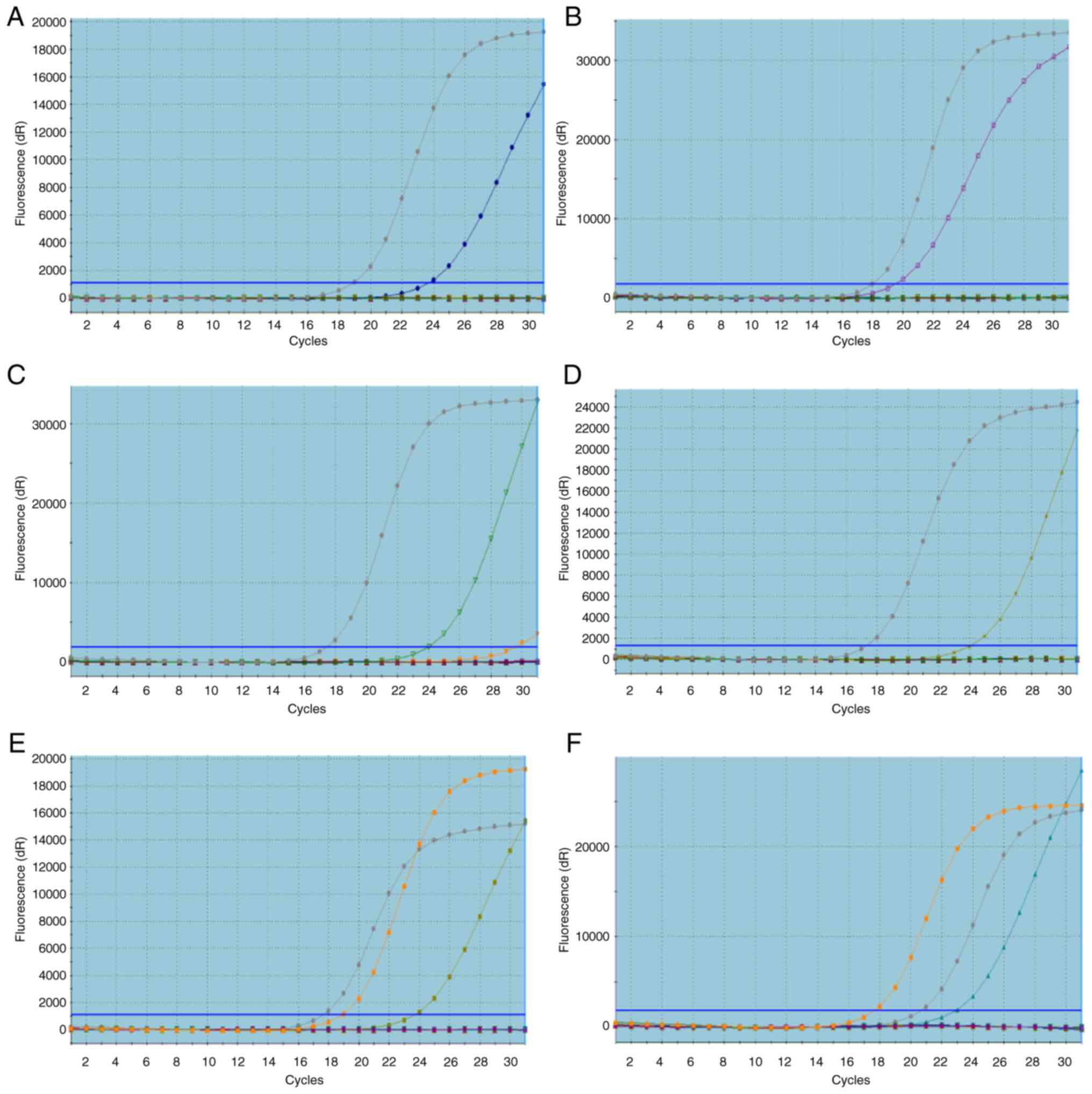
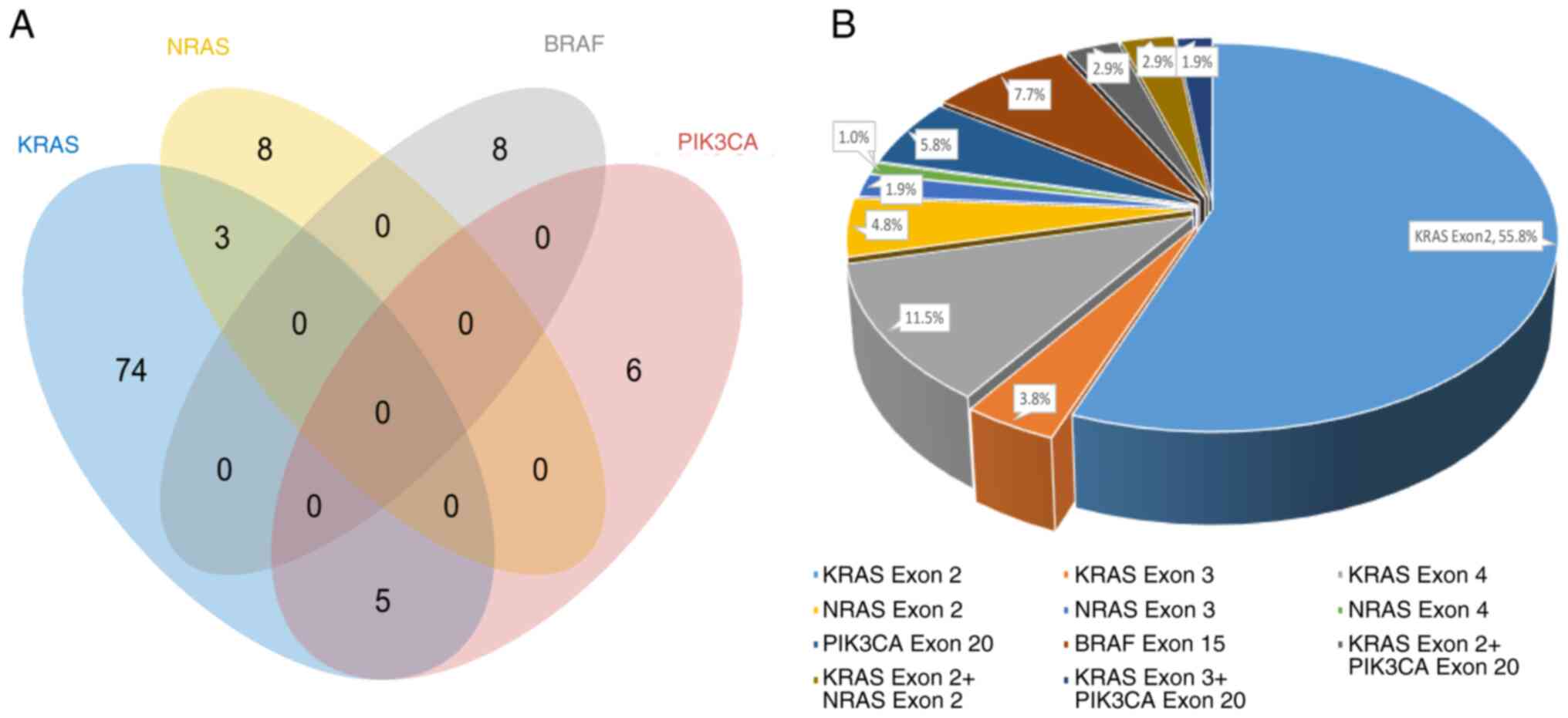
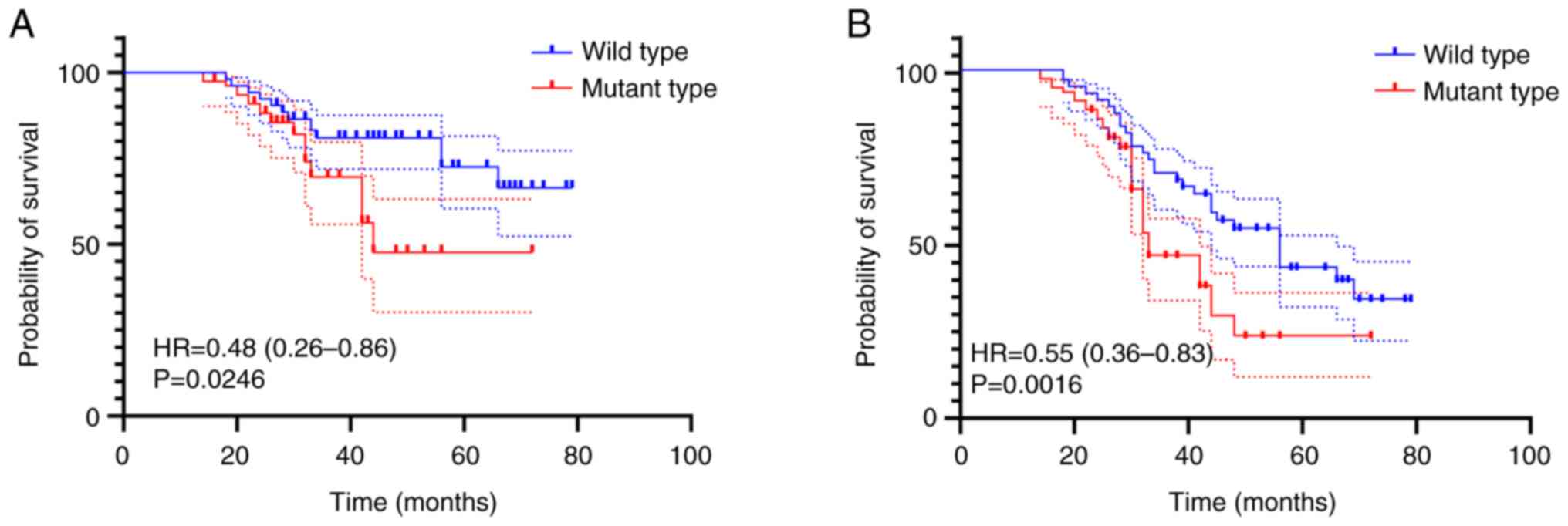
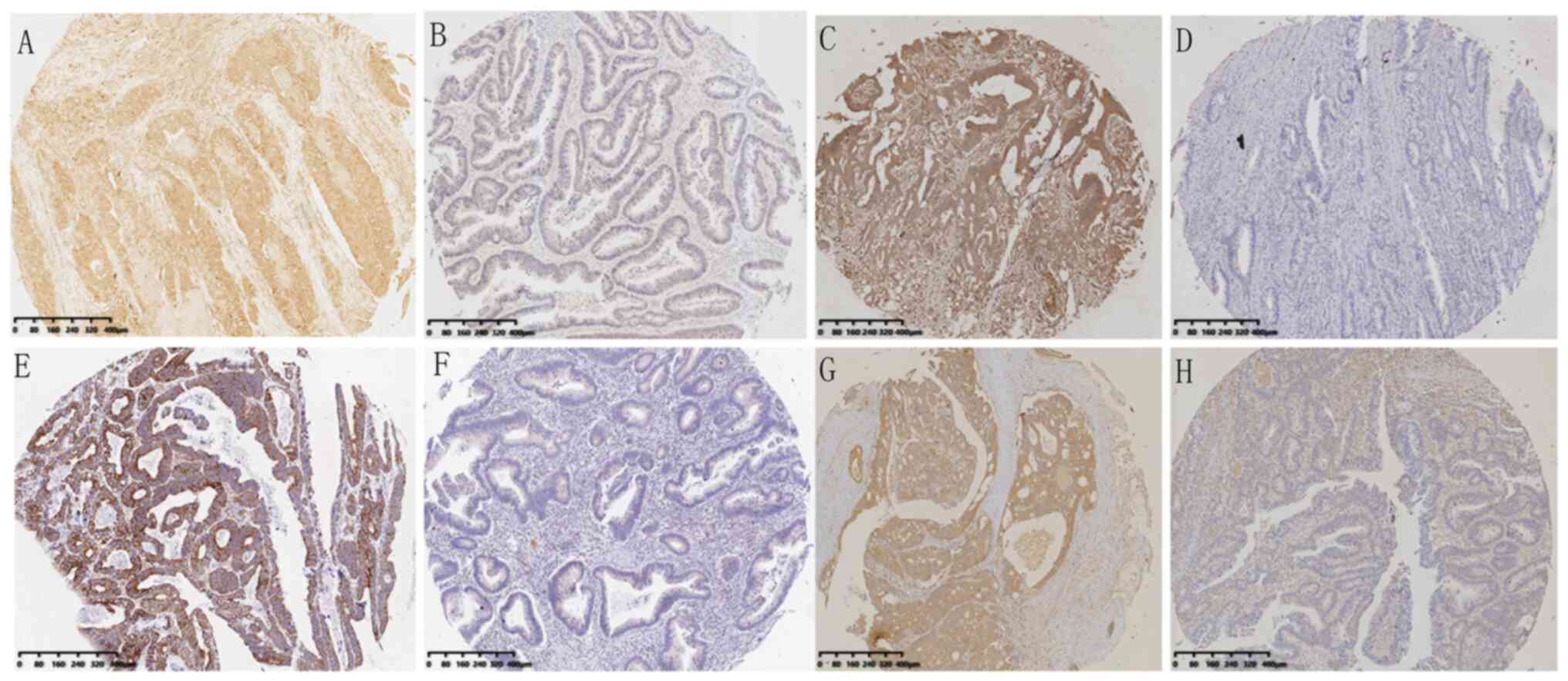
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36
Cancers in 185 Countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Li N, Lu B, Luo C, Cai J, Lu M, Zhang Y,
Chen H and Dai M: Incidence, mortality, survival, risk factor and
screening of colorectal cancer: A comparison among China, Europe,
and northern America. Cancer Lett. 522:255–268. 2021.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zheng ZX, Zheng RS, Zhang SW and Chen WQ:
