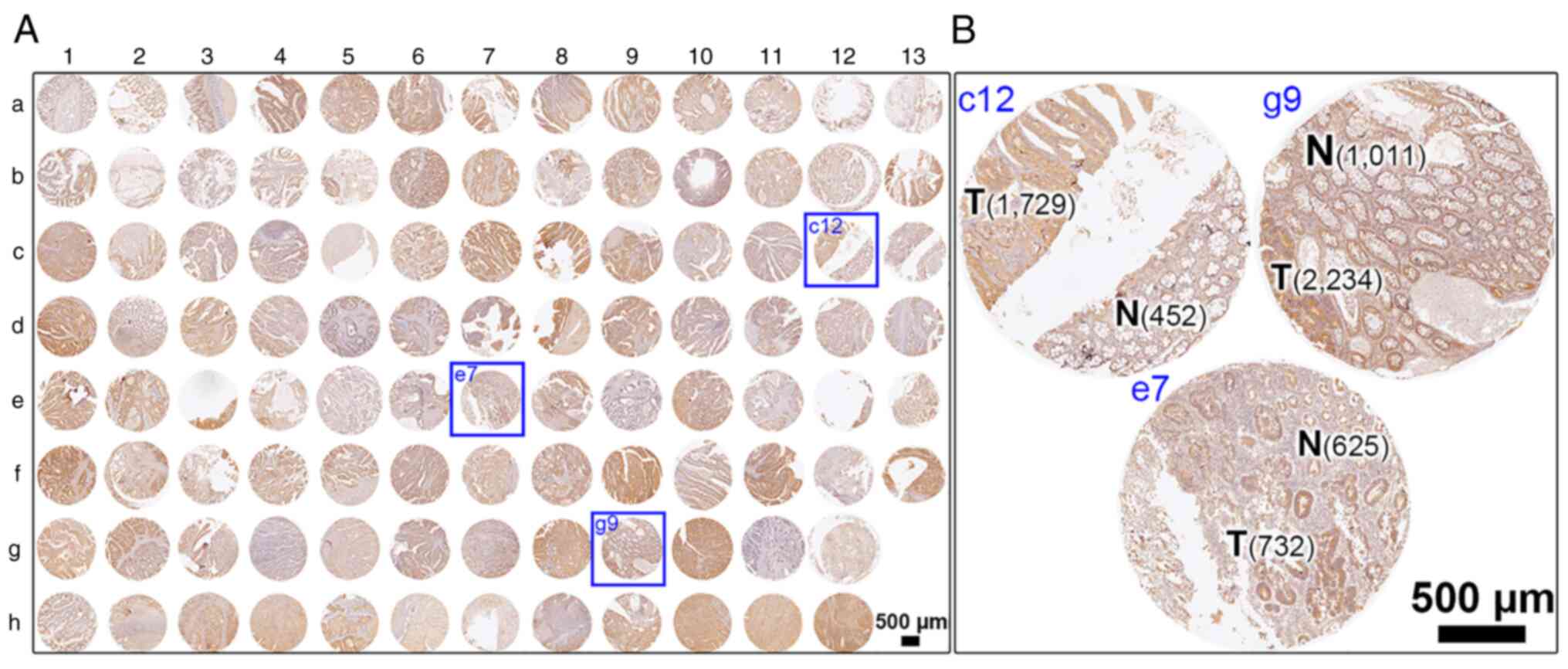
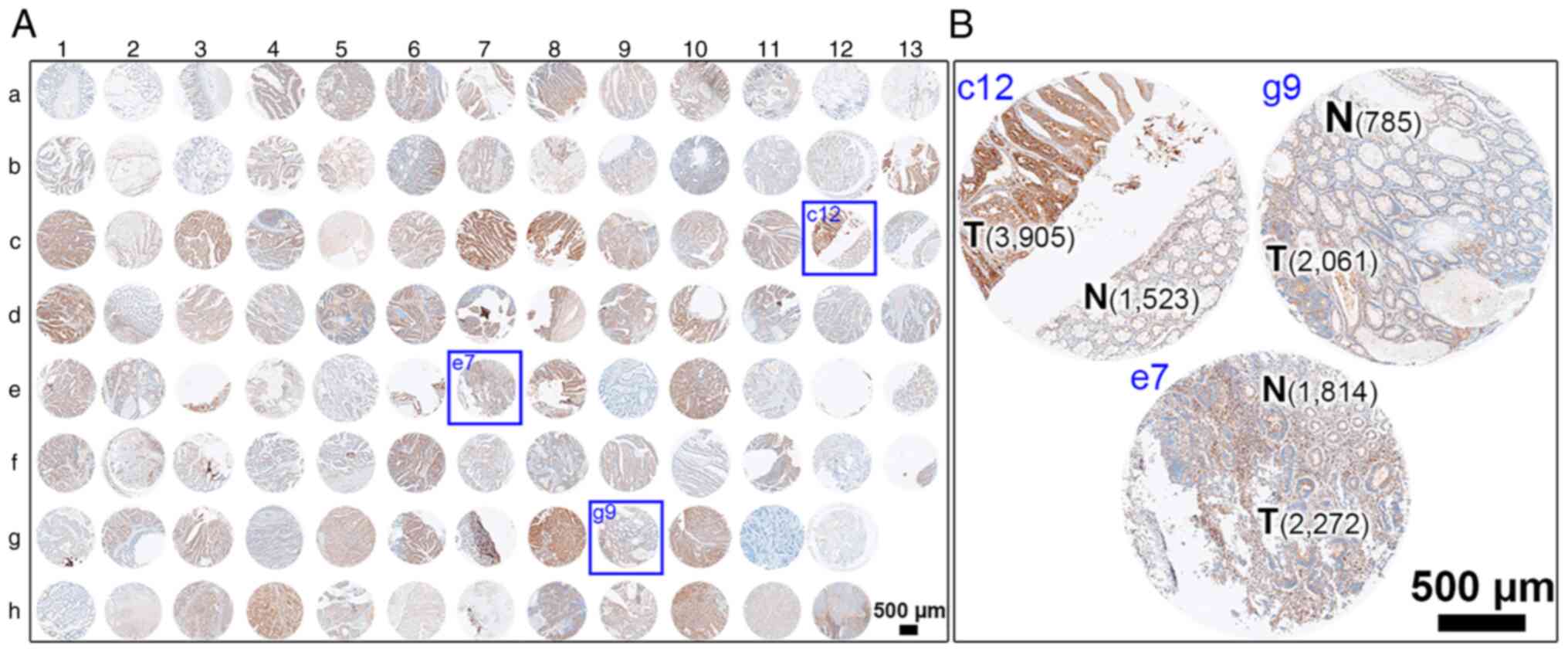
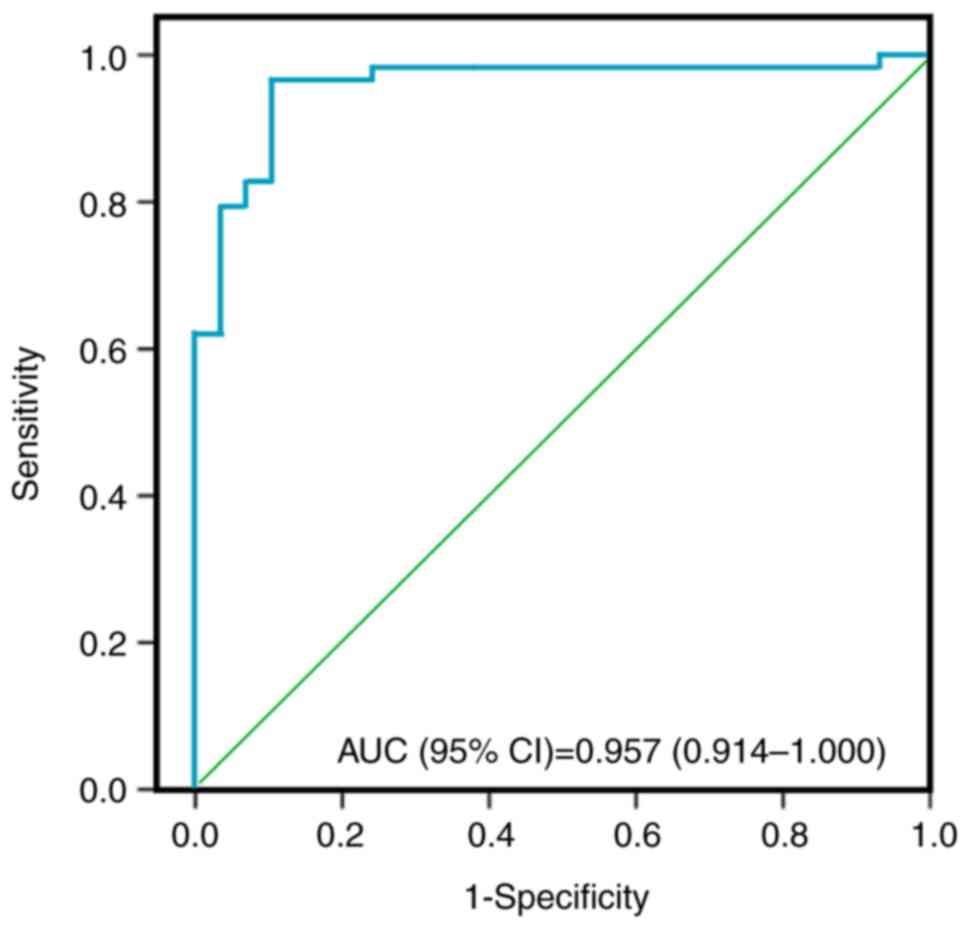
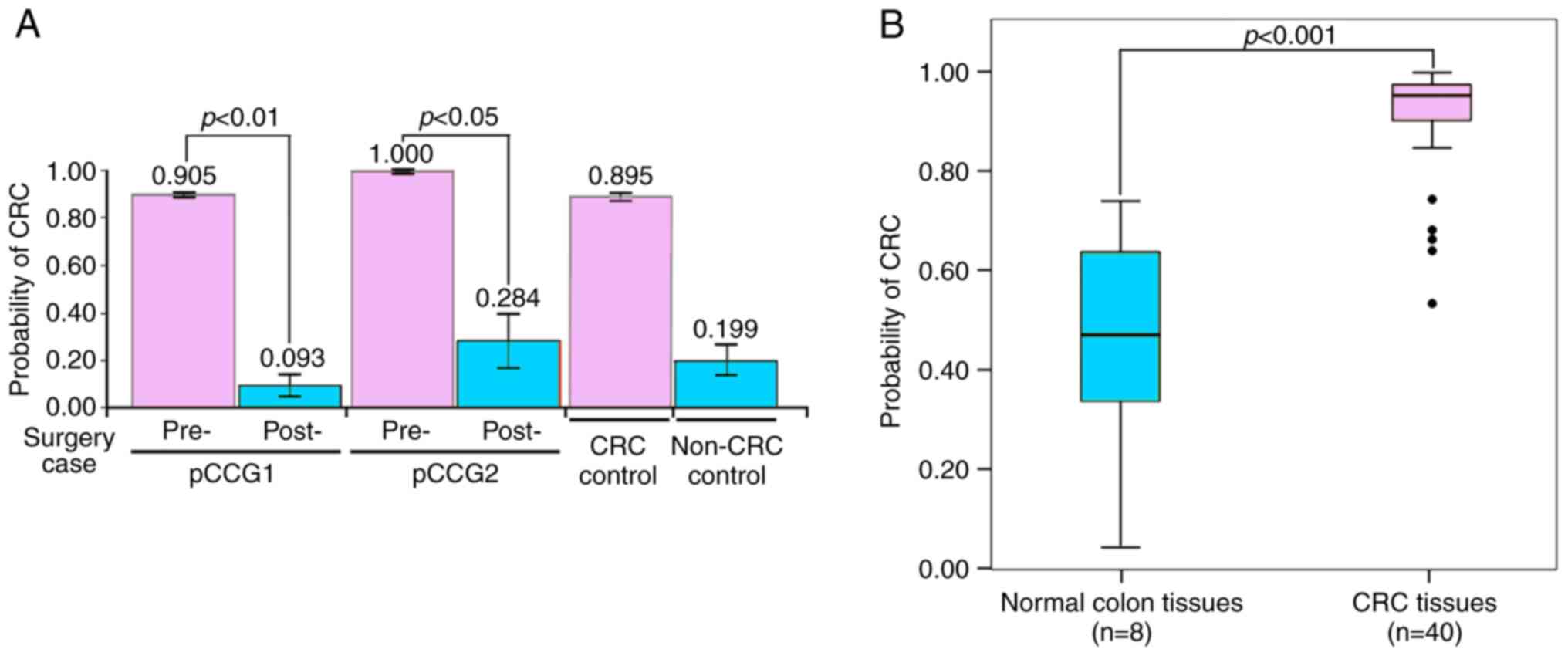
|
1
|
Taniguchi H, Moriya C, Igarashi H, Saitoh
A, Yamamoto H, Adachi Y and Imai K: Cancer stem cells in human
gastrointestinal cancer. Cancer Sci. 107:1556–1562. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Miyake M, Takemasa I, Matoba R, Tanino M,
Niijima S, Ikeda M, Yamamoto H, Sekimoto M, Kuhara S, Okayama T, et
al: Heterogeneity of colorectal cancers and extraction of
discriminator gene signatures for personalized prediction of
prognosis. Int J Oncol. 39:781–789. 2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Pitroda SP and Weichselbaum RR: Integrated
molecular and clinical staging defines the spectrum of metastatic
cancer. Nat Rev Clin Oncol. 16:581–588. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang S, Guan X, Ma M, Zhuang M, Ma T, Liu
Z, Chen H, Jiang Z, Chen Y, Wang G and Wang X: Reconsidering the
prognostic significance of tumour deposit count in the TNM staging
system for colorectal cancer. Sci Rep. 10(89)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Das V, Kalita J and Pal M: Predictive and
prognostic biomarkers in colorectal cancer: A systematic review of
recent advances and challenges. Biomed Pharmacother. 87:8–19.
