Introduction
Colorectal cancer (CRC) is one of the most prevalent
gastrointestinal malignancies worldwide. In recent years, its
morbidity and mortality have gradually increased. A total of ~19
million new cases and 10 million cancer-related deaths were
estimated in 2020(1). As nearly as
65.0% of new cases and 63.6% of CRC-related deaths occurred in
China, Europe and North America in 2020(2). The 5-year survival rate of patients
with CRC after surgery, radiotherapy and chemotherapy is >30%,
and it has become a serious threat to human survival (3). It was estimated that in 2010, there
were 270,000 new patients with CRC diagnoses and 130,000
CRC-related deaths in China (4). By
2025, the numbers of new patient diagnoses and deaths with CRC in
China are expected to reach 624,300 and 221,100, respectively
(5). At present, the TNM stage of
CRC remains the most important prognostic factor, but even for
patients with the same TNM stage, the prognosis can differ.
Additionally, in some CRC cases, there is no association between
the pathological Tumor-Node-Metastasis (pTNM) stage and CRC
biological and clinical behavior (6,7).
Therefore, identifying additional factors that can more accurately
predict the clinical course of CRC regardless of the pTNM stage has
been a major research focus for numerous years.
Although CRC occurs sporadically, its occurrence has
been associated with genetic variations, including chromosomal
instability, microsatellite instability, and Ras/Raf/MAPK
mutations. In recent years, anti-epidermal growth factor receptor
(EGFR) monoclonal antibodies have been used for the treatment of
CRC because of their ability to block downstream intracellular EGFR
signaling (8). However, the
therapeutic efficacy is strictly dependent on the effect of the
RAS/MAPK and PI3K-PTEN-AKT pathways downstream of the EGFR pathway,
which is involved in genetic integrity. EGFR has emerged as a key
target in specific inhibitor therapy for CRC, and activating
mutations in KRAS/NRAS are considered to be strong predictors of
resistance to EGFR-targeted drugs (9). These mutations lead to the
constitutive phosphorylation of RAS proteins independent of the
active state of upstream EGFR proteins (10).
RAS (KRAS/NRAS) is a proto-oncogene that encodes a
protein with GTPase activity that plays a role in EGFR signal
transduction and self-inactivation (11). BRAF is an important component of the
RAS/RAF/MAPK pathway that mediates the binding of RAF and MAPK
kinase (MAPKK/MEK1/2) in signal transduction and the regulation of
cell proliferation. PIK3CA encodes P110 alpha, the catalytic
subunit of PI3K, which mediates the PI3K/AKT pathway and promotes
cell survival. Hence, changes in PIK3CA may lead to abnormal
activation of the PI3K pathway. According to De Roock et al
(12) KRAS-, NRAS-, BRAF-, and
PIK3CA-based molecular biomarkers may have prognostic value in CRC.
Chemotherapy combined with wild-type RAS- and EGFR-targeted therapy
can improve the prognosis of patients (13). However, mutations in the RAS factor
or BRAF may activate the downstream RAS/Raf/MAPK pathway, thereby
inhibiting the effects of anti-EGFR therapy (14). KRAS and NRAS mutations are
predictive of cetuximab and panitumumab therapy efficacy in
clinical practice, but PIK3CA mutations are not included in the
current guidelines. However, Liao et al (15) found that regular use of aspirin was
correlated with longer survival among PIK3CA-mutated patients with
CRC. Therefore, it is necessary to detect mutations in RAS, BRAF,
PIK3CA and other genes in patients with CRC. In addition, cetuximab
and panitumumab are the main molecular targeted drugs available for
patients with CRC, and they act by inhibiting an EGFR signaling
pathway (PI3K/AKT/mTOR or RAS/RAF/MAPK). The use of these drugs
requires analysis of the RAS mutation status, and only patients
with wild-type RAS are eligible for treatment. However, <40% of
KRAS wild-type patients do not respond to anti-EGFR monoclonal
antibody treatment (16). This
resistance can be explained by mutations in other signaling
effectors downstream of EGFR, such as the BRAF, PIK3CA and NRAS
genes.
Ring finger protein 215 (RNF215) is a multichannel
membrane protein, and its encoding gene is located at 22q12.2. At
present, to the best of the authors' knowledge, only a few studies
of the RNF215 protein have been performed. Wu et al
(17) indicated that RNF215 can
bind to the NF-kB p65 subunit by partially inhibiting type I
interferon (IFN) production and limiting the accumulation of NF-kB
in the promoter region of IFNB1. Ma et al (18) suggested that high RNF215 expression
was associated with poor overall survival (OS) of head/neck
squamous cell carcinoma. The study by McIntosh et al
(19) indicated that
single-nucleotide polymorphism variations near RNF215 were
correlated with the expression levels of the neighboring gene
MTP18/SF3A1. The present preliminary study showed that the
expression of RNF215 was significantly higher in CRC tumor tissues
than in normal tissues. RNF215 may contribute to the development
and progression of CRC by participating in CRC-associated signaling
pathways, such as the Kyoto Encyclopedia of Genes and Genomes MAPK
signaling pathway, the WP RAS signaling pathway, and the WP PI3KAKT
signaling pathway (20). However,
the associations between RNF215 expression and KRAS, NRAS, BRAF and
PIK3CA mutations in patients with CRC have not been reported.
Therefore, there is a need to further investigate the role of
RNF215 in patients with CRC with KRAS, NRAS, BRAF and PIK3CA
mutations.
According to CRC guidelines (https://www.nccn.org/guidelines/nccn-guidelines),
KRAS/NRAS/BRAF gene mutation detection is recommended for patients
with primary or metastatic CRC before treatment to clarify the
status and guide treatment. Although PIK3CA mutation analysis is
not yet recommended, PIK3CA exon 20 mutations are associated with
worse prognosis in metastatic CRC patients than in wild-type
patients (21). Therefore, in the
present study, the occurrence of gene mutations in KRAS, NRAS, BRAF
and PIK3CA in CRC patients with CRC was assessed and the
correlation between mutation incidence and clinicopathological
features was estimated. Furthermore, immunohistochemistry (IHC) was
performed to determine whether KRAS/NRAS/BRAF/PIK3CA gene mutations
in patients with CRC could be detected by IHC. In addition, RNF215
expression in patients with mutations in KRAS, NRAS, BRAF and
PIK3CA was also investigated. The results of the present study
suggested that drug resistance in KRAS wild-type patients may be
associated with PIK3CA gene mutations, and for the first time, BRAF
mutations were found to be possibly associated with RNF215
expression.
Materials and methods
Clinical data
A total of 182 CRC resection specimens from Shanghai
Fifth People's Hospital affiliated with Fudan University (Shanghai,
China) from January 2012 to December 2016 were included. The
inclusion criteria for patients with CRC were as follows: i)
Patients with pathological and imaging examination data met the CRC
diagnostic criteria; ii) patients had no family history of CRC; and
iii) patients had favorable mental health. The exclusion criteria
were as follows: i) Patients did not meet the standard CRC
diagnostic criteria; ii) Patients had serious complications
affecting the heart, lung, or other important organs; and iii)
patients were not conscious or unable to communicate normally.
Patients with drug resistance were defined as KRAS wild-type
patients for whom anti-EGFR therapy (cetuximab and panitumumab) was
ineffective. The present study was reviewed and approved (approval
no. 2021071) by the Ethical Committee of Shanghai Fifth People's
Hospital, Fudan University (Shanghai, China). Written informed
consent was provided by the patients/participants who participated
in the present study.
Samples were collected and evaluated by two
professional pathologists according to the standard protocols of
the Department of Pathology. CRC tumors were staged according to
the Staging Manual of the American Joint Committee on Cancer
(Eighth Edition). The pathological characteristics of patients with
CRC were extracted from medical records and pathology reports.
Mutation detection
Archival CRC tumor tissue in formalin-fixed
paraffin-embedded (FFPE) blocks was available from all patients
included in the present study. All tissues were fixed with 10%
formalin at room temperature (20˚C) for more than 24 h. The FFPE
blocks were archived at the Department of Pathology, Shanghai Fifth
People's Hospital, Fudan University (Shanghai, China). Mutation
analysis was performed when the pathologist observed >10% of
tumor cells under the hematoxylin and eosin staining slide. DNA was
extracted using a DNA FFPE Tissue kit (cat no. 20150079; Amoy
Diagnostics Co., Ltd.), according to the instructions for dewaxing,
lysis, digestion, repair, adsorption, elution and other steps. DNA
quality and concentration were determined by spectrophotometry
(OD260/OD280 ratio 1.8-2.0). Mutations in the KRAS/NRAS/BRAF/PIK3CA
genes were confirmed by an amplification-refractory mutation system
(ARMS) (Human KRAS/NRAS/PIK3CA/BRAF Gene Mutation Combination
Detection kit; (cat. no. 20153401124; Amoy Diagnostics Co., Ltd.).
According to the manufacturer's instructions, the sample DNA
concentration was adjusted to an appropriate concentration (10
ng/ml) for sample preparation. The real-time quantitative PCR
amplification procedure is shown in Table I. The primers used in the study were
part of the kit. The fluorescence channel signal was collected
during the third stage of annealing at 60°C. Real-time
quantitative PCR was performed, and the files were saved. Hotspot
gene mutations, including those in exons 2, 3 and 4 of the human
KRAS gene, exons 2 and 3 of the NRAS gene, exon 15 of the BRAF
gene, and exon 20 of the PIK3CA gene, were detected (as shown in
Table II). The mutational analyses
were performed compared with the amplification levels of positive
and negative control tests provided by the manufacturer and
according to the relevant protocols.
 | Table IReverse transcription-quantitative
PCR amplification procedures. |
Table I
Reverse transcription-quantitative
PCR amplification procedures.
| Stage | Procedure | Temperature
(˚C) | Time | Cycles (n) |
|---|
| First stage | Denaturation | 95 | 5 min | 1 |
| Second stage | Denaturation | 95 | 25 sec | 15 |
| | Annealing | 64 | 20 sec | |
| | Extension | 72 | 20 sec | |
| Third stage | Denaturation | 93 | 25 sec | 31 |
| | Annealing | 60 | 35 sec | |
| | Extension | 72 | 20 sec | |
 | Table IIGene mutation detection site. |
Table II
Gene mutation detection site.
