Introduction
Aging is a biological process attributed to the
functional decline of an organism, and it has been the subject of
extensive research. It contributes to the pathogenesis of several
degenerative diseases, such as cancer, diabetes, atherosclerosis,
hypertension and Alzheimer's disease, which affect the quality of
life of individuals and lead to functional decline and subsequent
death (1). The key to reversing
aging lies in understanding its pathophysiology and underlying
mechanisms. Several theories have been proposed to describe the
phenomenon of aging, including increased reactive oxygen species
levels, mitochondrial dysfunction and telomere attrition (2,3).
Epigenetic alterations have been identified as a contributing
factor and have been proposed as one of the mechanisms underlying
aging. These alteration have garnered the attention of researchers
in recent years (4).
DNA methylation is a crucial epigenetic mechanism.
The role of DNA methylation varies depending on its position in the
genome. The loss or gain of methylation at particular positions can
result in altered gene expression, the basis for various diseases
such as Beckwith-Wiedemann syndrom and various type of cancers
(5,6). In addition to the coding region, DNA
methylation occurs in interspersed repetitive sequences (IRS) of
the genome such as LINE-1 and Alu. Alu is a short IRS that accounts
for 13.7% of the human genome (7).
The Alu element is a transposable element capable of replicating
and inserting itself into different areas of the genome. It
contains a high proportion of CpG islands which are sites of DNA
methylation. As a result, 25% of the methylation in the genome is
located in Alu (7). Alu methylation
suppresses transposon activity, while Alu hypomethylation can lead
to high transposon activity, resulting in genomic instability and
aging (8). The methylation level of
Alu is dynamic during normal cell differentiation and tumorigenesis
(7,8). With regards to aging, previous studies
have illustrated the association between the methylation of the Alu
elements and aging, but others have reported conflicting results
(8-10).
Alu hypomethylation is associated with age-related, noncommunicable
diseases such as osteoporosis, diabetes and Alzheimer's disease
(11-13).
The skin is the largest organ of the human body. The
aging process in the skin can be observed externally through
morphological changes, such as wrinkles, pigmentation and abnormal
hyperplasia. Owing to its location, the skin is the most
susceptible organ to external aging factors, such as ultraviolet
radiation and pollution. Skin aging can cause delayed wound healing
and loss of functional barriers to microbes (14,15).
Dermal fibroblasts, which are responsible for
various physiological functions of the skin, play a central role in
skin aging. As dermal fibroblasts rarely proliferate, they are more
susceptible to age-related damage, thus compromising cell function
(14). In skin aging, collagen, the
most abundant component of the extracellular matrix (ECM) secreted
by dermal fibroblasts, is fragmented and decreasing in collagen
production (15-17).
This is due to an increase in matrix metalloproteinase activity and
impaired growth factor signal transduction, a change that occurs as
a result of fibroblast aging (18).
Other components of the ECM also change during aging. Elastin,
another major component of the ECM produced by fibroblasts, is
selectively degraded during the intrinsic skin aging process
(19). Such alterations in
fibroblast function and ECM remodeling can be observed externally
in the aging phenotypes of the skin, such as wrinkles and loss of
elasticity (20). Most prominently,
these changes can be visualized in the facial area, as fibroblasts
in this region are thin and subjected to photoaging and intrinsic
chronological aging. Age-related phenotypes of the skin in the
periorbital area are accurate predictors of biological aging
(21).
In addition to the direct effects of aging, dermal
fibroblasts are affected by hypertension, an important age-related
disease with a high prevalence rate of 30-45% (22,23).
In vitro studies have shown that dermal fibroblasts
subjected to increased pressure exhibit accelerated aging
phenotypes (24,25). This is clinically associated with
ulcers in chronic venous hypertension, wherein fibroblasts lose
their ability to proliferate resulting in aberrant wound healing
(26). The effect of hypertension
on dermal fibroblasts is speculated, given the results of a
previous study on fibroblasts from other organs (26-28).
Hypertension has been linked to the altered activity of cardiac
fibroblasts, such as collagenase dysregulation and cellular
dedifferentiation (27,28). Although hypertension alters the gene
expression profile and cellular phenotypes of dermal fibroblasts,
the epigenetic events taking place remain poorly understood
(29,30). Understanding epigenetic events will
provide useful insights into whether and how hypertension
accelerates aging in dermal fibroblasts.
Several studies have investigated the mechanisms and
pathophysiology of fibroblast aging, including the role of
epigenetic alterations. Changes in the DNA methylation status of
human dermal fibroblasts at specific loci have been observed during
aging (31,32). It is proposed that rather than
occurring randomly during ontogenetic development, these
site-specific changes are specifically regulated through unknown
mechanisms. Moreover, most of the identified loci were located in
the gene and promoter regions, resulting in a lack of extensive
research on IRS (32). These
specific modifications in the methylation status cannot explain the
global hypomethylation phenomenon, which is considered to be the
result of passive random loss of methylation, a phenomenon which
the methylation status of the IRS is more reflective (9,33,34).
