greatly enhance screening
acceptance and be implemented in public health services due to its
relative ease of use and cost-effectiveness (52).
Specific gene expression profiles have been used to
classify tumors or predict prognoses (70,71).
In CRC research, whole-genome analyses of human stool specimens
have been performed by several groups (14,72-74).
Miyake et al (2) revealed
that a discriminator gene set could predict metastases from tissues
of patients with early-stage CRC. As an example of translation from
basic science to clinical care, these results suggested that
understanding the molecular mechanisms of CRC tumorigenesis may
lead to new approaches for the diagnosis of CRC. The results of the
present study provide evidence to support this concept (75). Actually, eight differentially
expressed genes (UBE2N, AKIRIN1, IMPDH, SLC15A4, DYNC1LI1, HRASLS2,
APOA1 and STK17B) were originally selected. However, six genes
(UBE2N, IMPDH, SLC15A4, DYNC1LI1, HRASLS2 and STK17B) were used for
the final LOOCV analysis in the present study, after validating all
corresponding relative expression using a series of RT-qPCR assays
for sample set II. Finally, only four genes (UBE2N, IMPDH1,
DYNC1LI1 and HRASLS2) presented with high sensitivity (96.6%) and
high sensitivity (89.7%). As a result, it was ultimately decided to
include only four, rather than eight or six genes, for subsequent
experiments in the present study. Stool-based assays were used to
detect CRC and proposed this specific gene panel (four-gene panel:
UBE2N, IMPDH1, DYNC1LI1 and HRASLS2) that exhibited the best
predictive power. Furthermore, apart from achieving a high
sensitivity for the detection of CRC in patients using this
four-gene panel, it also identified cases with a high risk for CRC,
including cases presenting with inflammation (e.g., colitis). This
finding may indicate that individuals with colitis are at an
increased risk of developing CRC (76). In addition, the panel used in the
present study may be applied to disease tracking and treatment
strategies due to the reduced positive predicted values obtained
for the post-surgical stool specimens. This implies that the
results of the present study may be used for the evaluation and
selection of the best treatment option for individual patients and
may allow for more intensive follow-up examinations for
post-surgical or post-chemotherapeutic patients with CRC.
Varying levels of gene expression in stool specimens
were reflected at the respective protein and mRNA levels in the
tissues using immunohistochemistry and the CRC cDNA array,
respectively. As previously reported by the authors, DYNC1LI1 was
expressed at a significantly higher level in cDNA samples from
patients with distant metastasis than in cDNA samples from
non-metastatic patients and in normal colonic tissues (77). In addition, increased (UBE2N and
IMPDH1) and decreased (HRASLS2) cDNA levels were observed to be
statistically significant for CRC tissues of all stages (P=0.020
for UBE2N, P=0.032 for IMPDH1 and P<0.001 for HRASLS2) compared
with those detected in eight cDNA samples of non-CRC tissues in the
HCRT104 cDNA array (Table SIV).
The densities of positive signaling cells for HRASLS2 (Fig. S1A), UBE2N (Fig. S1B), IMPDH1 (Fig. S1C), and DYNC1LI1 (Fig. S1D) varied with the grade of
differentiation. Briefly, poorly differentiated (G3) CRC had
relatively low HRASLS2- and high UBE2N-expressing cell densities,
compared to that of normal/benign tissues or well-differentiated
(G1) and moderately differentiated (G2) CRCs with statistical
significance (P<0.05 for HRASLS2, G2 vs. G3; P<0.0001 for
UNE2N, G1 vs. G3 and G2 vs. G3). Although there was no
statistically significant difference in the expression of IMPDH1
and DYNC1LI1 among the groups, their expression levels in
differently differentiated CRC tissues still exhibited a tendency
to increase gradually with a poorer differentiation. However,
further analyses using a greater number of samples are required to
confirm these results. The product of UBE2N, also known as UBC13,
is a member of the E2 ubiquitin-conjugating enzyme family (78). These ubiquitin ligase components may
function as oncogenes in several malignancies, including CRC
(79,80). This may also apply to diffuse large
B-cell lymphoma, in which the reduced expression of UBE2N inhibits
tumor cell survival (81).