2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sudoyo AW: Biomolecular markers as
determinants of patients selection for adjuvant chemotherapy of
sporadic colorectal cancers. Acta Med Indones. 42:45–50.
2010.PubMed/NCBI
|
|
7
|
Dong L and Ren H: Blood-based DNA
methylation biomarkers for early detection of colorectal cancer. J
Proteomics Bioinform. 11(120)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wilhelmsen M, Christensen IJ, Rasmussen L,
Jørgensen LN, Madsen MR, Vilandt J, Hillig T, Klaerke M, Nielsen
KT, Laurberg S, et al: Detection of colorectal neoplasia:
Combination of eight blood-based, cancer-associated protein
biomarkers. Int J Cancer. 140:1436–1446. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Marshall KW, Mohr S, Khettabi FE, Nossova
N, Chao S, Bao W, Ma J, Li XJ and Liew CC: A blood-based biomarker
panel for stratifying current risk for colorectal cancer. Int J
Cancer. 126:1177–1186. 2010.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Rodriguez-Casanova A, Costa-Fraga N,
Bao-Caamano A, López-López R, Muinelo-Romay L and Diaz-Lagares A:
Epigenetic landscape of liquid biopsy in colorectal cancer. Front
Cell Dev Biol. 9(622459)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Xu Y, Xu Q, Yang L, Ye X, Liu F, Wu F, Ni
S, Tan C, Cai G, Meng X, et al: Identification and validation of a
blood-based 18-gene expression signature in colorectal cancer. Clin
Cancer Res. 19:3039–3049. 2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Huang CC, Shen MH, Chen SK, Yang SH, Liu
CY, Guo JW, Chang KW and Huang CJ: Gut butyrate-producing organisms
correlate to Placenta specific 8 protein: Importance to colorectal
cancer progression. J Adv Res. 22:7–20. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Huang CJ, Lee CL, Yang SH, Chien CC, Huang
CC, Yang RN and Chang CC: Upregulation of the growth
arrest-specific-2 in recurrent colorectal cancers, and its
susceptibility to chemotherapy in a model cell system. Biochim
Biophys Acta. 1862:1345–1353. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lee CL, Huang CJ, Yang SH, Chang CC, Huang
CC, Chien CC and Yang RN: Discovery of genes from feces correlated
with colorectal cancer progression. Oncol Lett. 12:3378–3384.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Corbo C, Cevenini A and Salvatore F:
Biomarker discovery by proteomics-based approaches for early
detection and personalized medicine in colorectal cancer.
Proteomics Clin Appl. 11(1600072)2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Imperiale TF, Kisiel JB, Itzkowitz SH,
Scheu B, Duimstra EK, Statz S, Berger BM and Limburg PJ:
Specificity of the multi-target stool DNA test for colorectal
cancer screening in average-risk 45-49 year-olds: A cross-sectional
study. Cancer Prev Res (Phila). 14:489–496. 2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lin J, Zhang L, Chen M, Chen J, Wu Y, Wang
T, Lu Y, Ba Z, Cheng X, Xu R, et al: Evaluation of combined
detection of multigene mutation and SDC2/SFRP2 methylation in stool
specimens for colorectal cancer early diagnosis. Int J Colorectal
Dis. 37:1231–1238. 2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Vega-Benedetti AF, Loi E, Moi L, Orrù S,
Ziranu P, Pretta A, Lai E, Puzzoni M, Ciccone L, Casadei-Gardini A,
et al: Colorectal cancer early detection in stool samples tracing
CpG islands methylation alterations affecting gene expression. Int
J Mol Sci. 21(4494)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chen J, Sun H, Tang W, Zhou L, Xie X, Qu
Z, Chen M, Wang S, Yang T, Dai Y, et al: DNA methylation biomarkers
in stool for early screening of colorectal cancer. J Cancer.