The role of DNA methylation in IRS, specifically that of Alu
methylation in the process of skin cell aging, whether by the
chronological aspect of aging itself or the effect of aging through
intermediary age-related diseases such as hypertension, is yet to
be investigated. In the current study, the aim was to explore the
correlation between the Alu methylation status in human dermal
fibroblasts and the chronological age of patients whose specimens
were analyzed. The weather age-related diseases, namely
hypertension, affect the dermal fibroblast methylation profile was
also investigated.
Materials and methods
Study design, sample size and
population
The present study is an analytical cross-sectional
study. A total of 39 samples were obtained from patients who
visited the plastic and reconstructive surgery clinic at King
Chulalongkorn Memorial Hospital (Bangkok, Thailand) for surgical
procedures taken place between September 2020 and September 2021.
All patients were recruited following the inclusion and exclusion
criteria. The inclusion criteria included: i) Indication for skin
surgery with the excision of normal tissues; ii) ≥18 years old; and
iii) ability to make an informed decision. The exclusion criteria
included low quality or quantity of DNA from the cultured cells and
the presence of active skin disease at the site of operation. The
patients were then divided into two groups according to their age
at the date of tissue resection: The young age group (<60 years
old; n=22) and the old age group (>60 years old; n=17). Patients
were also divided into three age groups: Young (<45 years old;
n=8), middle (45-60 years old; n=14), and old (>60 years old;
n=17). The patients were then categorized into three groups
according to their systolic blood pressure (SBP) and diastolic
blood pressure (DBP) as: Normal (SBP<130 mmHg; DBP<85 mmHg),
high-normal (SBP range, 130-139 mmHg; DBP range, 85-89 mmHg) and
hypertensive (SBP≥140 mmHg; DBP≥90 mmHg) (22); based on SBP, there were a total of
14, 14 and 11 patients in every group, respectively, and based on
DBP, there were 17, 12 and 10 patients in every group,
respectively. Tissue samples were grouped according to the surgical
site as eyelid or non-eyelid (Table
SI). There was a total of 29 eyelid and 10 non-eyelid samples
(three brow samples, four sub-brow samples, one ear sample and two
alar samples). The lifestyle factors of the patients were not
recorded, therefore, the samples were grouped randomly for these
factors.
Ethical statement
The current study was reviewed and approved by the
institutional review board of the Faculty of Medicine,
Chulalongkorn University (approval no. 353/63). All 39 samples were
acquired from patients who underwent surgery at the plastic and
reconstructive surgery clinic at King Chulalongkorn Memorial
Hospital from September 2020 to September 2021. Written informed
consent was obtained from all patients before they participated in
the study. The establishment of primary cell lines from patients in
the present study was approved by the same institutional review
board.
Cells and culture
Dermal tissues were collected from patients during
surgical procedures. The participants had not received systemic or
topical treatment in the month before the surgery. After excision,
specimens were cut into pieces of size range 0.5-1 cm2
and immersed in Dulbecco's Modified Eagle medium (DMEM)
(Sigma-Aldrich; Merck KGaA) for transportation to the laboratory.
The dermis was explanted from the surgical specimen and cut into
several 5-mm2 pieces in the laboratory. The pieces were
then placed in Roux culture bottles and cultured in DMEM high
glucose (Sigma-Aldrich; Merck KGaA) mixed with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.). The bottles were
placed in an incubator at 37˚C in humidified air containing 5%
CO2. The cells were subcultured every 7 days.
DNA preparation and measurement of the
CpG methylation level of the Alu repetitive sequence
Cells in passages I, III and V were harvested by
trypsinization. The suspension was centrifuged at 1,500 x g at 4˚C
for 3 min to separate the cells from the solution. The supernatants
were discarded, and 500 µl Lysis Buffer II (0.75 M NaCl and 0.024 M
EDTA at pH 8.0), 50 µl 10% sodium dodecyl sulfate (Sigma-Aldrich;
Merck KGaA) and 50 µl Proteinase K (United States Biological) were
then added for DNA extraction. The suspension was incubated
overnight at 50˚C until the cells were lysed.
Phenol/chloroform was then added to the cell lysate,
and the mixture was centrifuged to separate DNA from the organic
compounds. The DNA-containing supernatant was removed and
precipitated with 100% isopropanol. The combined bisulfite
restriction analysis (COBRA) technique was used to identify
methylation sites (35). The DNA
was treated with sodium bisulfite using the EZ DNA Methylation-Gold
kit (Zymo Research Corp.), according to the manufacturer's
protocol.