| Gene name | Test section | Mutant name | Base change |
|---|
| KRAS | Exon 2 | G12S | 34G>A |
| | | G12D | 35G>A |
| | | G12C | 34G>T |
| | | G12R | 34G>C |
| | | G12V | 35G>T |
| | | G12A | 35G>C |
| | | G13C | 37G>T |
| | | G13D | 38G>A |
| | Exon 3 | Q61L | 182A>T |
| | | Q61R | 182A>G |
| | | Q61H | 183A>C |
| | | Q61 | 183A>T |
| | Exon 4 | K117N | 351A>C |
| | | K117N | 351A>T |
| | | A146T | 436G>A |
| | | A146V | 437C>T |
| | | A146P | 436G>C |
| NRAS | Exon 2 | G12D | 35G>A |
| | | G12S | 34G>A |
| | | G13R | 37G>C |
| | | G12C | 34G>T |
| | | G12V | 35G>T |
| | | G12A | 35G>C |
| | | G13V | 38G>T |
| | Exon 3 | Q61R | 182A>G |
| | | Q61K | 181C>A |
| | | Q61L | 182A>T |
| | | Q61H | 183A>C |
| | Exon 4 | A146T | 436G>A |
| PIK3CA | Exon 20 | H1047R | 3140A>G |
| | | H1047L | 3140A>T |
| BRAF | Exon 15 | V600E1 | 1799T>A |
| | | V600K | 1798,1799GT>AA
(complex) |
| | | V600E2 | 1799,1800TG>AA
(complex) |
| | | V600R | 1798,1799GT>AG
(complex) |
| | | V600D1 | 1799,1800TG>AC
(complex) |
| | | V600D2 | 1799,1800TG>AT
(complex) |
IHC
All 182 FFPE colorectal tumor tissues were used for
tissue chip (tissue microarray, TMA) construction, with each tumor
tissue consisting of three 1.5-mm representative punches as
previously described by the Kononen et al (22). According to the manufacturer's
protocol, 3-µm-thick TMA slides were tested on an automated Ventana
benchmark machine (Roche Tissue Diagnostics; Roche Diagnostics,
Ltd.). Commercially available antibodies against KRAS polyclonal,
(cat no. 12063-1-AP; 1:400; Proteintech Group, Inc.), NRAS clone
sp174 (cat no. ab227658; 1:100; Abcam), BRAF clone VE1 (cat no.
ab228461; 1:100; Abcam), PIK3CA clone SP139 (cat no. ab135384;
1:100; Abcam) and RNF215 polyclonal, (1:300; cat. no. Ys-9264R;
Shanghai Yaji Biological Technology Co., Ltd.) were used for IHC.
Sections were incubated with primary antibody for 16 h at 4˚C,
followed by the application of a secondary antibody [ultraView
Universal HRP Multimer (55 µg/ml); cat. no. (92)760-500; Ventana
Medical Systems, Inc.] for 40 min at 37˚C. Finally,
3,3'-diaminobenzidine (DAB) was used as the chromogenic substrate
and the sections were counterstained with hematoxylin for 1 min at
20˚C. Appropriate positive and negative slide controls were
included for each antibody. The sections were blocked with 5% BSA
(cat. no. SW3015; Beijing Solarbio Science & Technology Co.,
Ltd.) for 1 h in room temperature. Full slide images were reviewed
and evaluated by two gastrointestinal pathologists under a light
microscope (BX45; Olympus Corporation). For KRAS, NRAS, BRAF,
PIK3CA and RNF215, samples were labeled positive if >10% of
tumor cells in each batch showed cytoplasmic staining similar in
intensity to that of the positive controls. Any isolated nuclear
staining without cytoplasmic staining was determined to be
negative. IHC staining was performed on the corresponding large
tumor sections of the resected specimens by the same method to
verify the TMA protein expression.
Association between gene mutations and
RNF215 expression
RNF215 expression in KRAS-, NRAS-, BRAF-, and
PIK3CA-mutated cases was first evaluated with Tumor Immune
Estimation Resource (TIMER) 2.0 (http://timer.comp-genomics.org/) (23). Subsequently, the association between
RNF215 expression and KRAS, NRAS, BRAF and PIK3CA gene mutations in
the 182 CRC cases of the present study was further verified using
RNF215 IHC staining.
Statistical analysis
Descriptive statistics were employed for the
clinicopathological features of the patients. The statistical
results are summarized as percentages (%). The chi-square test or
Fisher's exact test was used for comparisons of clinicopathological
features, IHC expression, and gene mutation results. The survival
rate and statistical significance analyses were determined by the
Kaplan-Meier (KM) method with the log-rank test and
Gehan-Breslow-Wilcoxon test. All statistical analysis was performed
using the GraphPad Prism 9.0 statistical program (GraphPad
Software, Inc.). The RNF215 expression differences between
wild-type and mutated CRC cases in the TIMER 2.0 database
(http://timer.comp-genomics.org/) were
determined using the Wilcoxon rank sum test. All tests were
two-sided, and P<0.05 was considered to indicate a statistically
significant difference.
Results
Clinicopathological features
The clinicopathological characteristics and gene
mutation results of the study population are shown in Table III. Among the 182 patients with
CRC, 130 (71.4%) were males, and 52 (28.6%) were females. The age
of these patients ranged from 48-95 years, with an average of 69.1
years. There were 142 patients >60 years-old and 40 patients ≤60
years-old. The tumor sites were as follows: 66 cases were right
colon cancer (including ascending colon cancer and transverse colon
cancer), and 116 cases were left colon cancer (including descending
colon cancer, sigmoid colon cancer, rectal and anal cancer). The
general tumor type was endophytic in 116 patients and exophytic in
66 patients. There were 136 cases of well to moderate
differentiation and 46 cases of poor to no differentiation.
Additionally, 150 patients had a tumor diameter >3 cm, and 32
patients had a tumor diameter ≤3 cm. Lymph node metastasis was
found in 46 cases, and no lymph node metastasis was identified in
136 cases. In addition, 90 patients had distant metastasis, and 92
patients did not have distant metastasis. Neurovascular invasion
was identified in 76 patients, and no neurovascular invasion was
identified in 106 patients. The TNM stage distribution was as
follows: 12 cases of Stage I and 170 cases of Stage II-IV
disease.
 | Table IIIRelationship between KRAS, NRAS,
BRAF, and PIK3CA gene mutations and clinicopathological features in
182 patients with CRC [n (%)]. |
Table III
Relationship between KRAS, NRAS,
BRAF, and PIK3CA gene mutations and clinicopathological features in
182 patients with CRC [n (%)].
| Clinicopathological
features | n | KRAS (n=74) | P-value | NRAS (n=8) | P-value | BRAF (n=8) | P-value | PIK3CA (n=6) | P-value | ≥2 mutations
(n=8) | P-value | Total mutations
(n=104) | P-value |
|---|
| Sex | | | 0.1047 | | 0.6904 | | >0.9999 | | 0.6757 | | 0.6904 | | 0.0977 |
|
Male | 130 | 48 (36.9) | | 5 (3.9) | | 6 (4.6) | | 5 (3.9) | | 5 (3.9) | | 69 (53.1) | |