10:5264–5271. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
De Wijkerslooth T, Bossuyt P and Dekker E:
Strategies in screening for colon carcinoma. Neth J Med.
69:112–119. 2011.PubMed/NCBI
|
|
21
|
Young GP and Bosch LJW: Fecal tests: From
blood to molecular markers. Curr Colorectal Cancer Rep. 7:62–70.
2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ahlquist DA, Zou H, Domanico M, Mahoney
DW, Yab TC, Taylor WR, Butz ML, Thibodeau SN, Rabeneck L, Paszat
LF, et al: Next-generation stool DNA test accurately detects
colorectal cancer and large adenomas. Gastroenterology.
142:248–256.e25-e26. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Miller S and Steele S: Novel molecular
screening approaches in colorectal cancer. J Surg Oncol.
105:459–467. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Pox C: Colon cancer screening: Which
non-invasive filter tests? Dig Dis. 29:56–59. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Komor MA, Bosch LJ, Coupé VM, Rausch C,
Pham TV, Piersma SR, Mongera S, Mulder CJ, Dekker E, Kuipers EJ, et
al: Proteins in stool as biomarkers for non-invasive detection of
colorectal adenomas with high risk of progression. J Pathol.
250:288–298. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Choi HH, Cho YS, Choi JH, Kim HK, Kim SS
and Chae HS: Stool-based miR-92a and miR-144* as noninvasive
biomarkers for colorectal cancer screening. Oncology. 97:173–179.
2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Steele RJ, Kostourou I, McClements P,
Watling C, Libby G, Weller D, Brewster DH, Black R, Carey FA and
Fraser C: Effect of repeated invitations on uptake of colorectal
cancer screening using faecal occult blood testing: Analysis of
prevalence and incidence screening. BMJ. 341(c5531)2010.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Tonus C, Sellinger M, Koss K and Neupert
G: Faecal pyruvate kinase isoenzyme type M2 for colorectal cancer
screening: A meta-analysis. World J Gastroenterol. 18:4004–4011.
2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Osborn NK and Ahlquist DA: Stool screening
for colorectal cancer: Molecular approaches. Gastroenterology.
128:192–206. 2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Hamaya Y, Yoshida K, Takai T, Ikuma M,
Hishida A and Kanaoka S: Factors that contribute to faecal
cyclooxygenase-2 mRNA expression in subjects with colorectal
cancer. Br J Cancer. 102:916–921. 2010.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Mojtabanezhad Shariatpanahi A, Yassi M,
Nouraie M, Sahebkar A, Varshoee Tabrizi F and Kerachian MA: The
importance of stool DNA methylation in colorectal cancer diagnosis:
A meta-analysis. PLoS One. 13(e0200735)2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Robertson DJ and Imperiale TF: Stool
testing for colorectal cancer screening. Gastroenterology.
149:1286–1293. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Yang SH, Huang CJ, Lee CL, Liu CC, Chien
CC and Chen SH: Fecal RNA detection of cytokeratin 19 and ribosomal
protein L19 for colorectal cancer. Hepatogastroenterology.
57:710–715. 2010.PubMed/NCBI
|
|
34
|
Song L, Lu H and Yin J: Preliminary study
on discriminating HER2 2+ amplification status of breast cancers
based on texture features semi-automatically derived from pre-,
post-contrast, and subtraction images of DCE-MRI. PLoS One.
15(e0234800)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Juhola M, Joutsijoki H, Penttinen K, Shah
D and Aalto-Setälä K: On computational classification of genetic
cardiac diseases applying iPSC cardiomyocytes. Comput Methods
Programs Biomed. 210(106367)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yu Z, Yu H, Zou Q, Huang Z, Wang X, Tang
G, Bai L, Zhou C, Zhuang Z, Xie Y, et al: Nomograms for prediction
of molecular phenotypes in colorectal cancer. Onco Targets Ther.
13:309–321. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Liu CY, Huang CS, Huang CC, Ku WC, Shih HY
and Huang CJ: Co-occurrence of differentiated thyroid cancer and
second primary malignancy: Correlation with expression profiles of
mismatch repair protein and cell cycle regulators. Cancers (Basel).
13(5486)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Weiser MR: AJCC 8th edition: Colorectal
cancer. Ann Surg Oncol. 25:1454–1455. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Treanor D and Quirke P: Pathology of
colorectal cancer. Clin Oncol (R Coll Radiol). 19:769–776.