Colorectal cancer incidence and mortality in China, 2010. Asian Pac
J Cancer Prev. 15:8455–8460. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang L, Cao F, Zhang G, Shi L, Chen S,
Zhang Z, Zhi W and Ma T: Trends in and predictions of colorectal
cancer incidence and mortality in China from 1990 to 2025. Front
Oncol. 9(98)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Reggiani Bonetti L, Barresi V, Bettelli S,
Caprera C, Manfredini S and Maiorana A: Analysis of KRAS, NRAS,
PIK3CA, and BRAF mutational profile in poorly differentiated
clusters of KRAS-mutated colon cancer. Hum Pathol. 62:91–98.
2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Barresi V, Reggiani Bonetti L, Vitarelli
E, Di Gregorio C, Ponz de Leon M and Barresi G: Immunohistochemical
assessment of lymphovascular invasion in stage I colorectal
carcinoma: Prognostic relevance and correlation with nodal
micrometastases. Am J Surg Pathol. 36:66–72. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hossain MS, Karuniawati H, Jairoun AA,
Urbi Z, Ooi J, John A, Lim YC, Kibria KMK, Mohiuddin AKM, Ming LC,
et al: Colorectal cancer: A review of carcinogenesis, global
epidemiology, current challenges, risk factors, preventive and
treatment strategies. Cancers (Basel). 14(1732)2022.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liu J, Hu J, Cheng L, Ren W, Yang M, Liu
B, Xie L and Qian X: Biomarkers predicting resistance to epidermal
growth factor receptor-targeted therapy in metastatic colorectal
cancer with wild-type KRAS. Onco Targets Ther. 9:557–565.