Accordingly, it was inferred that this aberrant expression in CRC
may upregulate the UBC13-p53 complex, which may impede the normal
function of the p53 tumor suppressor (82,83).
The second upregulated gene, IMPDH1, is also a crucial factor in
p53-dependent growth regulation (84). IMPDH1 upregulation is tightly
associated with proliferative phenotypes, including malignancy
(85). Taken together with the
results of the present study, it may be suggested that the
increased expression of IMPDH1 in colonic cells is closely related
to the occurrence of CRC. Moreover, the overexpression of the
protein products of UBE2N and IMPDH1 in the mucosa and submucosa of
the colonic tract were also observed. This may imply that the
invasion of CRC cells into the inner cellular layers can also be
encountered in stool specimens. The results of the present study
may provide evidence that human stool specimens are suitable for
CRC detection (14,86). In addition to the genes that were
upregulated, HRASLS2, which is the only gene that was downregulated
in this four-gene panel, was reported to suppress the growth of
cancer cells and might be a tumor suppressor (87,88).
Nevertheless, the present study is the first report, to the best of
our knowledge, discussing the association between the
downregulation of HRASLS2 and CRC. Moreover, the loss of repressing
RAS activities may lead to CRC tumorigenesis, as HRASLS2 was
downregulated in colonic cells (89).
It is hoped that new insights into colorectal
carcinogenesis and personalized prediction can be achieved in a
non-invasive manner. One of the key elements in CRC therapy is the
ability to detect the disease easily, including using non-invasive
screening methods (90,91). The results of the present study
suggest that stool specimens may faithfully reproduce the actual
health status of the colonic tract. Using this four-gene panel, it
was demonstrated that CRC screening or detection in stool specimens
can be performed without the inclusion of an excessive number of
genes and that this type of sample, which is collected
non-invasively, being also able to reflect colonic defects via the
determination of aberrant protein expression in the colonic mucosa
or submucosa. It is possible to evaluate chemotherapeutic efficacy
or further elucidate the selection of the optimal treatment option
from stool specimens of individual cases, anytime and anywhere.
However, since RNA quality is critical for obtaining strong
prognostic and predictive values, fresh stool specimens or stool in
qualified storage buffers are of utmost importance and a limitation
of this assay. By contrast, to improve the accuracy of this
analysis, further studies are necessary to expand the sample size
of training set.
In conclusion, this specific four-gene panel,
consisting of UBE2N, IMPDH1, DYNC1LI1 and HRASLS2, represents a
clinical tool as potential prognostic and predictive targets in
stool specimens for CRC. The overexpression of UBE2N, IMPDH1 and
DYNC1LI1, and the downregulation of HRASLS2 was also detected in
CRC tissues. This molecular panel can be non-invasively and easily
detected from stool specimens and may allow for the assessment of
CRC screening, treatments, and follow-ups. The present study
comprehensively demonstrates the translation of basic science into
clinical practice.
Supplementary Material
Cell densities of immunoreactive cells
per mm2 from COC1021 colon cancer tissue array. (A)
HRASLS2, (B) UBE2N, (C) IMPDH1, (D) DYNC1LI1. QuPath (Version
0.3.0; https://qupath.github.io) was employed
to produce the cell densities of immunoreactive cells per
mm2 for all target proteins. One-way analysis of
variance was performed to compare the densities of positive
signaling cells forimmunohistochemical staining of target proteins
and followed by the Bonferroni post hoc test. *P<0.05
and ****P<0.0001. HRASLS2, phospholipase A and
acyltransferase 2; UBE2N, ubiquitin-conjugating enzyme E2N; IMPDH1,
inosine monophosphate dehydrogenase 1; DYNC1LI1, dynein,
cytoplasmic 1 light intermediate chain 1. Normal/Benign, two normal
colonic tissues, one congenital megacolon, and two adenomas; G1,
well-differentiated CRC cells; G2, moderately differentiated CRC
cells; G3, poorly differentiated CRC cells.