|
Female | 52 | 26 (50.0) | | 3 (5.8) | | 2 (3.9) | | 1 (1.9) | | 3 (5.8) | | 35 (67.3) | |
| Age, years | | | 0.7173 | | 0.3763 | | 0.0717 | | 0.6079 | | >0.9999 | | 0.2556 |
|
>60 | 142 | 59 (41.6) | | 5 (3.5) | | 4 (2.8) | | 4 (2.8) | | 6 (4.2) | | 78 (54.9) | |
|
≤60 | 40 | 15 (37.5) | | 3 (7.5) | | 4 (10.0) | | 2 (5.3) | | 2 (5.0) | | 26 (65.0) | |
| Localization | | | 0.3735 | | >0.9999 | | >0.9999 | | 0.0243 | | 0.4635 | | 0.9291 |
|
Right | 66 | 24 (36.4) | | 2 (4.3) | | 3 (4.6) | | 5 (7.6) | | 4 (6.1) | | 38 (48.1) | |
|
Left | 116 | 50 (43.1) | | 6 (4.6) | | 5 (4.3) | | 1 (0.9) | | 4 (3.5) | | 66 (61.9) | |
| Configuration | | | 0.3488 | | >0.9999 | | 0.4635 | | >0.9999 | | >0.9999 | | 0.1818 |
|
Endophytic | 116 | 44 (37.9) | | 5 (5.2) | | 4 (3.6) | | 4 (3.5) | | 5 (4.3) | | 62 (53.5) | |
|
Exophytic | 66 | 30 (45.5) | | 3 (3.0) | | 4 (6.1) | | 2 (3.1) | | 3 (4.6) | | 42 (63.6) | |
|
Differentiation | | | 0.1196 | | 0.4188 | | 0.0037 | | 0.1705 | | 0.1128 | | 0.2005 |
|
Well to
moderate | 136 | 60 (44.1) | | 5 (3.7) | | 2 (1.5) | | 3 (2.2) | | 4 (2.9) | | 74 (54.4) | |
|
Poor to
undifferentiated | 46 | 14 (30.4) | | 3 (6.5) | | 6 (13.0) | | 3 (6.5) | | 4 (8.7) | | 30 (65.2) | |
| Size | | | 0.3217 | | 0.3042 | | 0.1482 | | 0.2846 | | 0.3042 | | 0.8456 |
|
>3
cm | 150 | 64 (42.7) | | 6 (4.0) | | 5 (3.3) | | 4 (2.7) | | 6 (4.0) | | 85 (56.7) | |
|
≤3 cm | 32 | 10 (31.3) | | 2 (8.3) | | 3 (9.4) | | 2 (6.3) | | 2 (8.3) | | 19 (59.4) | |
| Lymph node
metastasis | | | 0.0826 | | 0.4188 | | >0.9999 | | 0.1705 | | 0.4188 | | 0.0027 |
|
Positive | 46 | 24 (52.2) | | 3 (6.5) | | 2 (4.4) | | 3 (6.5) | | 3 (6.5) | | 35 (76.1) | |
|
Negative | 136 | 50 (35.8) | | 5 (3.7) | | 6 (4.4) | | 3 (2.2) | | 5 (3.7) | | 69 (50.8) | |
| Distant
metastasis | | | 0.3028 | | 0.7205 | | 0.1665 | | 0.6824 | | 0.4943 | | 0.1708 |
|
Positive | 90 | 40 (44.4) | | 3 (3.3) | | 6 (6.7) | | 2 (2.2) | | 5 (5.6) | | 56 (62.2) | |
|
Negative | 92 | 34 (37.0) | | 5 (5.4) | | 2 (2.2) | | 4 (4.4) | | 3 (3.3) | | 48 (52.2) | |
| Neurovascular
invasion | | | 0.7828 | | 0.7211 | | 0.2813 | | 0.2399 | | >0.9999 | | 0.5392 |
|
Positive | 76 | 30 (39.5) | | 4 (5.3) | | 5 (6.6) | | 4 (5.3) | | 3 (4.0) | | 46 (44.2) | |
|
Negative | 106 | 44 (41.5) | | 4 (3.8) | | 3 (2.8) | | 2 (1.9) | | 5 (4.7) | | 58 (48.3) | |
| TNM stage | | | 0.7641 | | 0.4270 | | 0.0895 | | 0.3398 | | 0.0895 | | 0.0727 |
|
I | 12 | 4 (33.3) | | 1 (8.3) | | 2 (16.7) | | 1 (8.3) | | 2 (16.7) | | 10 (83.3) | |
|
II + III +
IV | 170 | 70 (41.2) | | 7 (4.1) | | 6 (3.5) | | 5 (2.9) | | 6 (3.5) | | 94 (55.3) | |
KRAS, NRAS, BRAF and PIK3CA gene
mutations
As shown in Table
IV, the total KRAS/NRAS/BRAF/PIK3CA mutation rate in 182
patients with CRC was 57.1% (104/182), the single mutation rates of
KRAS, NRAS, BRAF and PIK3CA were 40.7% (74/182), 4.4% (8/182), 4.4%
(8/182), and 3.3% (6/182), respectively. The concomitant mutation
rate of KRAS/PIK3CA was 2.7% (5/182) and that of KRAS/NRAS was 1.6%
(3/182). Representative KRAS, NRAS, BRAF, and PIK3CA gene mutation
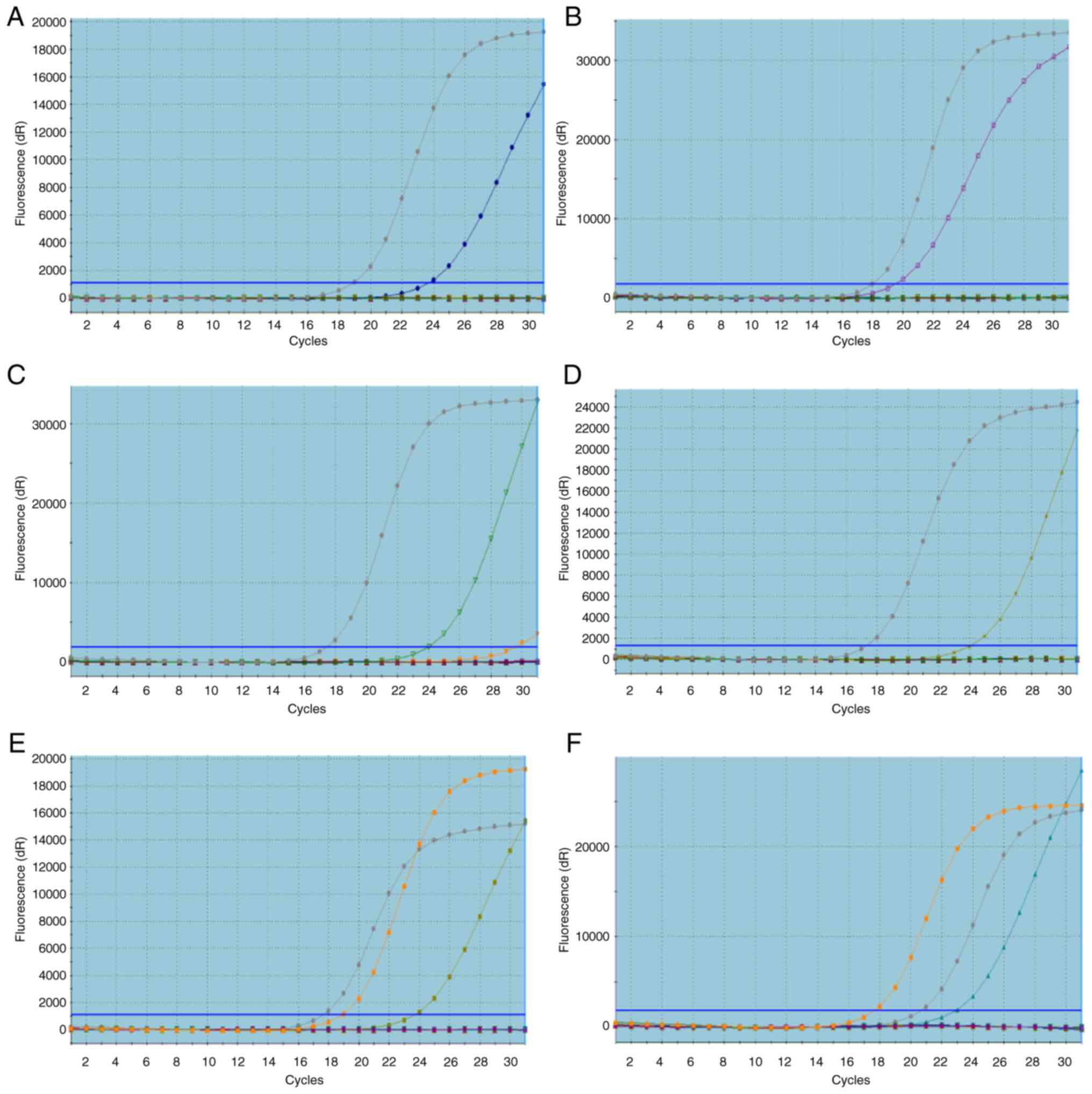
amplification curves in patients with CRC were generated by ARMS
(Fig. 1). There were no patients
with 3 or more gene mutations. The KRAS exon 2 mutation rate was
the highest (35.2%, 64/182), and KRAS exon 2 mutations mainly
occurred at codons 12 and 13. The KRAS exon 4, PIK3CA exon 20, NRAS
exon 2, BRAF exon 15, KRAS exon 3, NRAS exon 3 and NRAS exon 4
mutation rates were 6.6% (12/182), 6.0% (11/182), 4.4% (8/182),
4.4% (8/182), 3.3% (6/182), 1.1% (2/182) and 0.5% (1/182),
respectively.
 | Table IVMutations in the KRAS, NRAS, BRAF,
and PIK3CA genes in 182 patients with CRC. |
Table IV
Mutations in the KRAS, NRAS, BRAF,
and PIK3CA genes in 182 patients with CRC.
| Test location | Mutant name | Number | Mutation rate
(%) |
|---|
| KRAS Exon2 | G12S, G12D | 35 | 19.8 |
| KRAS Exon2 | G12C, G12R, G12V,
G12A, G13C | 17 | 9.3 |
| KRAS Exon2 | G13D | 12 | 6.6 |
| KRAS Exon3 | Q61L, Q61R,
Q61H(183A>C), Q61H(183A>T) | 6 | 3.3 |
| KRAS Exon4 | K117N(351A>C),
K117N(351A>T), A146T, A146V, A146P | 12 | 6.6 |
| NRAS Exon2 | G12D, G12S | 5 | 2.7 |
| NRAS Exon2 | G13R, G12C, G12V,
G12A, G13V | 3 | 1.6 |
| NRAS Exon3 | Q61R, Q61K, Q61L,
Q61H | 2 | 1.1 |
| NRAS Exon4 | A146T | 1 | 0.5 |
| PIK3CA Exon20 | H1047R, H1047L | 11 | 6.0 |
| BRAF Exon15 | V600E1, V600K,
V600E2, V600R, V600D1, V600D2 | 8 | 4.4 |
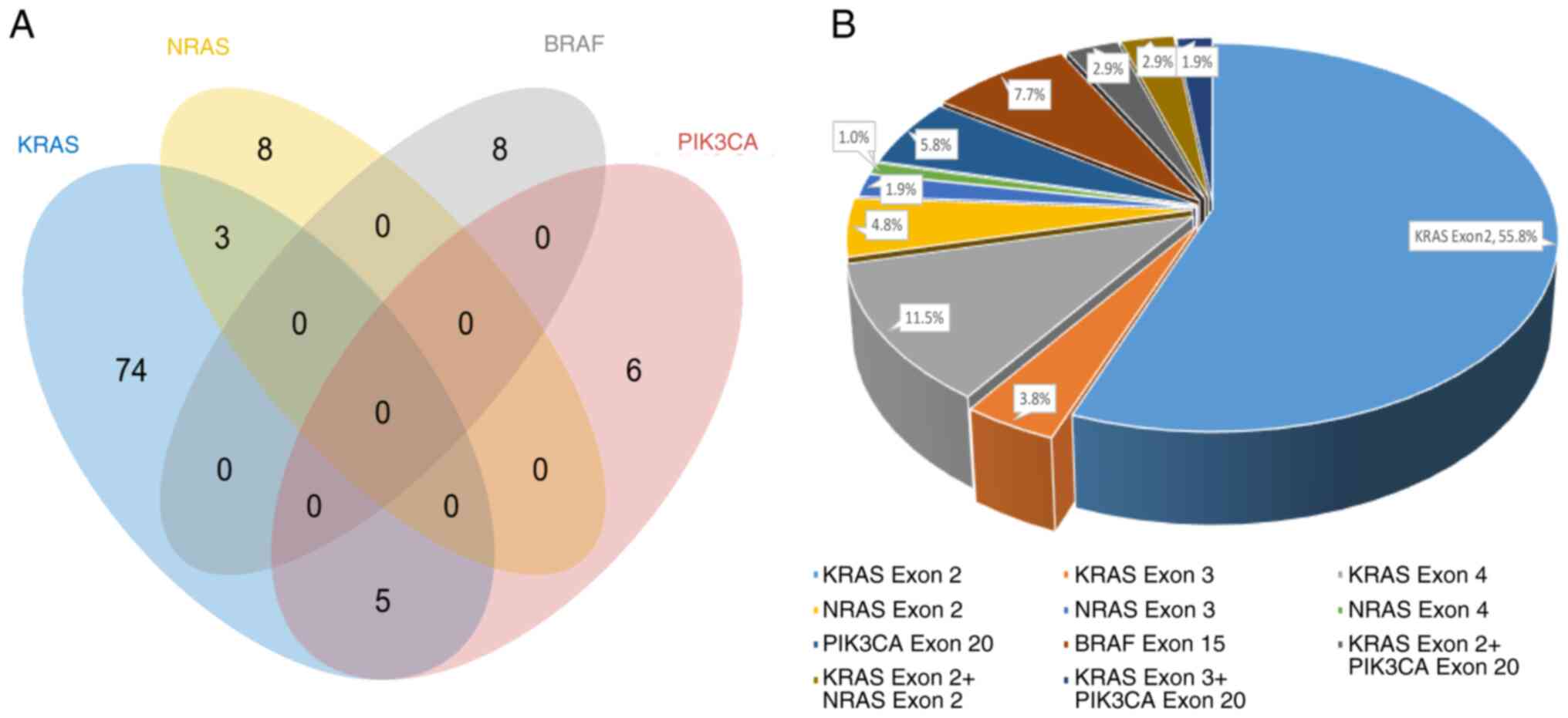
Among the 104 patients with gene mutations, the
single KRAS exon 2 mutation frequency was the highest (55.8%,
58/104), followed by the KRAS exon 4 mutation frequency was the
second highest (11.5%, 12/104), and the frequency of concomitant
mutations was 7.7% (8/104). The distribution of single and
concomitant KRAS, NRAS, BRAF and PIK3CA gene mutations is shown in
a Venn diagram (Fig. 2A). All other
mutation types are shown in Fig.
2B.
Associations of KRAS, NRAS, BRAF and
PIK3CA gene mutations with CRC patient clinical
characteristics
There were no statistically significant differences
in the incidences of analyzed mutations between patients with
different clinical characteristics, including sex, age, tumor
differentiation, tumor size, distant metastasis, neurovascular
invasion, postoperative recurrence, TNM stage and gene mutations
(P>0.05). However, the BRAF gene mutation rate in patients with
poorly differentiated cancer was significantly higher than that in
patients with well to moderately differentiated cancer (P=0.0037,
Table III), and no significant
differences in the mutation rates of other genes were found between
patients with different degrees of tumor differentiation
(P>0.05). The PIK3CA mutation rate in the right colon subgroup
was significantly higher than that in the left colon subgroup
(P=0.0243); however, no significant differences in other gene
mutations were identified between patients with different primary
tumor sites (P>0.05). The gene mutation rate of patients with
lymph node metastasis (76.1%, 35/46) was higher than that of
patients without lymph node metastasis (50.8%, 69/136), and the
difference was statistically significant (P=0.0027). However, no
significant difference in the rate of single-gene mutations was
identified between patients with and without lymph node metastasis
(P>0.05).