2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Palomba G, Doneddu V, Cossu A,
Paliogiannis P, Manca A, Casula M, Colombino M, Lanzillo A, Defraia
E, Pazzola A, et al: Prognostic impact of KRAS, NRAS, BRAF, and
PIK3CA mutations in primary colorectal carcinomas: A
population-based study. J Transl Med. 14(292)2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li QH, Wang YZ, Tu J, Liu CW, Yuan YJ, Lin
R, He WL, Cai SR, He YL and Ye JN: Anti-EGFR therapy in metastatic
colorectal cancer: Mechanisms and potential regimens of drug
resistance. Gastroenterol Rep (Oxf). 8:179–191. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
De Roock W, De Vriendt V, Normanno N,
Ciardiello F and Tejpar S: KRAS, BRAF, PIK3CA, and PTEN mutations:
Implications for targeted therapies in metastatic colorectal
cancer. Lancet Oncol. 12:594–603. 2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cremolini C, Benelli M, Fontana E, Pagani
F, Rossini D, Fucà G, Busico A, Conca E, Di Donato S, Loupakis F,
et al: Benefit from anti-EGFRs in RAS and BRAF wild-type metastatic
transverse colon cancer: A clinical and molecular proof of concept
study. ESMO Open. 4(e000489)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
McCubrey JA, Steelman LS, Abrams SL, Lee
JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA,
D'Assoro AB, et al: Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT
pathways in malignant transformation and drug resistance. Adv
Enzyme Regul. 46:249–279. 2006.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Liao X, Lochhead P, Nishihara R, Morikawa
T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, et
al: Aspirin use, tumor PIK3CA mutation, and
colorectal-cancer survival. N Engl J Med. 367:1596–1606.
2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wojas-Krawczyk K, Kalinka-Warzocha E,
Reszka K, Nicoś M, Szumiło J, Mańdziuk S, Szczepaniak K, Kupnicka
D, Lewandowski R, Milanowski J and Krawczyk P: Analysis of KRAS,
NRAS, BRAF, and PIK3CA mutations could predict metastases in
colorectal cancer: A preliminary study. Adv Clin Exp Med. 28:67–73.
2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wu Y, Chen D, Hu Y, Zhang S, Dong X, Liang
H, Liang M, Zhu Y, Tan C, An S, et al: Ring finger protein 215
negatively regulates type I IFN production via blocking NF-κB p65
activation. J Immunol. 209:2012–2021. 2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ma J, Li R and Wang J: Characterization of
a prognostic fourgene methylation signature associated with
radiotherapy for head and neck squamous cell carcinoma. Mol Med
Rep. 20:622–632. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
McIntosh LA, Marion MC, Sudman M, Comeau
ME, Becker ML, Bohnsack JF, Fingerlin TE, Griffin TA, Haas JP,
Lovell DJ, et al: Genome-Wide association meta-analysis reveals
novel juvenile idiopathic arthritis Susceptibility Loci. Arthritis
Rheumatol. 69:2222–2232. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wu JB, Li XJ, Liu H and Liu XP: Ring
finger protein 215 is a potential prognostic biomarker involved in
immune infiltration and angiogenesis in colorectal cancer.
Biomedical Reports. 19(50)2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lindner AU, Carberry S, Monsefi N, Barat
A, Salvucci M, O'Byrne R, Zanella ER, Cremona M, Hennessy BT,
Bertotti A, et al: Systems analysis of protein signatures
predicting cetuximab responses in KRAS, NRAS, BRAF and PIK3CA
wild-type patient-derived xenograft models of metastatic colorectal
cancer. Int J Cancer. 147:2891–2901. 2020.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kononen J, Bubendorf L, Kallioniemi A,
Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G
and Kallioniemi OP: Tissue microarrays for high-throughput
molecular profiling of tumor specimens. Nat Med. 4:844–847.