Analysis of eight differentially
expressed genes in stool specimensof 11 cases using theMann-Whitney
U test.
ProbCRC in the HCRT104 cDNA
array.
Numbers of cases with positive and
negative predictive values from the HCRT104 cDNA array.
Analysis of four genes for non-CRC and
CRC in the HCRT104 cDNA arrayusing theMann-Whitney U test.
Acknowledgements
The authors would like to thank Dr John Chung-Liang
Lee (Vie Longue Biotech, New Taipei 22102, Taiwan) for his
assistance with total RNA preparation from stool specimens in the
present study.
Funding
Funding: This research was funded by the Ministry of Science and
Technology, Taiwan, R.O.C., grant no. MOST109-2314-B-281-001.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
The conceptualization of the present study was
performed by CCH and YNC. CYS, CYL, MHS and CJH curated all data
analyzed and used in the present study. SCC, WCK and CJH performed
the gene quantifications. SLG performed the statistical analysis.
CCH, CYL and CJH performed and analyzed the pathological data from
the experiments. CCH and SCC acquired resources. CJH supervised the
study. CYS, CYL and MHS drafted the original manuscript. CCH, YNC,
CYS and CJH reviewed and edited the manuscript. All authors have
read and approved the final manuscript. CCH and CJH confirmed the
authenticity of all the raw data.
Ethics approval and consent to
participate
The study was conducted in accordance with the
Declaration of Helsinki and approved by the Institutional Review
Board of Cathay General Hospital, Taiwan, R.O.C. (approval no.
CGH-P101014). All the participating subjects provided their written
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Taniguchi H, Moriya C, Igarashi H, Saitoh
A, Yamamoto H, Adachi Y and Imai K: Cancer stem cells in human
gastrointestinal cancer. Cancer Sci. 107:1556–1562. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Miyake M, Takemasa I, Matoba R, Tanino M,
Niijima S, Ikeda M, Yamamoto H, Sekimoto M, Kuhara S, Okayama T, et
al: Heterogeneity of colorectal cancers and extraction of
discriminator gene signatures for personalized prediction of
prognosis. Int J Oncol. 39:781–789. 2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Pitroda SP and Weichselbaum RR: Integrated
molecular and clinical staging defines the spectrum of metastatic
cancer. Nat Rev Clin Oncol. 16:581–588. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang S, Guan X, Ma M, Zhuang M, Ma T, Liu
Z, Chen H, Jiang Z, Chen Y, Wang G and Wang X: Reconsidering the
prognostic significance of tumour deposit count in the TNM staging
system for colorectal cancer. Sci Rep. 10(89)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Das V, Kalita J and Pal M: Predictive and
prognostic biomarkers in colorectal cancer: A systematic review of
recent advances and challenges. Biomed Pharmacother. 87:8–19.
2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sudoyo AW: Biomolecular markers as
determinants of patients selection for adjuvant chemotherapy of
sporadic colorectal cancers. Acta Med Indones. 42:45–50.