2007.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chien CC, Chang CC, Yang SH, Chen SH and
Huang CJ: A homologue of the Drosophila headcase protein is a novel
tumor marker for early-stage colorectal cancer. Oncol Rep.
15:919–926. 2006.PubMed/NCBI
|
|
41
|
Dowling P, Clarke C, Hennessy K,
Torralbo-Lopez B, Ballot J, Crown J, Kiernan I, O'Byrne KJ, Kennedy
MJ, Lynch V and Clynes M: Analysis of acute-phase proteins, AHSG,
C3, CLI, HP and SAA, reveals distinctive expression patterns
associated with breast, colorectal and lung cancer. Int J Cancer.
131:911–923. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Ginestier C, Cervera N, Finetti P,
Esteyries S, Esterni B, Adélaïde J, Xerri L, Viens P, Jacquemier J,
Charafe-Jauffret E, et al: Prognosis and gene expression profiling
of 20q13-amplified breast cancers. Clin Cancer Res. 12:4533–4544.
2006.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Bair E and Tibshirani R: Semi-supervised
methods to predict patient survival from gene expression data. PLoS
Biol. 2(E108)2004.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Chou HL, Yao CT, Su SL, Lee CY, Hu KY,
Terng HJ, Shih YW, Chang YT, Lu YF, Chang CW, et al: Gene
expression profiling of breast cancer survivability by pooled cDNA
microarray analysis using logistic regression, artificial neural
networks and decision trees. BMC Bioinformatics.
14(100)2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Hughes G: On the binormal predictive
receiver operating characteristic curve for the joint assessment of
positive and negative predictive values. Entropy (Basel).
22(593)2020.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yang SH, Chien CC, Chen CW, Li SY and
Huang CJ: Potential of faecal RNA in diagnosing colorectal cancer.
Cancer Lett. 226:55–63. 2005.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Chang CC, Kao WY, Liu CY, Su HH, Kan YA,
Lin PY, Ku WC, Chang KW, Yang RN and Huang CJ: Butyrate
supplementation regulates expression of chromosome segregation
1-like protein to reverse the genetic distortion caused by p53
mutations in colorectal cancer. Int J Oncol. 60(64)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wei Q, Jiang H, Baker A, Dodge LK, Gerard
M, Young MR, Toledano MB and Colburn NH: Loss of sulfiredoxin
renders mice resistant to azoxymethane/dextran sulfate
sodium-induced colon carcinogenesis. Carcinogenesis. 34:1403–1410.
2013.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Bankhead P, Loughrey MB, Fernández JA,
Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ,
Coleman HG, et al: QuPath: Open source software for digital
pathology image analysis. Sci Rep. 7(16878)2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Huang CJ, Chien CC, Yang SH, Chang CC, Sun
HL, Cheng YC, Liu CC, Lin SC and Lin CM: Faecal ribosomal protein
L19 is a genetic prognostic factor for survival in colorectal
cancer. J Cell Mol Med. 12:1936–1943. 2008.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Bernal G: Use of RNA isolated from feces
as a promising tool for the early detection of colorectal cancer.
Int J Biol Markers. 27:e82–e89. 2012.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Grizzi F, Celesti G, Basso G and Laghi L:
Tumor budding as a potential histopathological biomarker in
colorectal cancer: Hype or hope? World J Gastroenterol.
18:6532–6536. 2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Belletti B, Nicoloso MS, Schiappacassi M,
Chimienti E, Berton S, Lovat F, Colombatti A and Baldassarre G:
p27(kip1) functional regulation in human cancer: A potential target
for therapeutic designs. Curr Med Chem. 12:1589–1605.
2005.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Blázquez C, Geelen MJ, Velasco G and
Guzmán M: The AMP-activated protein kinase prevents ceramide
synthesis de novo and apoptosis in astrocytes. FEBS Lett.
489:149–153. 2001.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Nikolouzakis TK, Vassilopoulou L,
Fragkiadaki P, Mariolis Sapsakos T, Papadakis GZ, Spandidos DA,
Tsatsakis AM and Tsiaoussis J: Improving diagnosis, prognosis and
prediction by using biomarkers in CRC patients (Review). Oncol Rep.
39:2455–2472. 2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Cheng YW and Li YC: Factors affecting the
follow-up time after a positive result in the fecal occult blood
test. PLoS One. 16(e0258130)2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Bardhan K and Liu K: Epigenetics and
colorectal cancer pathogenesis. Cancers (Basel). 5:676–713.