Prognostic significance of KRAS, NRAS,
BRAF and PIK3CA mutations in the entire cohort
The average length of follow-up for all 182 CRC
patients was 41.4 months (range, 14-79 months; 95% Confidence
interval, 38.04-44.79 months), and 72.5% (132/182) of patients were
alive when the present study was completed. In the
KRAS/NRAS/BRAF/PIK3CA mutation group, there were 25 deaths; 40
patients were alive with liver, lung, or bone metastasis, and 39
patients were alive without evidence of tumor metastasis. In the
KRAS wild-type group, 18 patients succumbed, and one of these
patients had liver or brain metastasis. A total of 34 patients were
alive with liver, lung, bone, or brain metastasis or tubular
adenoma. The remaining 36 patients were alive and without evidence
of tumor metastasis or recurrence. The majority of the KRAS
wild-type patients (71.6%, 53/74) received targeted anti-EGFR
therapy (cetuximab and panitumumab). However, no KRAS-mutant
patients received anti-EGFR therapy. OS was defined from the date
of pathological diagnosis of carcinoma to the date of death or the
date of examination of surviving patients in September 2022.
Disease-free survival (DFS) was calculated from the date of primary
colorectal carcinoma resection to the date of recurrence or
metastasis or the date of screening in September 2022. The
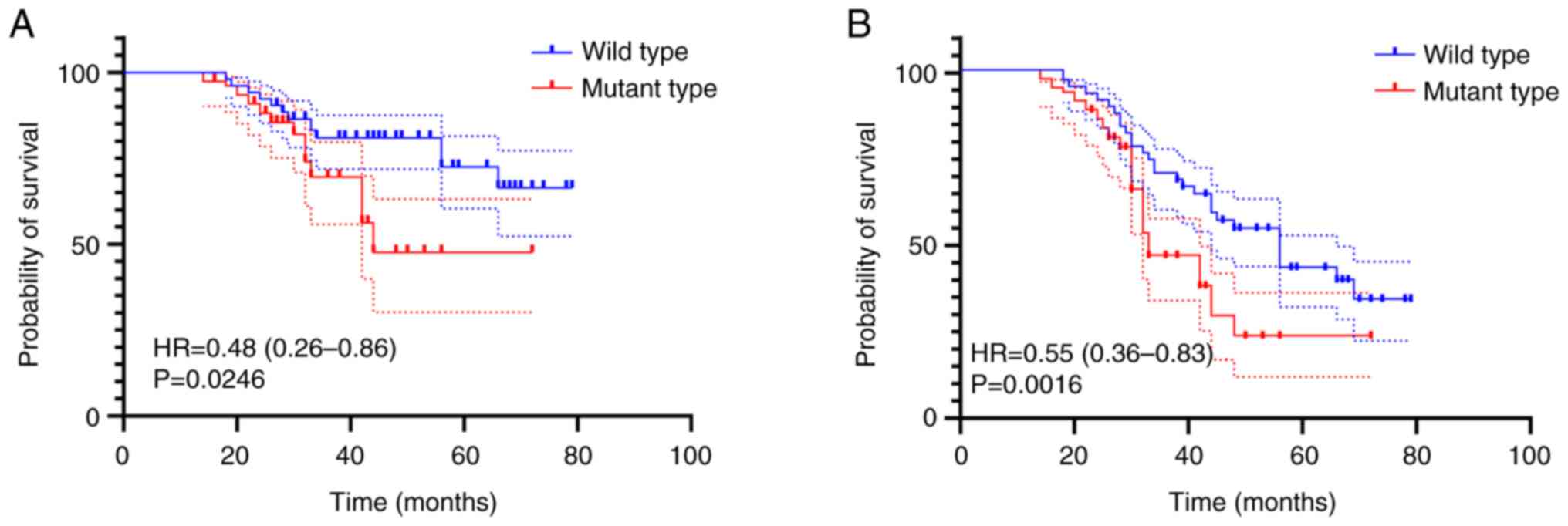
KRAS/NRAS/BRAF/PIK3CA mutation group had a significantly shorter OS
and DFS than the KRAS/NRAS/BRAF/PIK3CA wild-type group, as
demonstrated in Fig. 3.
IHC results
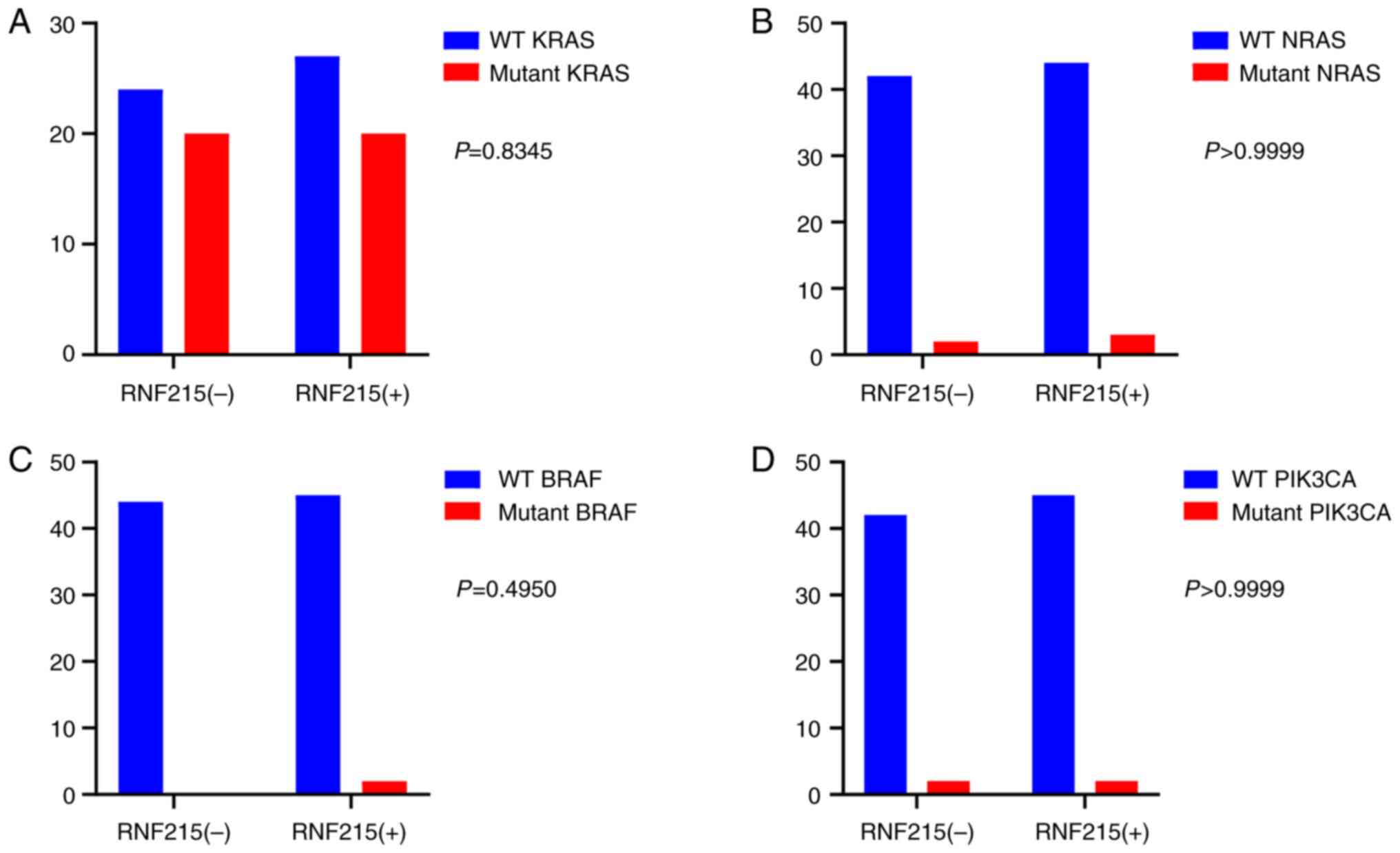
Among 182 CRC patients, the rate of positive KRAS
protein staining by IHC was 69.8% (127/182). The rate of positive
KRAS protein staining in patients with mutated KRAS was 65.9%
(54/82), while that in patients without KRAS mutation was 73.0%
(73/100). No significant difference was found between KRAS-mutant
patients and nonmutated patients (P=0.2692). The rate of positive
NRAS protein staining by IHC was 83.0% (151/182), and the rate of
positive NRAS protein staining in NRAS-mutant patients was 72.7%
(8/11). The rate of positive NRAS staining in non-mutated patients
was 83.6% (143/171), and no significant difference was identified
between NRAS-mutated and nonmutated patients (P=0.4024). The rate
of positive BRAF protein staining by IHC was 85.7% (156/182). The
rate of positive BRAF protein staining in BRAF-mutated patients was
62.5% (5/8), and that in patients without BRAF mutation was 87.3%
(151/173). No significant difference was found between BRAF-mutated
and nonmutated patients (P=0.0817). The rate of positive PIK3CA
protein staining by IHC was 84.1% (153/182). The rate of positive
PIK3CA protein staining in PIK3CA-mutated patients was 81.8%
(9/11), and that in patients without PIK3CA mutation was 84.8%
(145/173). No significant difference was identified between
PIK3CA-mutated and non-mutated patients (P=0.6785). The total rate
of positive of KRAS/NRAS/BRAF/PIK3CA protein staining by IHC was
not significantly different between KRAS/NRAS/BRAF/PIK3CA-mutated
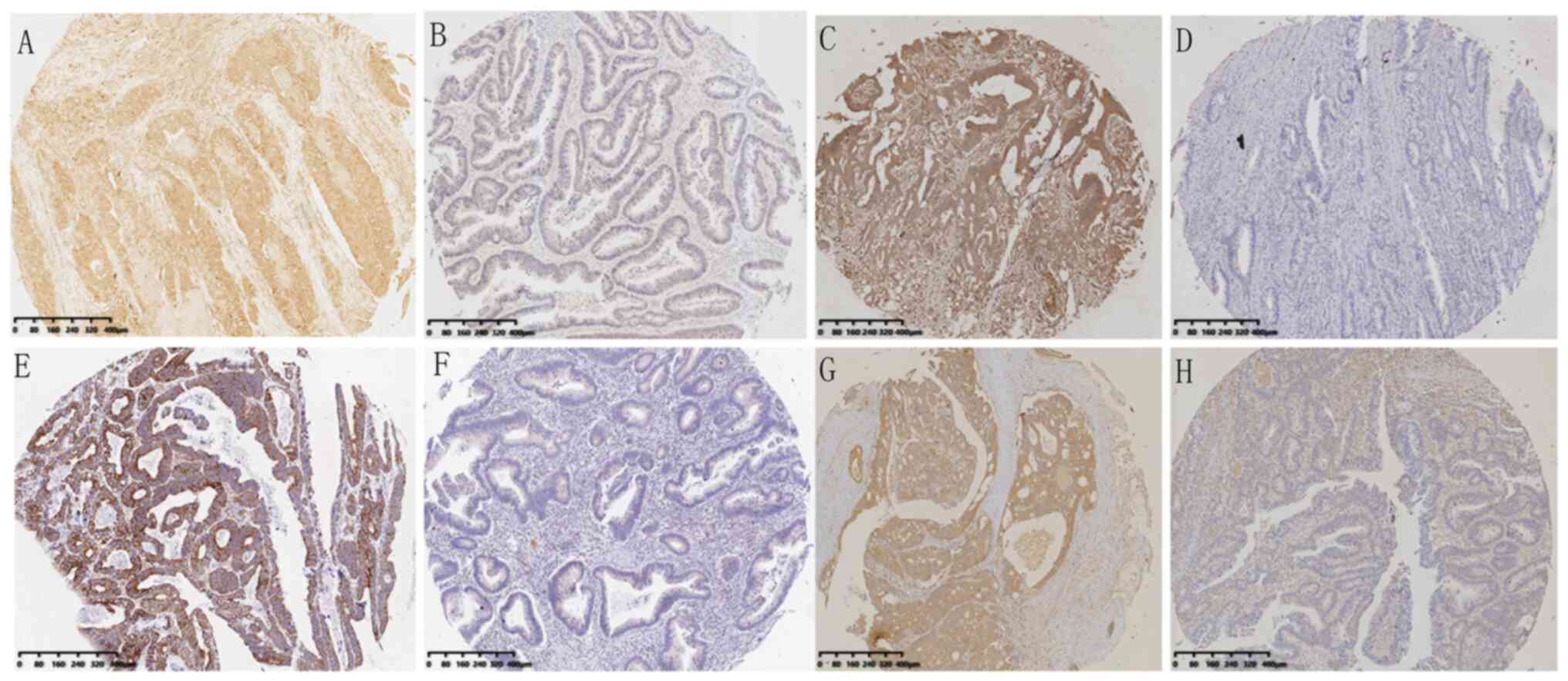
and nonmutated patients (P=0.5882), as shown in Table V. A typical sample with KRAS, NRAS,
BRAF and PIK3CA protein IHC staining is shown in Fig. 4A-H.