2010.PubMed/NCBI
|
|
7
|
Dong L and Ren H: Blood-based DNA
methylation biomarkers for early detection of colorectal cancer. J
Proteomics Bioinform. 11(120)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wilhelmsen M, Christensen IJ, Rasmussen L,
Jørgensen LN, Madsen MR, Vilandt J, Hillig T, Klaerke M, Nielsen
KT, Laurberg S, et al: Detection of colorectal neoplasia:
Combination of eight blood-based, cancer-associated protein
biomarkers. Int J Cancer. 140:1436–1446. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Marshall KW, Mohr S, Khettabi FE, Nossova
N, Chao S, Bao W, Ma J, Li XJ and Liew CC: A blood-based biomarker
panel for stratifying current risk for colorectal cancer. Int J
Cancer. 126:1177–1186. 2010.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Rodriguez-Casanova A, Costa-Fraga N,
Bao-Caamano A, López-López R, Muinelo-Romay L and Diaz-Lagares A:
Epigenetic landscape of liquid biopsy in colorectal cancer. Front
Cell Dev Biol. 9(622459)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Xu Y, Xu Q, Yang L, Ye X, Liu F, Wu F, Ni
S, Tan C, Cai G, Meng X, et al: Identification and validation of a
blood-based 18-gene expression signature in colorectal cancer. Clin
Cancer Res. 19:3039–3049. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Huang CC, Shen MH, Chen SK, Yang SH, Liu
CY, Guo JW, Chang KW and Huang CJ: Gut butyrate-producing organisms
correlate to Placenta specific 8 protein: Importance to colorectal
cancer progression. J Adv Res. 22:7–20. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Huang CJ, Lee CL, Yang SH, Chien CC, Huang
CC, Yang RN and Chang CC: Upregulation of the growth
arrest-specific-2 in recurrent colorectal cancers, and its
susceptibility to chemotherapy in a model cell system. Biochim
Biophys Acta. 1862:1345–1353. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lee CL, Huang CJ, Yang SH, Chang CC, Huang
CC, Chien CC and Yang RN: Discovery of genes from feces correlated
with colorectal cancer progression. Oncol Lett. 12:3378–3384.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Corbo C, Cevenini A and Salvatore F:
Biomarker discovery by proteomics-based approaches for early
detection and personalized medicine in colorectal cancer.
Proteomics Clin Appl. 11(1600072)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Imperiale TF, Kisiel JB, Itzkowitz SH,
Scheu B, Duimstra EK, Statz S, Berger BM and Limburg PJ:
Specificity of the multi-target stool DNA test for colorectal
cancer screening in average-risk 45-49 year-olds: A cross-sectional
study. Cancer Prev Res (Phila). 14:489–496. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lin J, Zhang L, Chen M, Chen J, Wu Y, Wang
T, Lu Y, Ba Z, Cheng X, Xu R, et al: Evaluation of combined
detection of multigene mutation and SDC2/SFRP2 methylation in stool
specimens for colorectal cancer early diagnosis. Int J Colorectal
Dis. 37:1231–1238. 2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Vega-Benedetti AF, Loi E, Moi L, Orrù S,
Ziranu P, Pretta A, Lai E, Puzzoni M, Ciccone L, Casadei-Gardini A,
et al: Colorectal cancer early detection in stool samples tracing
CpG islands methylation alterations affecting gene expression. Int
J Mol Sci. 21(4494)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen J, Sun H, Tang W, Zhou L, Xie X, Qu
Z, Chen M, Wang S, Yang T, Dai Y, et al: DNA methylation biomarkers
in stool for early screening of colorectal cancer. J Cancer.
10:5264–5271. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
De Wijkerslooth T, Bossuyt P and Dekker E:
Strategies in screening for colon carcinoma. Neth J Med.
69:112–119. 2011.PubMed/NCBI
|
|
21
|
Young GP and Bosch LJW: Fecal tests: From
blood to molecular markers. Curr Colorectal Cancer Rep. 7:62–70.
2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ahlquist DA, Zou H, Domanico M, Mahoney
DW, Yab TC, Taylor WR, Butz ML, Thibodeau SN, Rabeneck L, Paszat
LF, et al: Next-generation stool DNA test accurately detects
colorectal cancer and large adenomas. Gastroenterology.
142:248–256.e25-e26. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Miller S and Steele S: Novel molecular
screening approaches in colorectal cancer. J Surg Oncol.
105:459–467. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Pox C: Colon cancer screening: Which
non-invasive filter tests? Dig Dis. 29:56–59. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Komor MA, Bosch LJ, Coupé VM, Rausch C,
Pham TV, Piersma SR, Mongera S, Mulder CJ, Dekker E, Kuipers EJ, et
al: Proteins in stool as biomarkers for non-invasive detection of
colorectal adenomas with high risk of progression. J Pathol.