2013.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Mundade R, Imperiale TF, Prabhu L, Loehrer
PJ and Lu T: Genetic pathways, prevention, and treatment of
sporadic colorectal cancer. Oncoscience. 1:400–406. 2014.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Ahlquist DA: Molecular detection of
colorectal neoplasia. Gastroenterology. 138:2127–2139.
2010.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Bailey JR, Aggarwal A and Imperiale TF:
Colorectal cancer screening: Stool DNA and other noninvasive
modalities. Gut Liver. 10:204–211. 2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Youssef O, Sarhadi V, Ehsan H, Böhling T,
Carpelan-Holmström M, Koskensalo S, Puolakkainen P, Kokkola A and
Knuutila S: Gene mutations in stool from gastric and colorectal
neoplasia patients by next-generation sequencing. World J
Gastroenterol. 23:8291–8299. 2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Januszewicz W and Fitzgerald RC: Early
detection and therapeutics. Mol Oncol. 13:599–613. 2019.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Pickhardt PJ: Emerging stool-based and
blood-based non-invasive DNA tests for colorectal cancer screening:
The importance of cancer prevention in addition to cancer
detection. Abdom Radiol (NY). 41:1441–1444. 2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Shin HY, Suh M, Choi KS, Hwang SH, Jun JK,
Han DS, Lee YK, Oh JH, Lee CW and Lee DH: Higher satisfaction with
an alternative collection device for stool sampling in colorectal
cancer screening with fecal immunochemical test: A cross-sectional
study. BMC Cancer. 18(365)2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
de Wit M, Fijneman RJ, Verheul HM, Meijer
GA and Jimenez CR: Proteomics in colorectal cancer translational
research: Biomarker discovery for clinical applications. Clin
Biochem. 46:466–479. 2013.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Montrose DC, Zhou XK, Kopelovich L,
Yantiss RK, Karoly ED, Subbaramaiah K and Dannenberg AJ: Metabolic
profiling, a noninvasive approach for the detection of experimental
colorectal neoplasia. Cancer Prev Res (Phila). 5:1358–1367.
2012.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Slaby O: Non-coding RNAs as biomarkers for
colorectal cancer screening and early detection. Adv Exp Med Biol.
937:153–170. 2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Tsai TJ, Lim YP, Chao WY, Chen CC, Chen
YJ, Lin CY and Lee YR: Capping actin protein overexpression in
human colorectal carcinoma and its contributed tumor migration.
Anal Cell Pathol (Amst). 2018(8623937)2018.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Pan F, Chen T, Sun X, Li K, Jiang X,
Försti A, Zhu Y and Lai M: Prognosis prediction of colorectal
cancer using gene expression profiles. Front Oncol.
9(252)2019.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Xu G, Zhang M, Zhu H and Xu J: A 15-gene
signature for prediction of colon cancer recurrence and prognosis
based on SVM. Gene. 604:33–40. 2017.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Eckmann JD, Ebner DW and Kisiel JB:
Multi-target stool DNA testing for colorectal cancer screening:
Emerging learning on real-world performance. Curr Treat Options
Gastroenterol: Jan 21, 2020 (Epub ahead of print).
|
|
73
|
Kisiel JB, Klepp P, Allawi HT, Taylor WR,
Giakoumopoulos M, Sander T, Yab TC, Moum BA, Lidgard GP, Brackmann
S, et al: Analysis of DNA methylation at specific loci in stool
samples detects colorectal cancer and high-grade dysplasia in
patients with inflammatory bowel disease. Clin Gastroenterol
Hepatol. 17:914–921.e5. 2019.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Naber SK, Knudsen AB, Zauber AG, Rutter
CM, Fischer SE, Pabiniak CJ, Soto B, Kuntz KM and Lansdorp-Vogelaar
I: Cost-effectiveness of a multitarget stool DNA test for
colorectal cancer screening of medicare beneficiaries. PLoS One.
14(e0220234)2019.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Jung G, Hernández-Illán E, Moreira L,
Balaguer F and Goel A: Epigenetics of colorectal cancer: Biomarker
and therapeutic potential. Nat Rev Gastroenterol Hepatol.