 | Table VImmunohistochemical results of the
KRAS, NRAS, BRAF, and PIK3CA genes in 182 patients with CRC. |
Table V
Immunohistochemical results of the
KRAS, NRAS, BRAF, and PIK3CA genes in 182 patients with CRC.
| | Positive (n) | Negative (n) | P-value |
|---|
| KRAS | | | 0.2692 |
|
Mutated | 54 | 28 | |
|
Unmutated | 73 | 27 | |
| NRAS | | | 0.4024 |
|
Mutated | 8 | 3 | |
|
Unmutated | 143 | 28 | |
| BRAF | | | 0.0817 |
|
Mutated | 5 | 3 | |
|
Unmutated | 151 | 22 | |
| PIK3CA | | | 0.6785 |
|
Mutated | 9 | 2 | |
|
Unmutated | 145 | 26 | |
| Total (KRAS + NRAS
+ BRAF + PIK3CA) | | | 0.5882 |
|
Mutated | 94 | 10 | |
|
Unmutated | 73 | 5 | |
Association between gene mutation and
RNF215 expression
According to the results obtained from TIMER 2.0
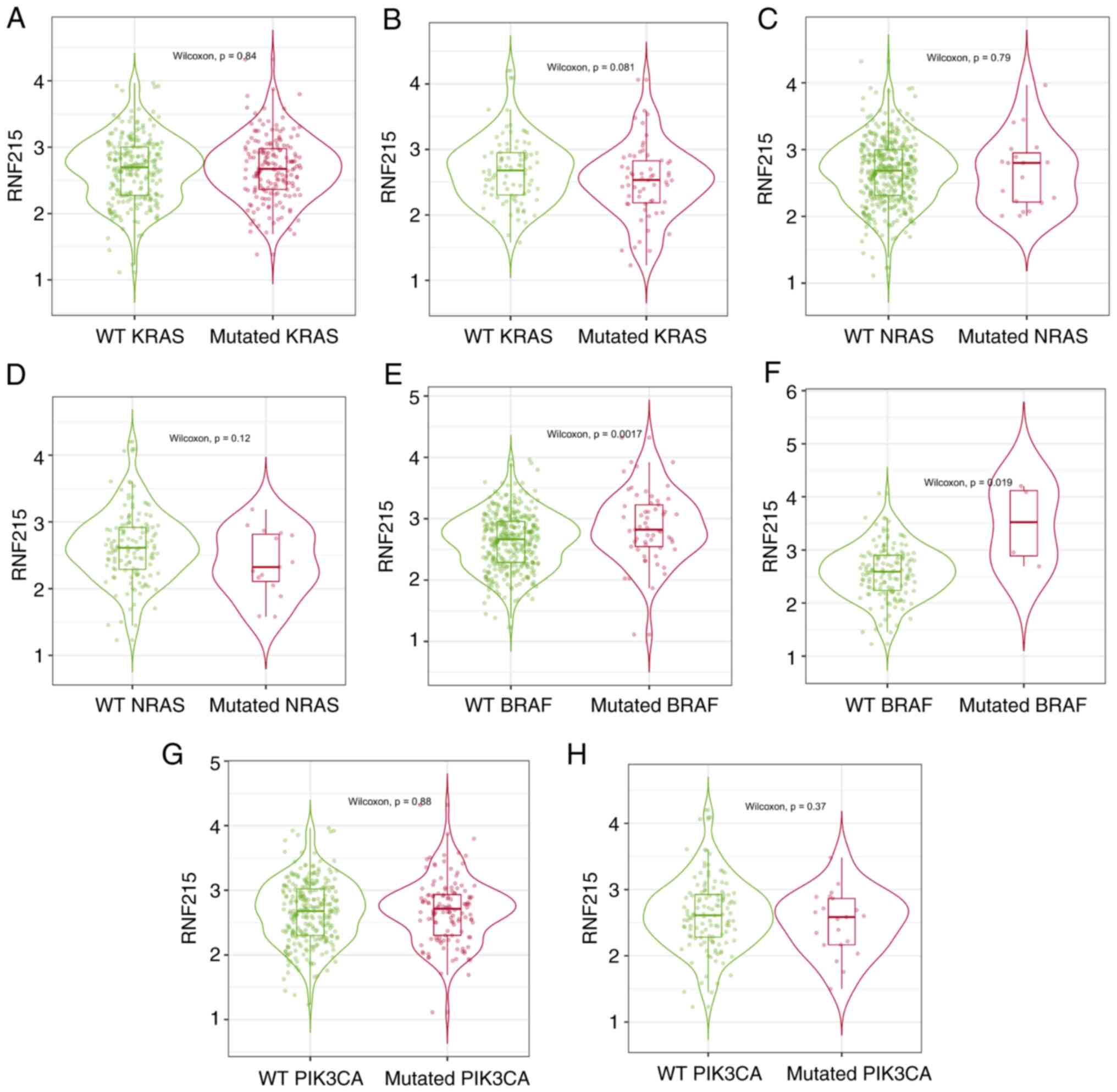
database (Fig. 5), RNF215
expression was significantly higher in the mutated BRAF group than
in the wild-type BRAF group in CRC (P<0.05). However, no
significant differences were identified between the mutated KRAS,
NRAS and PIK3CA groups and their corresponding wild-type groups
(P>0.05) in patients with CRC. To further validate the
expression level of RNF215 in CRC samples with KRAS, NRAS, BRAF and
PIK3CA mutations, RNF215 immunoassays with 182 CRC samples were
performed. Interestingly, the IHC results revealed no significant
differences between the mutated and corresponding wild-type groups
(all P>0.05) (Fig. 6).
Discussion
CRC is one of the most prevalent malignant tumors
worldwide (3-5).
In the past few decades, great advances have been made in the
clinical treatment of CRC through improvements in the understanding
of its pathophysiology and molecular mechanisms. However, CRC is a
heterogeneous disease with different treatment responses and
prognoses (24). Therefore, it is
necessary to identify molecular markers with predictive or
prognostic value. EGFR has been suggested as a target for the
treatment of CRC, and KRAS gene mutations play a dominant role in
resistance to EGFR inhibitors. However, there are other possible
mechanisms underlying this resistance; these mechanisms include
ligand expression, increased EGFR copy number, BRAF gene mutations
and activation of other signaling pathways (25). Therefore, gene mutations in exons 2,
3 and 4 of KRAS and NRAS, exon 15 of BRAF, and exon 20 of PIK3CA
have been recognized for their predictive value in
anti-EGFR-targeted therapy (26).
Thus, in the present study, these exons of the aforementioned genes
were analyzed in 182 patients with CRC, aiming to provide reference
data for clinical treatment.
In CRC, the RAS family is the most frequently
researched malignant gene family. In this family, the KRAS gene is
the most commonly researched family member. KRAS can activate the
downstream PI3K pathway and affect cell proliferation and
differentiation. Reportedly, the KRAS mutation rate in patients
with CRC is 30-50% (27,28). KRAS mutations most commonly occur in
codons 12 and 13 of exon 2 and codons 59 and 61 of exon 3(21). In the present study, the mutation
rate of the KRAS gene alone was 40.7% (74/182), and the concomitant
mutation rate of KRAS and other genes was 4.4% (8/182). These
mutations mainly occurred in the 12 and 13th codons of exon 2,
which demonstrated the highest mutation rate (61.5%, 64/104). The
mutation rate of the Q61 codon of exon 3 was 3.3% (6/182). The
mutation rate of the K117 and A146 codons in exon 4 was 6.6%
(12/182), which was consistent with the results in related studies
(29). Numerous studies have
claimed that KRAS gene mutations are more likely to occur in women
and right-sided colon cancer patients (27,30).
Chang et al (31) suggested
that KRAS gene mutations are associated with pathological
differentiation and the number of metastatic lymph nodes. A
previous study also suggested correlations among patient age, tumor
site and KRAS mutations (32).
However, no significant association between KRAS gene mutations and
the clinicopathological characteristics of patients was found in
the present study (P>0.05), which may be related to ethnic and
regional differences and the sample size. Therefore, this finding
requires further validation with a larger sample size.
NRAS is a common oncogene in human tumors and an
important member of the RAS gene family. The NRAS mutation rate in
CRC has been reported to be 2.2-7% (33,34).
In the present study, the total NRAS mutation rate was 6.0%
(11/182). The rate of mutation in codons 12 and 13 in exon 2 was
4.4% (8/182), that in codon Q61 of exon 3 was 1.1% (2/182), and
that in codon A146T of exon 4 was 0.5% (1/182), which was
consistent with the findings of a previous study (33). A previous study by Russo et
al (35) revealed that NRAS
gene mutations were more likely to occur in older adults. However,
no significant association was found between the
clinicopathological features of CRC and NRAS mutations in the
present study (P>0.05). The differences may be associated with
sample size, detection methods, race and geographical scope.