To determine the methylation level of the CpG site,
specifically at the Alu element, polymerase chain reaction (PCR)
using the Alu forward primer 5'-GGYGUGGTGGTTTAYGTTTGTAA-3' and the
Alu reverse primer 5'-CTAACTTTTTATATTTTTAATAAAAACRAAATTTCACCA-3'
with the following conditions was performed: Initial denaturation
at 95˚C for 15 min and 35 cycles of denaturation at 95˚C for 45
sec, annealing at 57˚C for 45 sec and extension at 72˚C for 45 sec
followed by a final extension at 72˚C for 15 min. The Alu sequence
primers were based on the Alu nucleotide sequences from the
accession number NM_031483.7(36),
and the aforementioned set of primers was used in our previous
study (9,11,35,37,38). The PCR products were subjected to sodium
bisulfite treatment (35). The PCR
product was digested with TaqI cut enzyme (Thermo Fisher
Scientific, Inc.) and incubated at 65˚C for 16 h. The product was
analyzed by gel electrophoresis on an 8% non-denaturing
polyacrylamide gel. The gels were submerged in SYBR Green (Lonza
Group, Ltd.) for 30 min for staining. Band intensity was observed
and measured using Strom840 and ImageQuanNT Software (Amersham;
Cytiva). DNA from HeLa cells was used as a positive control for
agarose gel electrophoresis and band intensity measurement
(Fig. S1).
In the COBRA method, the methylation pattern of two
CpG loci is reflected in the band length, which can be classified
as follows: Alu loci with two unmethylated CpGs
(uCuC; 133 bp); Alu loci with two methylated
CpGs (mCmC; 58 and 32 bp, respectively); Alu
loci with a 5'-unmethylated CpG and a 3'-methylated CpG
(uCmC; 75 bp); and Alu loci with a
5'-methylated CpG and a 3'-unmethylated CpG
(mCuC; 90 bp).
The intensity of each band was calculated by
dividing the measured intensity (arbitrary unit according to the
ImageQuanNT) by the length of each band as follows: A) 133 bp/133;
B) 58 bp/58; C) 75 bp/73; D) 90 bp/90; and E) 43 bp/41. The
percentage of Alu methylation was calculated by comparing the
number of methylated CpG loci to all the CpG loci located in Alu
([B +E]/[2A + B + C + D + E]) x100.
Statistical analysis
Statistical analysis was performed using SPSS
(version 25.0; IBM Corp.). The Alu methylation status was assessed,
and compared between the young and old age groups, two categories
of surgical sites and two sexes using an unpaired Student's t-test,
with the effect of sex regressed out in the first two analyses due
to the high skewness between sexes. Owing to the high skewness, the
homogeneity of variance and normality of distribution were also
investigated, both of which met the assumption of an unpaired
t-test (Levene's test, P>0.05; Shapiro-Wilk test, P>0.05).
Age and Alu methylation were evaluated using Spearman's rho.
Subgroup analysis was performed for each surgical site group to
determine the relationship between age and Alu methylation in
eyelid and non-eyelid tissues using an unpaired t-test. The
comparisons of the Alu methylation overall level and
uCuC, uCmC and
mCuC patterns between the SBP and the DBP
groups were performed using one-way analysis of variance (ANOVA),
assuming the variances were unequal (Welch's) and homogeneity of
variance was equal (Levene's test, P>0.05) followed by Tukey's
post hoc test. An unpaired t-test was also performed to compare the
Alu methylation status between the SBP and DBP groups. Two-way
ANOVA was performed to assess the interaction between age groups,
DBP groups and Alu methylation status. Homogeneity of variance and
normality of distribution were determined (Levene's test,
P>0.05; Shapiro-Wilk test, P>0.05). Sex could not be included
in these analyses because of multicollinearity. P<0.05 was
considered to indicate a statistically significant difference.
Results
Methylation level and sexes
The differences in Alu methylation levels between
male and female patients were not statistically significant
(P>0.05; Fig. 1).