1998.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q,
Li B and Liu XS: TIMER2.0 for analysis of tumor-infiltrating immune
cells. Nucleic Acids Res. 48:W509–W514. 2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Peng J, Huang D, Poston G, Ma X, Wang R,
Sheng W, Zhou X, Zhu X and Cai S: The molecular heterogeneity of
sporadic colorectal cancer with different tumor sites in Chinese
patients. Oncotarget. 8:49076–49083. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yao S, Wang X, Li C, Zhao T, Jin H and
Fang W: Kaempferol inhibits cell proliferation and glycolysis in
esophagus squamous cell carcinoma via targeting EGFR signaling
pathway. Tumour Biol. 37:10247–10256. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang B, Wu S, Huang F, Shen M, Jiang H, Yu
Y, Yu Q, Yang Y, Zhao Y, Zhou Y, et al: Analytical and clinical
validation of a novel amplicon-based NGS assay for the evaluation
of circulating tumor DNA in metastatic colorectal cancer patients.
Clin Chem Lab Med. 57:1501–1510. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ye ZL, Qiu MZ, Tang T, Wang F, Zhou YX,
Lei MJ, Guan WL and He CY: Gene mutation profiling in Chinese
colorectal cancer patients and its association with
clinicopathological characteristics and prognosis. Cancer Med.
9:745–756. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Jang S, Hong M, Shin MK, Kim BC, Shin HS,
Yu E, Hong SM, Kim J, Chun SM, Kim TI, et al: KRAS and PIK3CA
mutations in colorectal adenocarcinomas correlate with aggressive
histological features and behavior. Hum Pathol. 65:21–30.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li ZZ, Wang F, Zhang ZC, Wang F, Zhao Q,
Zhang DS, Wang FH, Wang ZQ, Luo HY, He MM, et al: Mutation
profiling in Chinese patients with metastatic colorectal cancer and
its correlation with clinicopathological features and anti-EGFR
treatment response. Oncotarget. 7:28356–28368. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Fan JZ, Wang GF, Cheng XB, Dong ZH, Chen
X, Deng YJ and Song X: Relationship between mismatch repair
protein, RAS, BRAF, PIK3CA gene expression and clinicopathological
characteristics in elderly colorectal cancer patients. World J Clin
Cases. 9:2458–2468. 2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chang XN, Shang FM, Jiang HY, Chen C, Zhao
ZY, Deng SH, Fan J, Dong XC, Yang M, Li Y, et al:
Clinicopathological features and prognostic value of KRAS/NRAS/BRAF
mutations in colorectal cancer patients of central China. Curr Med
Sci. 41:118–126. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chang YY, Lin PC, Lin HH, Lin JK, Chen WS,
Jiang JK, Yang SH, Liang WY and Chang SC: Mutation spectra of RAS
gene family in colorectal cancer. Am J Surg. 212:537–544.e3.
2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zheng G, Tseng LH, Haley L, Ibrahim J,
Bynum J, Xian R, Gocke CD, Eshleman JR and Lin MT: Clinical
validation of coexisting driver mutations in colorectal cancers.
Hum Pathol. 86:12–20. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Bokemeyer C, Kohne CH, Ciardiello F, Lenz
HJ, Heinemann V, Klinkhardt U, Beier F, Duecker K, van Krieken JH
and Tejpar S: FOLFOX4 plus cetuximab treatment and RAS mutations in
colorectal cancer. Eur J Cancer. 51:1243–1252. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Russo AL, Borger DR, Szymonifka J, Ryan
DP, Wo JY, Blaszkowsky LS, Kwak EL, Allen JN, Wadlow RC, Zhu AX, et
al: Mutational analysis and clinical correlation of metastatic
colorectal cancer. Cancer. 120:1482–1490. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Knickelbein K and Zhang L: Mutant KRAS as
a critical determinant of the therapeutic response of colorectal
cancer. Genes Dis. 2:4–12. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Jass JR: Classification of colorectal
cancer based on correlation of clinical, morphological and
molecular features. Histopathology. 50:113–130. 2007.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zeng C, Wang M, Xie S, Wang N, Wang Z, Yi
D, Kong F and Chen L: Clinical research progress on BRAF
V600E-mutant advanced colorectal cancer. J Cancer Res Clin Oncol:
Aug 28, 2023 doi: 10.1007/s00432-023-05301-0 (Epub ahead of
print).
|
|
39
|
Zeng J, Fan W, Li J, Wu G and Wu H:
KRAS/NRAS mutations associated with distant metastasis and
BRAF/PIK3CA mutations associated with poor tumor differentiation in
colorectal cancer. Int J Gen Med. 16:4109–4120. 2023.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chen D, Huang JF, Liu K, Zhang LQ, Yang Z,
Chuai ZR, Wang YX, Shi DC, Huang Q and Fu WL: BRAFV600E mutation
and its association with clinicopathological features of colorectal
cancer: A systematic review and meta-analysis. PLoS One.