The BRAF gene is one of the most important
proto-oncogenes in humans. Reportedly, the BRAF mutation rate in
CRC is 1.8-20% (36,37). The mutation rate in the present
study was 4.4% (8/182), consistent with previous studies (38,39).
Numerous studies have suggested that BRAF gene mutations are
associated with certain clinicopathological features, including
right-sided tumor location, poor differentiation and peritoneal
metastasis (40). A study by Siena
et al (41) suggested that
the BRAF mutation rate in female CRC patients was significantly
higher than that in male patients. The present study found that
BRAF gene mutations were more likely to occur in poorly
differentiated CRC patients (P<0.05). This may suggest that
BRAF-targeted inhibitors (including vemurafenib, dabrafenib and
encorafenib) may be beneficial in poorly differentiated CRC
patients. However, the number of BRAF-mutant cases in the present
study was too small to allow any meaningful statistical analysis,
and more cases are needed for further confirmation. In addition,
according to Siena et al (41), BRAF mutations and KRAS and NRAS
mutations are mutually exclusive. In the present study, there were
no KRAS or NRAS mutations in the 8 patients with BRAF gene
mutations, which was consistent with the study by Siena et
al (41). Guo et al
(42) suggested that the mutation
rate of the BRAF gene was higher in CRC patients with lymph node
metastasis. EGFR-targeted therapy is recommended for patients
without driver mutations in KRAS/NRAS. Lymph node metastasis is not
a criterion for targeted therapy. However, interestingly, the
present study did not find that a single BRAF gene mutation was
associated with the presence of lymph node metastasis. However, the
total mutation rate of KRAS/NRAS/BRAF/PIK3CA in patients with lymph
node metastasis was 76.1% (35/46), which was significantly higher
than that in patients without lymph node metastasis (50.8%, 69/136)
(P<0.05). EGF selectively binds to EGFR and triggers the
receptor to form a dimer that activates RAS which can transmit
signals from the activated transmembrane receptor EGFR to effectors
in the MAPK and PI3K/AKT signaling pathways in the cytoplasm,
regulating cell survival and proliferation (43). This finding suggests that
EGFR-targeted therapy may be beneficial for patients with CRC and
lymph node metastasis, providing a meaningful reference index for
the clinical identification of these patients, although this
finding does need to be confirmed in a larger sample size.
As one of the common proto-oncogenes in CRC, PIK3CA
participates in regulating cell proliferation and differentiation,
apoptosis, and other functions by activating the PI3K-AKT-mTOR
pathway. When PIK3CA is mutated, it can cause continuous abnormal
activation of the aforementioned pathway, resulting in the
development of CRC. Chen et al (44) confirmed that aspirin enhances the
cytotoxic effect of RSL3 in PIK3CA-mutated CRC, and the combination
of aspirin and a ferroptosis inducer showed promising therapeutic
effects in CRC treatment. In recent years, it has been shown that
in addition to KRAS mutations, PIK3CA gene mutations are common in
patients with CRC, and the potential clinical value of PIK3CA
mutation as a tumor marker and molecular target has been studied.
Studies have shown that PIK3CA has a high mutation rate of 14-32%
in Western CRC patients (45,46).
In the present study, 11 mutations of the PIK3CA gene were
confirmed, a mutation rate of 6.0% (11/182). A total of five of the
samples with PIK3CA mutations also had mutations in the KRAS gene.
The low mutation rate of PIK3CA in the present study may be related
to the low mutation rate of PIK3CA in Chinese CRC patients and the
fact that the current analysis only assessed exon 20 (a high
mutation frequency region) of the PIK3CA gene. Studies have
suggested that mutations in PIK3CA are often accompanied by
mutations in other genes (8),
especially KRAS, and the present study found similar results. Ye
et al (27) found that the
most common mutation was in exon 9 in the PIK3CA gene, and the exon
9 mutation of PIK3CA depended on the RAS-GTP mode. The mutation of
exon 20 did not involve Ras. The study by Jang et al
(28) revealed an association
between certain KRAS and PIK3CA variants and aggressive
clinicopathological characteristics. The present study also found
that PIK3CA gene mutations were related to the site of tumor
occurrence, but because of the small sample size, it was not
possible to perform additional analyses. Moreover, Mao et al
(47) suggested that mutations in
exon 20 of the PIK3CA gene are a biomarker of anti-EGFR monoclonal
antibody resistance in patients with KRAS wild-type metastatic CRC,
and failure of anti-EGFR therapy in KRAS wild-type patients may be
caused by PIK3CA gene mutations.
In patients with CRC, EGFR-mediated signaling
pathway activation via ligand binding can lead to the activation of
the two major downstream signaling pathways (RAS/RAF/MAPK and
PI3K/AKT/mTOR). PIK3CA gene mutations are often accompanied by KRAS
mutations (the concomitant mutation rate in the present study was
45.4%), and studies have shown that EGFR-targeted therapy is
beneficial for wild-type CRC patients without KRAS and PIK3CA
mutations. In addition, KRAS wild-type patients with PIK3CA
mutations are more likely to develop resistance to EGFR-targeted
therapy, which may be due to EGFR-targeted drugs blocking the
RAS/RAF/MAPK pathway. However, cell proliferation can still be
caused by the activation of PI3K/AKT/mTOR. Hence, it is considered
that the failure of EGFR-targeted treatment in KRAS wild-type
patients may be associated with mutations in the PIK3CA gene, which
may represent a new mechanism underlying EGFR-targeted treatment
failure (10). Therefore, a
combined assessment of multiple genes is crucial for the selection
of treatment options, and PIK3CA gene mutations may become a new
target for the treatment of CRC. When choosing a molecularly
targeted therapy plan for patients with CRC, multiple genes
involved in the signaling pathways being targeted should be
assessed before supporting the development of a more precise
treatment plan (48).
Although the present study showed that the
KRAS/NRAS/BRAF/PIK3CA gene mutation in patients with lymph node
metastasis was significantly higher than that in patients without
lymph node metastasis, the present study also suggested that KRAS,
NRAS, BRAF and PIK3CA mutations were not significantly associated
with IHC protein expression. It is considered that this may be
attributed to the fact that the primary anti-KRAS antibody used in
the present study was a polyclonal antibody, and although the
antibodies used to detect NRAS, BRAF and PIK3CA were monoclonal
antibodies, only common hotspot mutation sites for gene mutations
were detected, and these hotspot mutation sites were not detected
in the IHC analysis (NRAS, clone sp174; BRAF, clone VE1; PIK3CA,
clone sp139). In addition, it is also considered that the
sensitivity and specificity of the IHC antibodies may also need to
be further improved. Another explanation may be the fact that only
182 Chinese CRC cases were analysed. Therefore, although IHC is
more convenient and inexpensive, the results of the present study,
suggested that KRAS, NRAS, BRAF and PIK3CA gene mutations in CRC
patients cannot be detected by IHC. DNA sequencing and ARMS are
still the most effective means of detecting genetic mutations.
In addition, 39 patients were alive without evidence
of tumor metastasis in the present cohort study. This could be
caused by the following reasons: First, some patients had a short
follow-up time, and when the follow-up time is extended, distant
metastasis may occur in these patients. Second, some patients were
early-stage patients, therefore, they may receive timely treatment
and did not develop metastasis. Third, the number of specimens in
the present study was relatively small, with only 182 patients, and
if the number of specimens is sufficiently large, metastasis might
occur in more patients. Finally, this might be related to race, as
all the patients selected for the present study, were Chinese
patients with CRC, and it is possible that more patients with
distant metastases would have developed if more racial patients
from multiple centers had been included.
In recent years, it has been postulated that genetic
mutations can be used for the early diagnosis of CRC. He et
al (49) suggested that fecal
TP53 and KRAS could be used as specific genes for CRC screening,
diagnosis, prognosis prediction and recurrence monitoring (49). Similar results were obtained by Lin
et al (50), who found that
the combined detection of fecal KRAS/BRAF/APC mutations and
SFRP2/SDC2 methylation had potential application value for the
auxiliary diagnosis of CRC. It has been also hypothesized that
independent clones with pathogenic KRAS and TP53 mutations are
common in individuals with CRC (51). Alizadeh-Sedigh et al
(52) found that a panel
identifying PIK3CA, KRAS and BRAF mutations had favorable
performance in detecting CRC DNA in plasma circulating-free DNA.
According to the results of the gene mutation analysis of the
present study and literature review, it was hypothesized that
KRAS/NRAS/BRAF/PIK3CA gene mutations could be biomarkers of
carcinogenesis in colorectal adenoma patients. However, this needs
to be further verified with further research.
Increasing attention has been given to personalized
targeted therapy for CRC. Some targeted drugs, such as cetuximab,
panitumumab and bevacizumab, have been shown to have a positive
effect in the treatment of CRC patients. KRAS mutations are a poor
prognostic factor in CRC patients (53), and mutations in codons 12 and 13
specifically are potentially associated with reduced efficacy of
anti-EGFR monoclonal antibodies (54,55).
The present study also suggested that KRAS wild-type CRC patients
had longer OS and DFS than KRAS-mutated CRC patients. This may be
related to the fact that the majority of the KRAS wild-type
patients received targeted therapy (cetuximab and panitumumab), and
numerous patients benefit greatly from these targeted drugs in the
clinic. However, some researchers have shown that KRAS G12C can be
targeted in KRAS-mutated patients by a covalent compound that locks
the mutant protein in its inactive GDP-bound state (56). Furthermore, NRAS, BRAF and PIK3CA
mutations may negatively affect the response to EGFR inhibitors.
Patients with BRAF (4.7%), PIK3CA exon 20 (3%), and NRAS mutations
(2%) had a lower response rate to cetuximab plus chemotherapy
(57). Cathomas (55) suggested that PIK3CA mutations were
associated with poorer clinical outcomes and poor response to
targeted therapy with anti-EGFR monoclonal antibodies in CRC
patients with wild-type RAS. Moreover, it may be possible to design
vaccines for RAS, BRAF, or PIK3CA mutant peptides or
immunotherapies using polyclonal T cells to target these gene
mutations in the future.
The present study assessed the relationship between
KRAS, NRAS, BRAF and PIK3A mutations and RNF215 expression for the
first time. No significant differences in RNF215 expression were
found between the mutated KRAS, NRAS and PIK3CA groups and their
corresponding wild-type groups (P>0.05) in CRC patients. RNF215
expression was significantly higher in the mutated BRAF group than
in the wild-type BRAF group according to the TIMER 2.0 database,
indicating that BRAF mutations may be associated with RNF215
expression. However, interestingly, the immunostaining results of
the present study did not show a significant difference. This may
be caused by the small number of patients in the present study,
with only three patients with BRAF mutations. Therefore, the
association between KRAS, NRAS, BRAF, and PIK3CA mutations and
RNF215 expression needs to be further investigated in more patients
in the future. According to a previous study by the authors
(20), RNF215 was associated with
several important pathways involved in CRC occurrence, including
MAPK signaling pathway and the RAS signaling pathway. Therefore, it
was considered that BRAF may regulate the RNF215 expression via the
MAPK signaling pathway. However, the detailed mechanism by which
BRAF regulates RNF215 expression needs to be further confirmed
because of the lack of relevant studies.