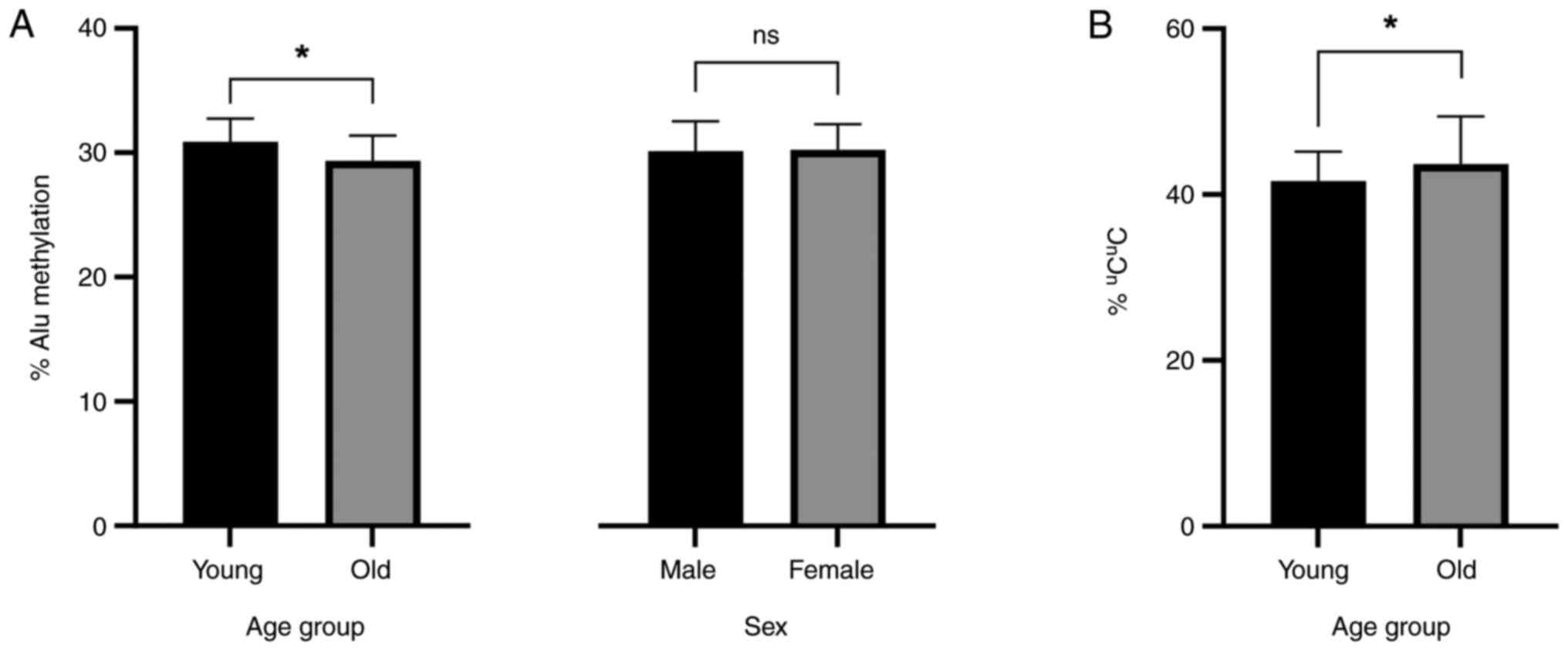 | Figure 1(A) Alu methylation levels in dermal
fibroblasts collected from patients of different ages and sexes.
The young age group was classified as ≤60 years, while the old age
group was defined as >60 years. Regarding mean Alu methylation
shown as mean ± SD, that was 30.9±1.66% for 22 samples in the young
group and 29.40±1.99% for 17 samples in the old group. The
statistical significance of the differences was calculated using
two-way ANOVA considering the surgical site, SBP and DBP groups as
co-independent variables (P=0.023, 0.036 and 0.013, respectively).
There were 34 females and five males with a mean ± SD Alu
methylation level of 30.20±2.06 and 30.19±2.38%, respectively. An
independent samples t-test was performed (P>0.05). (B)
Percentage of the unmethylated allele uCuC in
the young and old age groups. The mean ± SD Alu methylation levels
of the young and old groups were 41.60±3.56 and 43.67±5.74%,
respectively; P=0.016, calculated using two-way ANOVA with DBP
status as a covariate. *P<0.05. Ns, not significant;
ANOVA, analysis of variance; SBP, systolic blood pressure; DBP,
diastolic blood pressure. |
Alu methylation pattern and
chronological ages
The Alu methylation levels of human dermal
fibroblasts obtained from patients of different ages were analyzed.
Patients were classified using two criteria as follows, first
criteria into two age groups: Young age (≤60 years old) and old age
(>60 years old), and second criteria into three age groups:
Young age (<45 years old), middle age (45-60 years old) and old
age (>60 years old). The additional stratification in the three
groups criterion was done to assess subtle changes that occur
during the aging process and to enhance the clarity of the observed
trends.
In the two-group criteria, there were statistically
significant differences in the Alu methylation between young and
old age groups (30.90±1.66 and 29.40±1.99%, respectively; Fig. 1A) using two-way ANOVA considering
the surgical site, and SBP and DBP groups as co-independent
variables (P=0.023, 0.036 and 0.013, respectively). When sex was
used as a co-independent variable, a trend with an almost
statistically significant difference was observed (P=0.051).