250:288–298. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Choi HH, Cho YS, Choi JH, Kim HK, Kim SS
and Chae HS: Stool-based miR-92a and miR-144* as noninvasive
biomarkers for colorectal cancer screening. Oncology. 97:173–179.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Steele RJ, Kostourou I, McClements P,
Watling C, Libby G, Weller D, Brewster DH, Black R, Carey FA and
Fraser C: Effect of repeated invitations on uptake of colorectal
cancer screening using faecal occult blood testing: Analysis of
prevalence and incidence screening. BMJ. 341(c5531)2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tonus C, Sellinger M, Koss K and Neupert
G: Faecal pyruvate kinase isoenzyme type M2 for colorectal cancer
screening: A meta-analysis. World J Gastroenterol. 18:4004–4011.
2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Osborn NK and Ahlquist DA: Stool screening
for colorectal cancer: Molecular approaches. Gastroenterology.
128:192–206. 2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hamaya Y, Yoshida K, Takai T, Ikuma M,
Hishida A and Kanaoka S: Factors that contribute to faecal
cyclooxygenase-2 mRNA expression in subjects with colorectal
cancer. Br J Cancer. 102:916–921. 2010.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Mojtabanezhad Shariatpanahi A, Yassi M,
Nouraie M, Sahebkar A, Varshoee Tabrizi F and Kerachian MA: The
importance of stool DNA methylation in colorectal cancer diagnosis:
A meta-analysis. PLoS One. 13(e0200735)2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Robertson DJ and Imperiale TF: Stool
testing for colorectal cancer screening. Gastroenterology.
149:1286–1293. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yang SH, Huang CJ, Lee CL, Liu CC, Chien
CC and Chen SH: Fecal RNA detection of cytokeratin 19 and ribosomal
protein L19 for colorectal cancer. Hepatogastroenterology.
57:710–715. 2010.PubMed/NCBI
|
|
34
|
Song L, Lu H and Yin J: Preliminary study
on discriminating HER2 2+ amplification status of breast cancers
based on texture features semi-automatically derived from pre-,
post-contrast, and subtraction images of DCE-MRI. PLoS One.
15(e0234800)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Juhola M, Joutsijoki H, Penttinen K, Shah
D and Aalto-Setälä K: On computational classification of genetic
cardiac diseases applying iPSC cardiomyocytes. Comput Methods
Programs Biomed. 210(106367)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yu Z, Yu H, Zou Q, Huang Z, Wang X, Tang
G, Bai L, Zhou C, Zhuang Z, Xie Y, et al: Nomograms for prediction
of molecular phenotypes in colorectal cancer. Onco Targets Ther.
13:309–321. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liu CY, Huang CS, Huang CC, Ku WC, Shih HY
and Huang CJ: Co-occurrence of differentiated thyroid cancer and
second primary malignancy: Correlation with expression profiles of
mismatch repair protein and cell cycle regulators. Cancers (Basel).
13(5486)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Weiser MR: AJCC 8th edition: Colorectal
cancer. Ann Surg Oncol. 25:1454–1455. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Treanor D and Quirke P: Pathology of
colorectal cancer. Clin Oncol (R Coll Radiol). 19:769–776.
2007.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chien CC, Chang CC, Yang SH, Chen SH and
Huang CJ: A homologue of the Drosophila headcase protein is a novel
tumor marker for early-stage colorectal cancer. Oncol Rep.
15:919–926. 2006.PubMed/NCBI
|
|
41
|
Dowling P, Clarke C, Hennessy K,
Torralbo-Lopez B, Ballot J, Crown J, Kiernan I, O'Byrne KJ, Kennedy
MJ, Lynch V and Clynes M: Analysis of acute-phase proteins, AHSG,
C3, CLI, HP and SAA, reveals distinctive expression patterns
associated with breast, colorectal and lung cancer. Int J Cancer.
131:911–923. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Ginestier C, Cervera N, Finetti P,
Esteyries S, Esterni B, Adélaïde J, Xerri L, Viens P, Jacquemier J,
Charafe-Jauffret E, et al: Prognosis and gene expression profiling
of 20q13-amplified breast cancers. Clin Cancer Res. 12:4533–4544.
2006.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Bair E and Tibshirani R: Semi-supervised
methods to predict patient survival from gene expression data. PLoS
Biol. 2(E108)2004.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Chou HL, Yao CT, Su SL, Lee CY, Hu KY,
Terng HJ, Shih YW, Chang YT, Lu YF, Chang CW, et al: Gene
expression profiling of breast cancer survivability by pooled cDNA
microarray analysis using logistic regression, artificial neural
networks and decision trees. BMC Bioinformatics.