In addition, apart from CRC, through a search in
PubMed, three types of tumors that may be associated with the
KRAS/NRAS/BRAF/PIK3CA axis were identified, including lung cancer,
ameloblastoma and ovarian cancer. Seo et al (58) identified driver somatic mutations in
EGFR, KRAS, NRAS, BRAF and PIK3CA in lung adenocarcinoma. Nguyen
et al (59) revealed driver
mutations in FGFR2, KRAS, NRAS, PIK3CA and SMO in ameloblastoma.
Somatic mutations were identified in KRAS, NRAS, BRAF, PIK3CA, EGFR
and PTEN in ovarian cancer patients by Despierre et al
(60). Rachiglio et al
(61) investigated the presence of
hotspot mutations in genes, including KRAS, NRAS, BRAF, ERBB2,
PIK3CA and MET, in patients with non-small cell lung cancer.
It appears that the collection of samples in the
present study is quite biased to KRAS mutation and there are
numerous KRAS mutations. However, in fact, CRC cases were not
intentionally selected, and this result is a true reflection of the
KRAS, NRAS, BRAF and PIK3CA mutation status of these cases, with
similar findings to some recent studies (62,63).
The KRAS mutation rate was high, and it is considered that this may
be due to the limitations of the present study's analysis. First,
the present study was performed on a relatively small number of
patients at a single center, with only 182 patients, and the
limited sample size prevented the authors from drawing firm
conclusions. Second, ARMS was utilized to detect
KRAS/NRAS/BRAF/PIK3CA gene mutations and selected coding regions of
the four genes were only analyzed, and the KRAS hotspot (up to 17)
mutation was the highest, as shown in Table II. Therefore, all mutation sites
were not assessed with NGS or other sequencing analyses. Other
limitations in the present study include the retrospective nature
of the analysis, the small number of genes analyzed, the limited
number of exons tested for each gene, ethnic and regional
differences. In addition, some of the statistically significant
associations between the mutated genes and clinicopathological
features may be random effects based on the limited number of
cases. In summary, there are few overall data, and more research is
needed. Furthermore, there is very limited information regarding
the prognostic/predictive value of simultaneous detection of these
genetic alterations in the same tumor. Therefore, in the future
studies, the authors will collaborate with multiple research
centers to include CRC patients from more geographically diverse
populations to further validate the present findings.
In summary, in the present study, more than 50% of
the patients with CRC had one or more gene mutations. KRAS
mutations were the most common mutations, mainly in codons 12 and
13 of exon 2, while mutations in NRAS, BRAF and PIK3CA were
relatively rare. KRAS wild-type CRC patients could benefit from
EGFR-targeted drugs, and KRAS mutations may be a poor prognostic
factor in CRC patients. The simultaneous detection of
KRAS/NRAS/BRAF/PIK3CA gene mutations is conducive to the
development of the most suitable treatment regimen for CRC patients
and will help provide new targets for the development of new
therapeutic drugs for CRC. It was found that KRAS/NRAS/BRAF/PIK3CA
gene mutations were associated with certain clinicopathological
characteristics of CRC patients. The present study also showed that
PIK3CA mutations may often be accompanied by KRAS mutations.
Therefore, PIK3CA gene mutations may prevent patients with
wild-type KRAS from benefiting from EGFR-targeted therapy. In
addition, the present study also demonstrated that BRAF mutations
may be associated with RNF215 expression. However, these findings
need to be further verified in larger cohorts. In addition, the
results of the present study indicated that gene mutations in CRC
patients are best detected by DNA sequencing or ARMS.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the High-level
Professional Physician Training Program of Minhang, Shanghai (grand
no. 2020MZYS10) and the Natural Science Research Project of the
Science and Technology Commission in Minhang, Shanghai (grand no.
2022MHZ043).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JBW conceptualized the study, developed methodology
and wrote the original draft. HL and YJL conducted the research and
provided experimental suggestions. XJL performed data curation and
analysis. XPL designed the study and performed the final revision
of the manuscript. All authors have read and approved the final
version of the manuscript. JBW and XPL confirm the authenticity of
all the raw data.
Ethics approval and consent to
participate
The studies were reviewed and approved by the
Ethical Committee of Shanghai Fifth People's Hospital, Fudan
University (approval no. 2021071; Shanghai, China). Written
informed consent was provided by all patients/participants to
participate in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36
Cancers in 185 Countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Li N, Lu B, Luo C, Cai J, Lu M, Zhang Y,
Chen H and Dai M: Incidence, mortality, survival, risk factor and
screening of colorectal cancer: A comparison among China, Europe,
and northern America. Cancer Lett. 522:255–268. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zheng ZX, Zheng RS, Zhang SW and Chen WQ:
Colorectal cancer incidence and mortality in China, 2010. Asian Pac
J Cancer Prev. 15:8455–8460. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang L, Cao F, Zhang G, Shi L, Chen S,
Zhang Z, Zhi W and Ma T: Trends in and predictions of colorectal
cancer incidence and mortality in China from 1990 to 2025. Front
Oncol. 9(98)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Reggiani Bonetti L, Barresi V, Bettelli S,
Caprera C, Manfredini S and Maiorana A: Analysis of KRAS, NRAS,
PIK3CA, and BRAF mutational profile in poorly differentiated
clusters of KRAS-mutated colon cancer. Hum Pathol. 62:91–98.
2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Barresi V, Reggiani Bonetti L, Vitarelli
E, Di Gregorio C, Ponz de Leon M and Barresi G: Immunohistochemical
assessment of lymphovascular invasion in stage I colorectal
carcinoma: Prognostic relevance and correlation with nodal
micrometastases. Am J Surg Pathol. 36:66–72. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hossain MS, Karuniawati H, Jairoun AA,
Urbi Z, Ooi J, John A, Lim YC, Kibria KMK, Mohiuddin AKM, Ming LC,
et al: Colorectal cancer: A review of carcinogenesis, global
epidemiology, current challenges, risk factors, preventive and
treatment strategies. Cancers (Basel). 14(1732)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liu J, Hu J, Cheng L, Ren W, Yang M, Liu
B, Xie L and Qian X: Biomarkers predicting resistance to epidermal
growth factor receptor-targeted therapy in metastatic colorectal
cancer with wild-type KRAS. Onco Targets Ther. 9:557–565.
2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Palomba G, Doneddu V, Cossu A,
Paliogiannis P, Manca A, Casula M, Colombino M, Lanzillo A, Defraia
E, Pazzola A, et al: Prognostic impact of KRAS, NRAS, BRAF, and
PIK3CA mutations in primary colorectal carcinomas: A
population-based study. J Transl Med. 14(292)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li QH, Wang YZ, Tu J, Liu CW, Yuan YJ, Lin
R, He WL, Cai SR, He YL and Ye JN: Anti-EGFR therapy in metastatic
colorectal cancer: Mechanisms and potential regimens of drug
resistance. Gastroenterol Rep (Oxf). 8:179–191. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
De Roock W, De Vriendt V, Normanno N,
Ciardiello F and Tejpar S: KRAS, BRAF, PIK3CA, and PTEN mutations:
Implications for targeted therapies in metastatic colorectal
cancer. Lancet Oncol. 12:594–603. 2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cremolini C, Benelli M, Fontana E, Pagani
F, Rossini D, Fucà G, Busico A, Conca E, Di Donato S, Loupakis F,
et al: Benefit from anti-EGFRs in RAS and BRAF wild-type metastatic
transverse colon cancer: A clinical and molecular proof of concept
study. ESMO Open. 4(e000489)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
McCubrey JA, Steelman LS, Abrams SL, Lee
JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA,
D'Assoro AB, et al: Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT
pathways in malignant transformation and drug resistance. Adv
Enzyme Regul. 46:249–279. 2006.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Liao X, Lochhead P, Nishihara R, Morikawa
T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, et
al: Aspirin use, tumor PIK3CA mutation, and
colorectal-cancer survival. N Engl J Med. 367:1596–1606.
2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wojas-Krawczyk K, Kalinka-Warzocha E,
Reszka K, Nicoś M, Szumiło J, Mańdziuk S, Szczepaniak K, Kupnicka
D, Lewandowski R, Milanowski J and Krawczyk P: Analysis of KRAS,
NRAS, BRAF, and PIK3CA mutations could predict metastases in
colorectal cancer: A preliminary study. Adv Clin Exp Med. 28:67–73.
2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wu Y, Chen D, Hu Y, Zhang S, Dong X, Liang
H, Liang M, Zhu Y, Tan C, An S, et al: Ring finger protein 215
negatively regulates type I IFN production via blocking NF-κB p65
activation. J Immunol. 209:2012–2021. 2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ma J, Li R and Wang J: Characterization of
a prognostic fourgene methylation signature associated with
radiotherapy for head and neck squamous cell carcinoma. Mol Med
Rep. 20:622–632. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
McIntosh LA, Marion MC, Sudman M, Comeau
ME, Becker ML, Bohnsack JF, Fingerlin TE, Griffin TA, Haas JP,
Lovell DJ, et al: Genome-Wide association meta-analysis reveals
novel juvenile idiopathic arthritis Susceptibility Loci. Arthritis
Rheumatol. 69:2222–2232. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wu JB, Li XJ, Liu H and Liu XP: Ring
finger protein 215 is a potential prognostic biomarker involved in
immune infiltration and angiogenesis in colorectal cancer.
Biomedical Reports. 19(50)2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lindner AU, Carberry S, Monsefi N, Barat
A, Salvucci M, O'Byrne R, Zanella ER, Cremona M, Hennessy BT,
Bertotti A, et al: Systems analysis of protein signatures
predicting cetuximab responses in KRAS, NRAS, BRAF and PIK3CA
wild-type patient-derived xenograft models of metastatic colorectal
cancer. Int J Cancer. 147:2891–2901. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kononen J, Bubendorf L, Kallioniemi A,
Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G
and Kallioniemi OP: Tissue microarrays for high-throughput
molecular profiling of tumor specimens. Nat Med. 4:844–847.
1998.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Peng J, Huang D, Poston G, Ma X, Wang R,
Sheng W, Zhou X, Zhu X and Cai S: The molecular heterogeneity of
sporadic colorectal cancer with different tumor sites in Chinese
patients. Oncotarget. 8:49076–49083. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yao S, Wang X, Li C, Zhao T, Jin H and
Fang W: Kaempferol inhibits cell proliferation and glycolysis in
esophagus squamous cell carcinoma via targeting EGFR signaling
pathway. Tumour Biol. 37:10247–10256. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang B, Wu S, Huang F, Shen M, Jiang H, Yu
Y, Yu Q, Yang Y, Zhao Y, Zhou Y, et al: Analytical and clinical
validation of a novel amplicon-based NGS assay for the evaluation
of circulating tumor DNA in metastatic colorectal cancer patients.
Clin Chem Lab Med. 57:1501–1510. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ye ZL, Qiu MZ, Tang T, Wang F, Zhou YX,
Lei MJ, Guan WL and He CY: Gene mutation profiling in Chinese
colorectal cancer patients and its association with
clinicopathological characteristics and prognosis. Cancer Med.