Regarding the pattern of Alu methylation, the percentage of
uCuC was found to be significantly higher in
the old age group using two-way ANOVA with DBP status as covariate
(young age group, 41.60±3.56% and old age group: 43.70±5.74%;
P<0.05; Fig. 1B).
In the three-group criteria, there was a significant
difference between young, middle and old age groups using two-way
ANOVA with DBP status as a covariate (31.20±2.14, 30.70±1.79 and
29.40±2.03, respectively; P<0.01). Post-hoc analysis showed
statistically significant differences between the young and old
age, and the middle and old age groups with both the young age and
the middle group having significantly higher methylation than the
old age group (P<0.05; Fig.
2A).
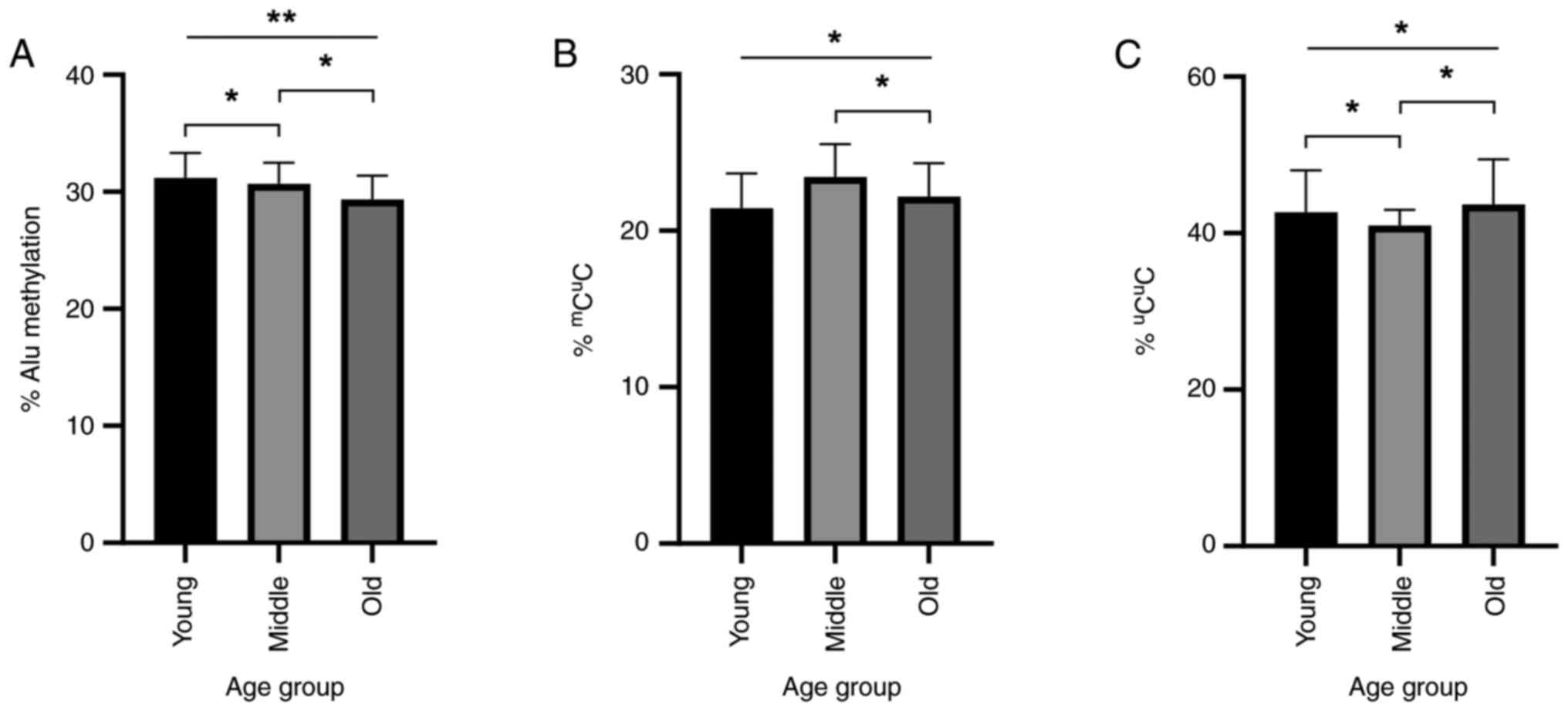 | Figure 2Alu methylation level and percentage
of methylation pattern alleles in dermal fibroblasts collected from
patients of different ages. In the three-group criteria, patients
were classified into the young age group (<45 years old; n=8),
middle age group (45-60 years old; n=14) and old age group (>60
years old; n=17). Statistical significance was calculated using
two-way analysis of variance with DBP status as a covariate. (A)
Alu methylation level in young, middle and old age groups. The mean
± SD Alu methylation levels were 31.20±2.14, 30.70±1.79 and
29.40±2.03%, respectively (P<0.01). Post-hoc analysis revealed
P<0.05 in both young-middle and middle-old comparisons. (B)
Percentage of the partial methylated allele
mCuC. The mean ± SD Alu methylation levels