14(100)2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Hughes G: On the binormal predictive
receiver operating characteristic curve for the joint assessment of
positive and negative predictive values. Entropy (Basel).
22(593)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yang SH, Chien CC, Chen CW, Li SY and
Huang CJ: Potential of faecal RNA in diagnosing colorectal cancer.
Cancer Lett. 226:55–63. 2005.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Chang CC, Kao WY, Liu CY, Su HH, Kan YA,
Lin PY, Ku WC, Chang KW, Yang RN and Huang CJ: Butyrate
supplementation regulates expression of chromosome segregation
1-like protein to reverse the genetic distortion caused by p53
mutations in colorectal cancer. Int J Oncol. 60(64)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wei Q, Jiang H, Baker A, Dodge LK, Gerard
M, Young MR, Toledano MB and Colburn NH: Loss of sulfiredoxin
renders mice resistant to azoxymethane/dextran sulfate
sodium-induced colon carcinogenesis. Carcinogenesis. 34:1403–1410.
2013.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Bankhead P, Loughrey MB, Fernández JA,
Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ,
Coleman HG, et al: QuPath: Open source software for digital
pathology image analysis. Sci Rep. 7(16878)2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Huang CJ, Chien CC, Yang SH, Chang CC, Sun
HL, Cheng YC, Liu CC, Lin SC and Lin CM: Faecal ribosomal protein
L19 is a genetic prognostic factor for survival in colorectal
cancer. J Cell Mol Med. 12:1936–1943. 2008.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Bernal G: Use of RNA isolated from feces
as a promising tool for the early detection of colorectal cancer.
Int J Biol Markers. 27:e82–e89. 2012.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Grizzi F, Celesti G, Basso G and Laghi L:
Tumor budding as a potential histopathological biomarker in
colorectal cancer: Hype or hope? World J Gastroenterol.
18:6532–6536. 2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Belletti B, Nicoloso MS, Schiappacassi M,
Chimienti E, Berton S, Lovat F, Colombatti A and Baldassarre G:
p27(kip1) functional regulation in human cancer: A potential target
for therapeutic designs. Curr Med Chem. 12:1589–1605.
2005.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Blázquez C, Geelen MJ, Velasco G and
Guzmán M: The AMP-activated protein kinase prevents ceramide
synthesis de novo and apoptosis in astrocytes. FEBS Lett.
489:149–153. 2001.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Nikolouzakis TK, Vassilopoulou L,
Fragkiadaki P, Mariolis Sapsakos T, Papadakis GZ, Spandidos DA,
Tsatsakis AM and Tsiaoussis J: Improving diagnosis, prognosis and
prediction by using biomarkers in CRC patients (Review). Oncol Rep.
39:2455–2472. 2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Cheng YW and Li YC: Factors affecting the
follow-up time after a positive result in the fecal occult blood
test. PLoS One. 16(e0258130)2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Bardhan K and Liu K: Epigenetics and
colorectal cancer pathogenesis. Cancers (Basel). 5:676–713.
2013.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Mundade R, Imperiale TF, Prabhu L, Loehrer
PJ and Lu T: Genetic pathways, prevention, and treatment of
sporadic colorectal cancer. Oncoscience. 1:400–406. 2014.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Ahlquist DA: Molecular detection of
colorectal neoplasia. Gastroenterology. 138:2127–2139.