9(e90607)2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Siena S, Sartore-Bianchi A, Di
Nicolantonio F, Balfour J and Bardelli A: Biomarkers predicting
clinical outcome of epidermal growth factor receptor-targeted
therapy in metastatic colorectal cancer. J Natl Cancer Inst.
101:1308–1324. 2009.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Guo F, Gong H, Zhao H, Chen J, Zhang Y,
Zhang L, Shi X, Zhang A, Jin H, Zhang J and He Y: Mutation status
and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese
colorectal cancer patients. Sci Rep. 8(6076)2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Li ZN, Zhao L, Yu LF and Wei MJ: BRAF and
KRAS mutations in metastatic colorectal cancer: Future perspectives
for personalized therapy. Gastroenterol Rep (Oxf). 8:192–205.
2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Chen H, Qi Q, Wu N, Wang Y, Feng Q, Jin R
and Jiang L: Aspirin promotes RSL3-induced ferroptosis by
suppressing mTOR/SREBP-1/SCD1-mediated lipogenesis in
PIK3CA-mutatnt colorectal cancer. Redox Biol.
55(102426)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Liao X, Morikawa T, Lochhead P, Imamura Y,
Kuchiba A, Yamauchi M, Nosho K, Qian ZR, Nishihara R, Meyerhardt
JA, et al: Prognostic role of PIK3CA mutation in colorectal cancer:
cohort study and literature review. Clin Cancer Res. 18:2257–2268.
2012.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Baldus SE, Schaefer KL, Engers R, Hartleb
D, Stoecklein NH and Gabbert HE: Prevalence and heterogeneity of
KRAS, BRAF, and PIK3CA mutations in primary colorectal
adenocarcinomas and their corresponding metastases. Clin Cancer
Res. 16:790–799. 2010.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Mao C, Yang ZY, Hu XF, Chen Q and Tang JL:
PIK3CA exon 20 mutations as a potential biomarker for resistance to
anti-EGFR monoclonal antibodies in KRAS wild-type metastatic
colorectal cancer: A systematic review and meta-analysis. Ann
Oncol. 23:1518–1525. 2012.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Li W, Li H, Liu R, Yang X, Gao Y, Niu Y,
Geng J, Xue Y, Jin X, You Q, et al: Comprehensive analysis of the
relationship between RAS and RAF mutations and MSI status of
colorectal cancer in Northeastern China. Cell Physiol Biochem.
50:1496–1509. 2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
He SY, Li YC, Wang Y, Peng HL, Zhou CL,
Zhang CM, Chen SL, Yin JF and Lin M: Fecal gene detection based on
next generation sequencing for colorectal cancer diagnosis. World J
Gastroenterol. 28:2920–2936. 2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Lin J, Zhang L, Chen M, Chen J, Wu Y, Wang
T, Lu Y, Ba Z, Cheng X, Xu R, et al: Evaluation of combined
detection of multigene mutation and SDC2/SFRP2 methylation in stool
specimens for colorectal cancer early diagnosis. Int J Colorectal
Dis. 37:1231–1238. 2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Matas J, Kohrn B, Fredrickson J, Carter K,
Yu M, Wang T, Gui X, Soussi T, Moreno V, Grady WM, et al:
Colorectal cancer is associated with the presence of cancer driver
mutations in normal colon. Cancer Res. 82:1492–1502.
2022.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Alizadeh-Sedigh M, Mahmoodzadeh H, Fazeli
MS, Haddadi-Aghdam M and Teimoori-Toolabi L: The potential of
PIK3CA, KRAS, BRAF, and APC hotspot mutations as a non-invasive
detection method for colorectal cancer. Mol Cell Probes.