9:745–756. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Jang S, Hong M, Shin MK, Kim BC, Shin HS,
Yu E, Hong SM, Kim J, Chun SM, Kim TI, et al: KRAS and PIK3CA
mutations in colorectal adenocarcinomas correlate with aggressive
histological features and behavior. Hum Pathol. 65:21–30.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li ZZ, Wang F, Zhang ZC, Wang F, Zhao Q,
Zhang DS, Wang FH, Wang ZQ, Luo HY, He MM, et al: Mutation
profiling in Chinese patients with metastatic colorectal cancer and
its correlation with clinicopathological features and anti-EGFR
treatment response. Oncotarget. 7:28356–28368. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Fan JZ, Wang GF, Cheng XB, Dong ZH, Chen
X, Deng YJ and Song X: Relationship between mismatch repair
protein, RAS, BRAF, PIK3CA gene expression and clinicopathological
characteristics in elderly colorectal cancer patients. World J Clin
Cases. 9:2458–2468. 2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chang XN, Shang FM, Jiang HY, Chen C, Zhao
ZY, Deng SH, Fan J, Dong XC, Yang M, Li Y, et al:
Clinicopathological features and prognostic value of KRAS/NRAS/BRAF
mutations in colorectal cancer patients of central China. Curr Med
Sci. 41:118–126. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chang YY, Lin PC, Lin HH, Lin JK, Chen WS,
Jiang JK, Yang SH, Liang WY and Chang SC: Mutation spectra of RAS
gene family in colorectal cancer. Am J Surg. 212:537–544.e3.
2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zheng G, Tseng LH, Haley L, Ibrahim J,
Bynum J, Xian R, Gocke CD, Eshleman JR and Lin MT: Clinical
validation of coexisting driver mutations in colorectal cancers.
Hum Pathol. 86:12–20. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Bokemeyer C, Kohne CH, Ciardiello F, Lenz
HJ, Heinemann V, Klinkhardt U, Beier F, Duecker K, van Krieken JH
and Tejpar S: FOLFOX4 plus cetuximab treatment and RAS mutations in
colorectal cancer. Eur J Cancer. 51:1243–1252. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Russo AL, Borger DR, Szymonifka J, Ryan
DP, Wo JY, Blaszkowsky LS, Kwak EL, Allen JN, Wadlow RC, Zhu AX, et
al: Mutational analysis and clinical correlation of metastatic
colorectal cancer. Cancer. 120:1482–1490. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Knickelbein K and Zhang L: Mutant KRAS as
a critical determinant of the therapeutic response of colorectal
cancer. Genes Dis. 2:4–12. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Jass JR: Classification of colorectal
cancer based on correlation of clinical, morphological and
molecular features. Histopathology. 50:113–130. 2007.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zeng C, Wang M, Xie S, Wang N, Wang Z, Yi
D, Kong F and Chen L: Clinical research progress on BRAF
V600E-mutant advanced colorectal cancer. J Cancer Res Clin Oncol:
Aug 28, 2023 doi: 10.1007/s00432-023-05301-0 (Epub ahead of
print).
|
|
39
|
Zeng J, Fan W, Li J, Wu G and Wu H:
KRAS/NRAS mutations associated with distant metastasis and
BRAF/PIK3CA mutations associated with poor tumor differentiation in
colorectal cancer. Int J Gen Med. 16:4109–4120. 2023.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chen D, Huang JF, Liu K, Zhang LQ, Yang Z,
Chuai ZR, Wang YX, Shi DC, Huang Q and Fu WL: BRAFV600E mutation
and its association with clinicopathological features of colorectal
cancer: A systematic review and meta-analysis. PLoS One.
9(e90607)2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Siena S, Sartore-Bianchi A, Di
Nicolantonio F, Balfour J and Bardelli A: Biomarkers predicting
clinical outcome of epidermal growth factor receptor-targeted
therapy in metastatic colorectal cancer. J Natl Cancer Inst.
101:1308–1324. 2009.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Guo F, Gong H, Zhao H, Chen J, Zhang Y,
Zhang L, Shi X, Zhang A, Jin H, Zhang J and He Y: Mutation status
and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese
colorectal cancer patients. Sci Rep. 8(6076)2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Li ZN, Zhao L, Yu LF and Wei MJ: BRAF and
KRAS mutations in metastatic colorectal cancer: Future perspectives
for personalized therapy. Gastroenterol Rep (Oxf). 8:192–205.
2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Chen H, Qi Q, Wu N, Wang Y, Feng Q, Jin R
and Jiang L: Aspirin promotes RSL3-induced ferroptosis by
suppressing mTOR/SREBP-1/SCD1-mediated lipogenesis in
PIK3CA-mutatnt colorectal cancer. Redox Biol.
55(102426)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Liao X, Morikawa T, Lochhead P, Imamura Y,
Kuchiba A, Yamauchi M, Nosho K, Qian ZR, Nishihara R, Meyerhardt
JA, et al: Prognostic role of PIK3CA mutation in colorectal cancer:
cohort study and literature review. Clin Cancer Res. 18:2257–2268.
2012.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Baldus SE, Schaefer KL, Engers R, Hartleb
D, Stoecklein NH and Gabbert HE: Prevalence and heterogeneity of
KRAS, BRAF, and PIK3CA mutations in primary colorectal
adenocarcinomas and their corresponding metastases. Clin Cancer
Res. 16:790–799. 2010.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Mao C, Yang ZY, Hu XF, Chen Q and Tang JL:
PIK3CA exon 20 mutations as a potential biomarker for resistance to
anti-EGFR monoclonal antibodies in KRAS wild-type metastatic
colorectal cancer: A systematic review and meta-analysis. Ann
Oncol. 23:1518–1525. 2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Li W, Li H, Liu R, Yang X, Gao Y, Niu Y,
Geng J, Xue Y, Jin X, You Q, et al: Comprehensive analysis of the
relationship between RAS and RAF mutations and MSI status of
colorectal cancer in Northeastern China. Cell Physiol Biochem.
50:1496–1509. 2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
He SY, Li YC, Wang Y, Peng HL, Zhou CL,
Zhang CM, Chen SL, Yin JF and Lin M: Fecal gene detection based on
next generation sequencing for colorectal cancer diagnosis. World J
Gastroenterol. 28:2920–2936. 2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Lin J, Zhang L, Chen M, Chen J, Wu Y, Wang
T, Lu Y, Ba Z, Cheng X, Xu R, et al: Evaluation of combined
detection of multigene mutation and SDC2/SFRP2 methylation in stool
specimens for colorectal cancer early diagnosis. Int J Colorectal
Dis. 37:1231–1238. 2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Matas J, Kohrn B, Fredrickson J, Carter K,
Yu M, Wang T, Gui X, Soussi T, Moreno V, Grady WM, et al:
Colorectal cancer is associated with the presence of cancer driver
mutations in normal colon. Cancer Res. 82:1492–1502.
2022.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Alizadeh-Sedigh M, Mahmoodzadeh H, Fazeli
MS, Haddadi-Aghdam M and Teimoori-Toolabi L: The potential of
PIK3CA, KRAS, BRAF, and APC hotspot mutations as a non-invasive
detection method for colorectal cancer. Mol Cell Probes.
63(101807)2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ma BB, Mo F, Tong JH, Wong A, Wong SC, Ho
WM, Wu C, Lam PW, Chan KF, Chan TS, et al: Elucidating the
prognostic significance of KRAS, NRAS, BRAF and PIK3CA mutations in
Chinese patients with metastatic colorectal cancer. Asia Pac J Clin
Oncol. 11:160–169. 2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Cremolini C, Rossini D, Dell'Aquila E,
Lonardi S, Conca E, Del Re M, Busico A, Pietrantonio F, Danesi R,
Aprile G, et al: Rechallenge for patients with RAS and BRAF
Wild-type metastatic colorectal cancer with acquired resistance to
First-line cetuximab and irinotecan: A phase 2 Single-arm clinical
trial. JAMA Oncol. 5:343–350. 2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Segelov E, Thavaneswaran S, Waring PM,
Desai J, Robledo KP, Gebski VJ, Elez E, Nott LM, Karapetis CS,
Lunke S, et al: Response to cetuximab with or without irinotecan in
patients with refractory metastatic colorectal cancer harboring the
KRAS G13D mutation: Australasian Gastro-intestinal trials group
ICECREAM study. J Clin Oncol. 34:2258–2264. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ostrem JM, Peters U, Sos ML, Wells JA and
Shokat KM: K-Ras(G12C) inhibitors allosterically control GTP
affinity and effector interactions. Nature. 503:548–551.
2013.PubMed/NCBI View Article : Google Scholar
|
|
57
|
De Roock W, Claes B, Bernasconi D, De
Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V,
Papamichael D, Laurent-Puig P, et al: Effects of KRAS, BRAF, NRAS,
and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy
in chemotherapy-refractory metastatic colorectal cancer: A
retrospective consortium analysis. Lancet Oncol. 11:753–762.
2010.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Seo JS, Ju YS, Lee WC, Shin JY, Lee JK,
Bleazard T, Lee J, Jung YJ, Kim JO, Shin JY, et al: The
transcriptional landscape and mutational profile of lung
adenocarcinoma. Genome Res. 22:2109–2119. 2012.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Nguyen J, Saffari PS, Pollack AS, Vennam
S, Gong X, West RB and Pollack JR: New ameloblastoma cell lines
enable preclinical study of targeted therapies. J Dent Res.
101:1517–1525. 2022.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Despierre E, Vergote I, Anderson R, Coens
C, Katsaros D, Hirsch FR, Boeckx B, Varella-Garcia M, Ferrero A,
Ray-Coquard I, et al: Epidermal growth factor receptor (EGFR)
pathway biomarkers in the randomized phase III trial of erlotinib
versus observation in ovarian cancer patients with No evidence of
disease progression after First-line platinum-based chemotherapy.
Target Oncol. 10:583–596. 2015.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Rachiglio AM, Fenizia F, Piccirillo MC,
Galetta D, Crinò L, Vincenzi B, Barletta E, Pinto C, Ferraù F,
Lambiase M, et al: The presence of concomitant mutations affects
the activity of EGFR tyrosine kinase inhibitors in EGFR-mutant
non-small cell lung cancer (NSCLC) patients. Cancers (Basel).
11(341)2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Sclafani F, Wilson SH, Cunningham D,
Gonzalez De Castro D, Kalaitzaki E, Begum R, Wotherspoon A,
Capdevila J, Glimelius B, Roselló S, et al: Analysis of KRAS, NRAS,
BRAF, PIK3CA and TP53 mutations in a large prospective series of
locally advanced rectal cancer patients. Int J Cancer. 146:94–102.
2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Mahdi Y, Khmou M, Souadka A, Agouri HE,
Ech-Charif S, Mounjid C and Khannoussi BE: Correlation between KRAS
and NRAS mutational status and clinicopathological features in 414
cases of metastatic colorectal cancer in Morocco: The largest North
African case series. BMC Gastroenterol. 23(193)2023.PubMed/NCBI View Article : Google Scholar
|