2010.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Bailey JR, Aggarwal A and Imperiale TF:
Colorectal cancer screening: Stool DNA and other noninvasive
modalities. Gut Liver. 10:204–211. 2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Youssef O, Sarhadi V, Ehsan H, Böhling T,
Carpelan-Holmström M, Koskensalo S, Puolakkainen P, Kokkola A and
Knuutila S: Gene mutations in stool from gastric and colorectal
neoplasia patients by next-generation sequencing. World J
Gastroenterol. 23:8291–8299. 2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Januszewicz W and Fitzgerald RC: Early
detection and therapeutics. Mol Oncol. 13:599–613. 2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Pickhardt PJ: Emerging stool-based and
blood-based non-invasive DNA tests for colorectal cancer screening:
The importance of cancer prevention in addition to cancer
detection. Abdom Radiol (NY). 41:1441–1444. 2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Shin HY, Suh M, Choi KS, Hwang SH, Jun JK,
Han DS, Lee YK, Oh JH, Lee CW and Lee DH: Higher satisfaction with
an alternative collection device for stool sampling in colorectal
cancer screening with fecal immunochemical test: A cross-sectional
study. BMC Cancer. 18(365)2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
de Wit M, Fijneman RJ, Verheul HM, Meijer
GA and Jimenez CR: Proteomics in colorectal cancer translational
research: Biomarker discovery for clinical applications. Clin
Biochem. 46:466–479. 2013.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Montrose DC, Zhou XK, Kopelovich L,
Yantiss RK, Karoly ED, Subbaramaiah K and Dannenberg AJ: Metabolic
profiling, a noninvasive approach for the detection of experimental
colorectal neoplasia. Cancer Prev Res (Phila). 5:1358–1367.
2012.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Slaby O: Non-coding RNAs as biomarkers for
colorectal cancer screening and early detection. Adv Exp Med Biol.
937:153–170. 2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Tsai TJ, Lim YP, Chao WY, Chen CC, Chen
YJ, Lin CY and Lee YR: Capping actin protein overexpression in
human colorectal carcinoma and its contributed tumor migration.
Anal Cell Pathol (Amst). 2018(8623937)2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Pan F, Chen T, Sun X, Li K, Jiang X,
Försti A, Zhu Y and Lai M: Prognosis prediction of colorectal
cancer using gene expression profiles. Front Oncol.
9(252)2019.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Xu G, Zhang M, Zhu H and Xu J: A 15-gene
signature for prediction of colon cancer recurrence and prognosis
based on SVM. Gene. 604:33–40. 2017.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Eckmann JD, Ebner DW and Kisiel JB:
Multi-target stool DNA testing for colorectal cancer screening:
Emerging learning on real-world performance. Curr Treat Options
Gastroenterol: Jan 21, 2020 (Epub ahead of print).
|
|
73
|
Kisiel JB, Klepp P, Allawi HT, Taylor WR,
Giakoumopoulos M, Sander T, Yab TC, Moum BA, Lidgard GP, Brackmann
S, et al: Analysis of DNA methylation at specific loci in stool
samples detects colorectal cancer and high-grade dysplasia in
patients with inflammatory bowel disease. Clin Gastroenterol
Hepatol. 17:914–921.e5. 2019.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Naber SK, Knudsen AB, Zauber AG, Rutter
CM, Fischer SE, Pabiniak CJ, Soto B, Kuntz KM and Lansdorp-Vogelaar
I: Cost-effectiveness of a multitarget stool DNA test for
colorectal cancer screening of medicare beneficiaries. PLoS One.
14(e0220234)2019.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Jung G, Hernández-Illán E, Moreira L,
Balaguer F and Goel A: Epigenetics of colorectal cancer: Biomarker
and therapeutic potential. Nat Rev Gastroenterol Hepatol.
17:111–130. 2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Olén O, Erichsen R, Sachs MC, Pedersen L,
Halfvarson J, Askling J, Ekbom A, Sørensen HT and Ludvigsson JF:
Colorectal cancer in ulcerative colitis: A Scandinavian
population-based cohort study. Lancet. 395:123–131. 2020.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Chang CC, Chao KC, Huang CJ, Hung CS and
Wang YC: Association between aberrant dynein cytoplasmic 1 light
intermediate chain 1 expression levels, mucins and chemosensitivity
in colorectal cancer. Mol Med Rep. 22:185–192. 2020.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Hodge CD, Ismail IH, Edwards RA, Hura GL,
Xiao AT, Tainer JA, Hendzel MJ and Glover JN: RNF8 E3 ubiquitin
ligase stimulates Ubc13 E2 conjugating activity that is essential
for DNA double strand break signaling and BRCA1 tumor suppressor
recruitment. J Biol Chem. 291:9396–9410. 2016.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Wu Z, Neufeld H, Torlakovic E and Xiao W:
Uev1A-Ubc13 promotes colorectal cancer metastasis through
regulating CXCL1 expression via NF-кB activation. Oncotarget.