were 21.40±2.21, 23.40±2.10 and 22.20±2.13 for the young, middle
and old groups, respectively (P<0.05). (C) Percentage of the
unmethylated allele uCuC. The mean ± SD Alu
methylated levels were 42.70±5.34, 41.00±2.00 and 43.70±5.74 for
the young, middle and old groups, respectively (P<0.05). DBP,
diastolic blood pressure. *P<0.05,
**P<0.01. |
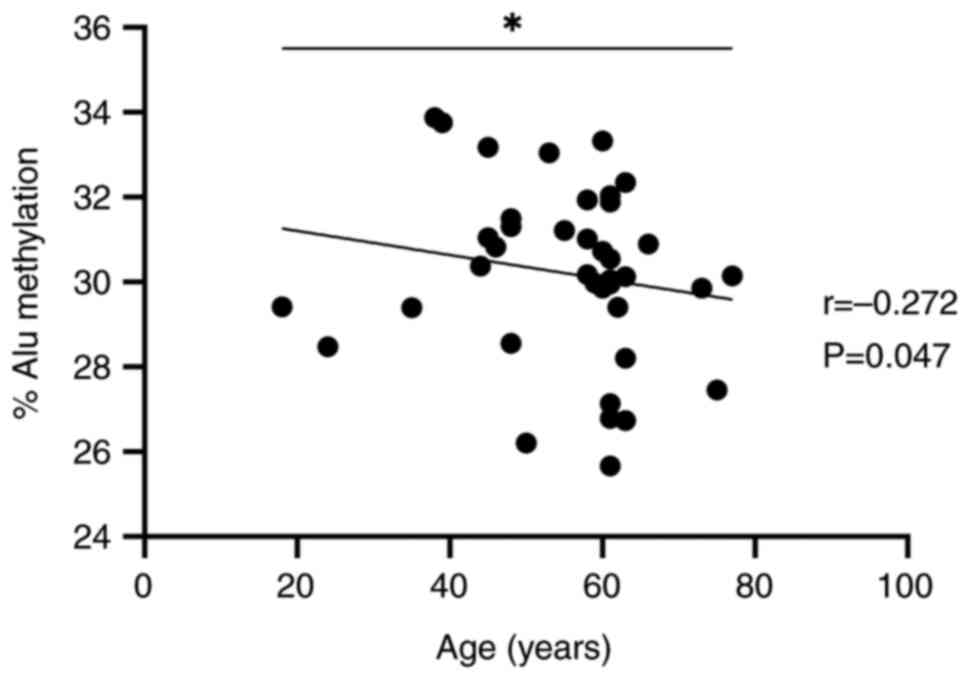
To avoid the effect of arbitrary grouping,
correlation matrix analysis was performed using Spearman's rho
since age was considered only in years and not as continuous
numbers. Assuming that the age and Alu methylation would be
negatively correlated, the correlation was statistically
significant (P=0.047; r=-0.272; Fig.
3).
Using two-way ANOVA with DBP status as a covariate
statistical test for the methylation pattern analysis, the
mCuC methylation was shown to be highest in
the middle age group, followed by the old and young age groups
(21.40±2.21, 23.40±2.10 and 22.20±2.13% in young, middle and old
respectively; P<0.05; Fig. 2B. A
similar but inverse trend was observed in the
uCuC methylation, where this pattern was
lowest in the middle age group, followed by young and old age
(42.70±5.34, 41.00±2.00 and 43.70±5.74% in young, middle and old
respectively; P<0.05; Fig. 2C).
Post-hoc analysis showed a difference in the
mCuC methylation percentage between the old
and middle age groups, and a difference in the
uCuC methylation percentage between the
middle and both the young and old age groups; all differences
described were statistically significant (P<0.05; Fig. 2B and C).