63(101807)2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Ma BB, Mo F, Tong JH, Wong A, Wong SC, Ho
WM, Wu C, Lam PW, Chan KF, Chan TS, et al: Elucidating the
prognostic significance of KRAS, NRAS, BRAF and PIK3CA mutations in
Chinese patients with metastatic colorectal cancer. Asia Pac J Clin
Oncol. 11:160–169. 2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Cremolini C, Rossini D, Dell'Aquila E,
Lonardi S, Conca E, Del Re M, Busico A, Pietrantonio F, Danesi R,
Aprile G, et al: Rechallenge for patients with RAS and BRAF
Wild-type metastatic colorectal cancer with acquired resistance to
First-line cetuximab and irinotecan: A phase 2 Single-arm clinical
trial. JAMA Oncol. 5:343–350. 2019.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Segelov E, Thavaneswaran S, Waring PM,
Desai J, Robledo KP, Gebski VJ, Elez E, Nott LM, Karapetis CS,
Lunke S, et al: Response to cetuximab with or without irinotecan in
patients with refractory metastatic colorectal cancer harboring the
KRAS G13D mutation: Australasian Gastro-intestinal trials group
ICECREAM study. J Clin Oncol. 34:2258–2264. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ostrem JM, Peters U, Sos ML, Wells JA and
Shokat KM: K-Ras(G12C) inhibitors allosterically control GTP
affinity and effector interactions. Nature. 503:548–551.
2013.PubMed/NCBI View Article : Google Scholar
|
|
57
|
De Roock W, Claes B, Bernasconi D, De
Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V,
Papamichael D, Laurent-Puig P, et al: Effects of KRAS, BRAF, NRAS,
and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy
in chemotherapy-refractory metastatic colorectal cancer: A
retrospective consortium analysis. Lancet Oncol. 11:753–762.
2010.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Seo JS, Ju YS, Lee WC, Shin JY, Lee JK,
Bleazard T, Lee J, Jung YJ, Kim JO, Shin JY, et al: The
transcriptional landscape and mutational profile of lung
adenocarcinoma. Genome Res. 22:2109–2119. 2012.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Nguyen J, Saffari PS, Pollack AS, Vennam
S, Gong X, West RB and Pollack JR: New ameloblastoma cell lines
enable preclinical study of targeted therapies. J Dent Res.
101:1517–1525. 2022.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Despierre E, Vergote I, Anderson R, Coens
C, Katsaros D, Hirsch FR, Boeckx B, Varella-Garcia M, Ferrero A,
Ray-Coquard I, et al: Epidermal growth factor receptor (EGFR)
pathway biomarkers in the randomized phase III trial of erlotinib
versus observation in ovarian cancer patients with No evidence of
disease progression after First-line platinum-based chemotherapy.
Target Oncol. 10:583–596. 2015.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Rachiglio AM, Fenizia F, Piccirillo MC,
Galetta D, Crinò L, Vincenzi B, Barletta E, Pinto C, Ferraù F,
Lambiase M, et al: The presence of concomitant mutations affects
the activity of EGFR tyrosine kinase inhibitors in EGFR-mutant
non-small cell lung cancer (NSCLC) patients. Cancers (Basel).
11(341)2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Sclafani F, Wilson SH, Cunningham D,
Gonzalez De Castro D, Kalaitzaki E, Begum R, Wotherspoon A,
Capdevila J, Glimelius B, Roselló S, et al: Analysis of KRAS, NRAS,
BRAF, PIK3CA and TP53 mutations in a large prospective series of
locally advanced rectal cancer patients. Int J Cancer. 146:94–102.
2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Mahdi Y, Khmou M, Souadka A, Agouri HE,
Ech-Charif S, Mounjid C and Khannoussi BE: Correlation between KRAS
and NRAS mutational status and clinicopathological features in 414
cases of metastatic colorectal cancer in Morocco: The largest North
African case series. BMC Gastroenterol. 23(193)2023.PubMed/NCBI View Article : Google Scholar
|