9:15952–15967. 2018.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Zhang P, Wang L, Rodriguez-Aguayo C, Yuan
Y, Debeb BG, Chen D, Sun Y, You MJ, Liu Y, Dean DC, et al: miR-205
acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13. Nat
Commun. 5(5671)2014.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Pulvino M, Liang Y, Oleksyn D, DeRan M,
Van Pelt E, Shapiro J, Sanz I, Chen L and Zhao J: Inhibition of
proliferation and survival of diffuse large B-cell lymphoma cells
by a small-molecule inhibitor of the ubiquitin-conjugating enzyme
Ubc13-Uev1A. Blood. 120:1668–1677. 2012.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Cheng J, Fan YH, Xu X, Zhang H, Dou J,
Tang Y, Zhong X, Rojas Y, Yu Y, Zhao Y, et al: A small-molecule
inhibitor of UBE2N induces neuroblastoma cell death via activation
of p53 and JNK pathways. Cell Death Dis. 5(e1079)2014.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Ishtiaq A, Bakhtiar A, Silas E, Saeed J,
Ajmal S, Mushtaq I, Ali T, Wahedi HM, Khan W, Khan U, et al:
Pistacia integerrima alleviated Bisphenol A induced toxicity
through Ubc13/p53 signalling. Mol Biol Rep. 47:6545–6559.
2020.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Pelletier J, Riaño-Canalias F, Almacellas
E, Mauvezin C, Samino S, Feu S, Menoyo S, Domostegui A,
Garcia-Cajide M, Salazar R, et al: Nucleotide depletion reveals the
impaired ribosome biogenesis checkpoint as a barrier against DNA
damage. EMBO J. 39(e103838)2020.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Ouchida M, Kanzaki H, Ito S, Hanafusa H,
Jitsumori Y, Tamaru S and Shimizu K: Novel direct targets of
miR-19a identified in breast cancer cells by a quantitative
proteomic approach. PLoS One. 7(e44095)2012.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Zhao G, Liu X, Liu Y, Li H, Ma Y, Li S,
Zhu Y, Miao J, Xiong S, Fei S and Zheng M: Aberrant DNA methylation
of SEPT9 and SDC2 in stool specimens as an integrated biomarker for
colorectal cancer early detection. Front Genet.
11(643)2020.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Gu Y, Lin X, Kapoor A, Chow MJ, Jiang Y,
Zhao K and Tang D: The oncogenic potential of the centromeric
border protein FAM84B of the 8q24.21 gene desert. Genes (Basel).
11(312)2020.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Uyama T, Jin XH, Tsuboi K, Tonai T and
Ueda N: Characterization of the human tumor suppressors TIG3 and
HRASLS2 as phospholipid-metabolizing enzymes. Biochim Biophys Acta.
1791:1114–1124. 2009.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Shyu RY, Hsieh YC, Tsai FM, Wu CC and
Jiang SY: Cloning and functional characterization of the HRASLS2
gene. Amino Acids. 35:129–137. 2008.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Bi F, Wang Q, Dong Q, Wang Y, Zhang L and
Zhang J: Circulating tumor DNA in colorectal cancer: Opportunities
and challenges. Am J Transl Res. 12:1044–1055. 2020.PubMed/NCBI
|
|
91
|
Lansdorp-Vogelaar I, Goede SL, Bosch LJW,
Melotte V, Carvalho B, van Engeland M, Meijer GA, de Koning HJ and
van Ballegooijen M: Cost-effectiveness of high-performance
biomarker tests vs fecal immunochemical test for noninvasive
colorectal cancer screening. Clin Gastroenterol Hepatol.
16:504–512.e11. 2018.PubMed/NCBI View Article : Google Scholar
|