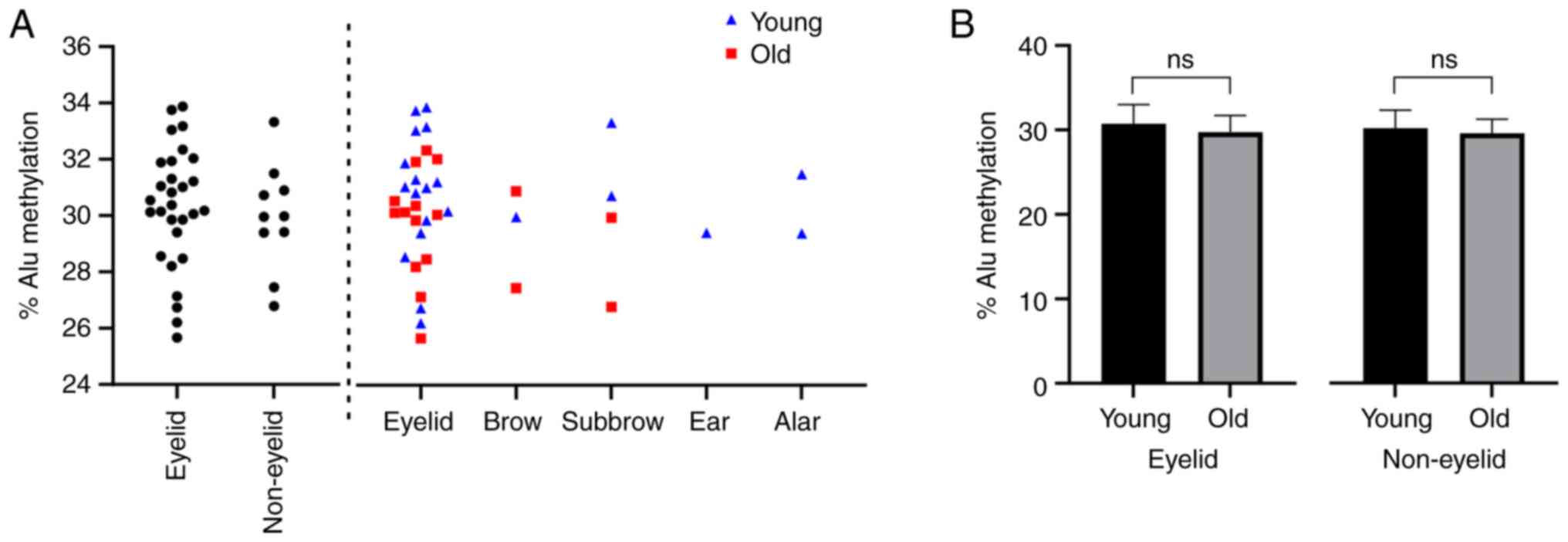
Surgical site and aging pattern
Tissue samples were also classified into two groups
according to the surgical site: Eyelid and non-eyelid. No
statistically significant differences were shown in the Alu
methylation between the two groups (Fig. 4A). Subgroup analysis revealed that
the correlation between Alu methylation and age was stronger in the
eyelid group than in the non-eyelid group (P=0.087 and P=0.112,
respectively; Fig. 4B). The number
of Alu loci with uCuC,
uCmC and mCuC
methylation patterns was not statistically different among age
groups in tissues from both eyelid and non-eyelid groups. The
methylation of samples from different surgical sites is shown in
Fig. 4. A general trend but non
statistically significant of lower Alu methylation levels can be
observed in the old age group compared with those in the young age
group in both eyelid and non-eyelid samples. Notably, some surgical
sites, namely the ear and alar, only have samples from the young
age group. This was attributed to the difficulty in recruiting
subjects as these areas tend to exhibit fewer features of aging
and, therefore, have fewer needs for reconstructive surgery.
Methylation level and blood
pressure
Patients were classified into three groups based on
their blood pressure measured on the day of the surgery: Normal,
high-normal and hypertensive, according to the ESC/ESH guidelines
on hypertension published in 2018(21). The Alu methylation levels in the
normal, high-normal and hypertensive groups according to SBP were
30.30±2.15, 30.20±2.15 and 30.10±2.06%, respectively (Fig. 5A), whereas the Alu methylation
levels in the DBP groups were 29.60±1.85, 31.10±1.78 and
30.20±2.48% for the normal, high-normal and hypertensive groups,
respectively (Fig. 5B). The
percentage of uCuC methylation was also
analysed for the normal, high-normal and hypertensive groups; for
SBP, these were 42.80±5.57, 42.70±3.84 and 41.90±4.84%,
respectively (Fig. 5C), and for DBP
these were 44.20±5.19, 40.40±2.95 and 42.30±4.79%, respectively
(Fig. 5D). Analysis of DBP showed a
statistically significant increase in the overall Alu methylation
in the high-normal group compared with that in the normal group
(P<0.05; Fig. 5B). The
uCuC Alu methylation pattern was also found
at lower proportions in the high-normal DBP group compared with
that in the normal group (P<0.05; Fig. 5D). Two-way ANOVA with the three age
groups as a covariate also revealed a statistically significant
difference in the uCuC methylation percentage
between the prehypertensive (diastolic high-normal) and
hypertensive groups. No statistically significant differences in
SBP were observed among the groups (P>0.05; Fig. 5A and D).