17:111–130. 2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Olén O, Erichsen R, Sachs MC, Pedersen L,
Halfvarson J, Askling J, Ekbom A, Sørensen HT and Ludvigsson JF:
Colorectal cancer in ulcerative colitis: A Scandinavian
population-based cohort study. Lancet. 395:123–131. 2020.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Chang CC, Chao KC, Huang CJ, Hung CS and
Wang YC: Association between aberrant dynein cytoplasmic 1 light
intermediate chain 1 expression levels, mucins and chemosensitivity
in colorectal cancer. Mol Med Rep. 22:185–192. 2020.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Hodge CD, Ismail IH, Edwards RA, Hura GL,
Xiao AT, Tainer JA, Hendzel MJ and Glover JN: RNF8 E3 ubiquitin
ligase stimulates Ubc13 E2 conjugating activity that is essential
for DNA double strand break signaling and BRCA1 tumor suppressor
recruitment. J Biol Chem. 291:9396–9410. 2016.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Wu Z, Neufeld H, Torlakovic E and Xiao W:
Uev1A-Ubc13 promotes colorectal cancer metastasis through
regulating CXCL1 expression via NF-кB activation. Oncotarget.
9:15952–15967. 2018.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Zhang P, Wang L, Rodriguez-Aguayo C, Yuan
Y, Debeb BG, Chen D, Sun Y, You MJ, Liu Y, Dean DC, et al: miR-205
acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13. Nat
Commun. 5(5671)2014.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Pulvino M, Liang Y, Oleksyn D, DeRan M,
Van Pelt E, Shapiro J, Sanz I, Chen L and Zhao J: Inhibition of
proliferation and survival of diffuse large B-cell lymphoma cells
by a small-molecule inhibitor of the ubiquitin-conjugating enzyme
Ubc13-Uev1A. Blood. 120:1668–1677. 2012.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Cheng J, Fan YH, Xu X, Zhang H, Dou J,
Tang Y, Zhong X, Rojas Y, Yu Y, Zhao Y, et al: A small-molecule
inhibitor of UBE2N induces neuroblastoma cell death via activation
of p53 and JNK pathways. Cell Death Dis. 5(e1079)2014.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Ishtiaq A, Bakhtiar A, Silas E, Saeed J,
Ajmal S, Mushtaq I, Ali T, Wahedi HM, Khan W, Khan U, et al:
Pistacia integerrima alleviated Bisphenol A induced toxicity
through Ubc13/p53 signalling. Mol Biol Rep. 47:6545–6559.
2020.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Pelletier J, Riaño-Canalias F, Almacellas
E, Mauvezin C, Samino S, Feu S, Menoyo S, Domostegui A,
Garcia-Cajide M, Salazar R, et al: Nucleotide depletion reveals the
impaired ribosome biogenesis checkpoint as a barrier against DNA
damage. EMBO J. 39(e103838)2020.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Ouchida M, Kanzaki H, Ito S, Hanafusa H,
Jitsumori Y, Tamaru S and Shimizu K: Novel direct targets of
miR-19a identified in breast cancer cells by a quantitative
proteomic approach. PLoS One. 7(e44095)2012.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Zhao G, Liu X, Liu Y, Li H, Ma Y, Li S,
Zhu Y, Miao J, Xiong S, Fei S and Zheng M: Aberrant DNA methylation
of SEPT9 and SDC2 in stool specimens as an integrated biomarker for
colorectal cancer early detection. Front Genet.
11(643)2020.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Gu Y, Lin X, Kapoor A, Chow MJ, Jiang Y,
Zhao K and Tang D: The oncogenic potential of the centromeric
border protein FAM84B of the 8q24.21 gene desert. Genes (Basel).
11(312)2020.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Uyama T, Jin XH, Tsuboi K, Tonai T and
Ueda N: Characterization of the human tumor suppressors TIG3 and
HRASLS2 as phospholipid-metabolizing enzymes. Biochim Biophys Acta.
1791:1114–1124. 2009.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Shyu RY, Hsieh YC, Tsai FM, Wu CC and
Jiang SY: Cloning and functional characterization of the HRASLS2
gene. Amino Acids. 35:129–137. 2008.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Bi F, Wang Q, Dong Q, Wang Y, Zhang L and
Zhang J: Circulating tumor DNA in colorectal cancer: Opportunities
and challenges. Am J Transl Res. 12:1044–1055. 2020.PubMed/NCBI
|
|
91
|
Lansdorp-Vogelaar I, Goede SL, Bosch LJW,
Melotte V, Carvalho B, van Engeland M, Meijer GA, de Koning HJ and
van Ballegooijen M: Cost-effectiveness of high-performance
biomarker tests vs fecal immunochemical test for noninvasive
colorectal cancer screening. Clin Gastroenterol Hepatol.
16:504–512.e11. 2018.PubMed/NCBI View Article : Google Scholar
|