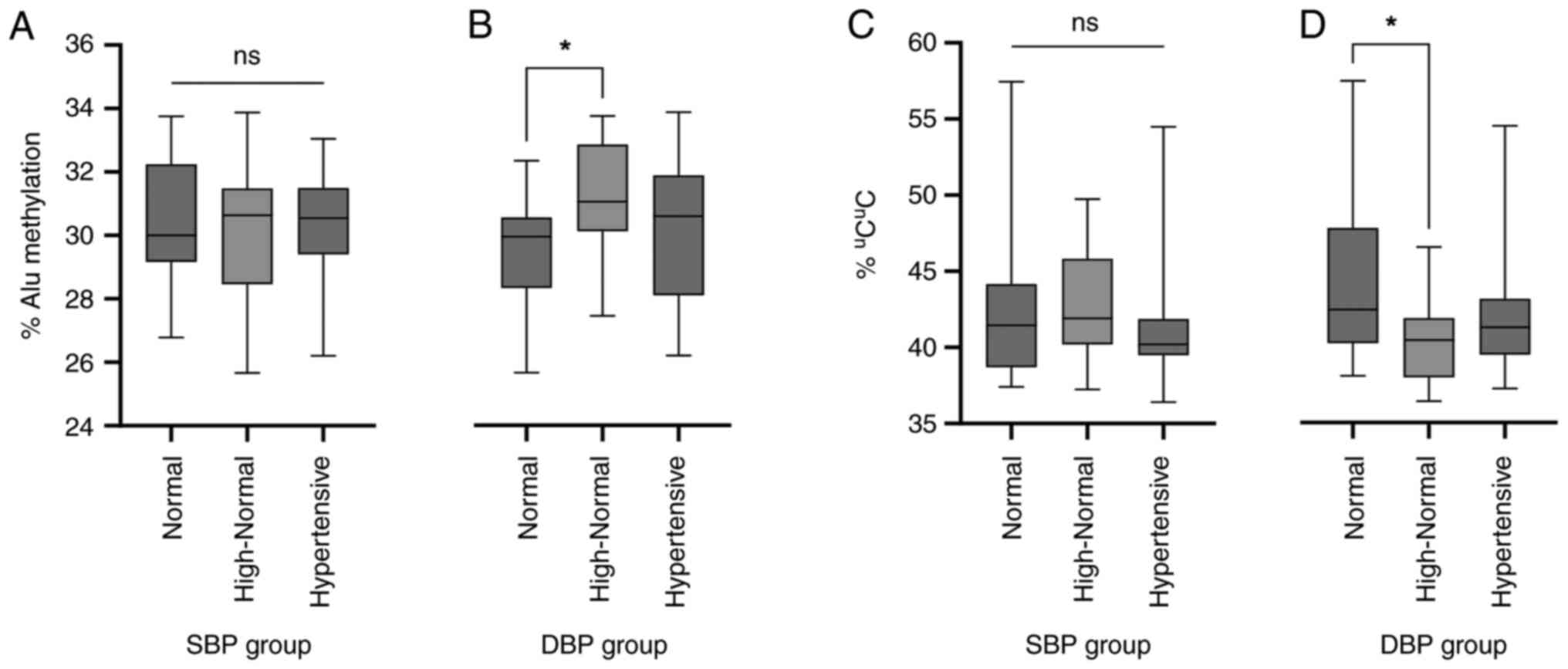 | Figure 5Alu methylation and blood pressure
status. SBP and DBP were used to categorize patients into three
groups: Normal (SBP<130 mmHg; DBP<85 mmHg), high-normal (SBP
range, 130-139 mmHg; DBP range, 85-89 mmHg) and hypertensive
(SBP≥140 mmHg; DBP≥90 mmHg). Based on SBP, there were 14, 14 and 11
patients in the normal, high-normal and hypertensive group,
respectively. Based on DBP, there were 17, 12 and 10 patients in
the normal, high-normal and hypertensive group, respectively. (A)
Alu methylation level and SBP status. No statistical difference was
found using ANOVA, assuming the variances were unequal (Welch's)
and the homogeneity of variance was equal (Levene's test,
P>0.05). (B) Alu methylation level and DBP status. A statistical
difference was observed between the high-normal and normal groups
using an independent samples t-test (P<0.05). (C) Percentage of
the unmethylated allele uCuC and SBP status.
No statistical difference was found using one-way ANOVA. (D)
Percentage of the unmethylated allele uCuC
and DBP status. A statistical difference was observed between the
high-normal and normal groups using independent samples t-test.
*P<0.05. SBP, systolic blood pressure; DBP, diastolic
blood pressure; ANOVA, one-way analysis of variance; ns, not
significant. |
Methylation pattern, age and DBP
Two-way ANOVA of the percentage of
uCuC patterns showed an interaction between
age and DBP when analyzing both factors as covariates (P=0.01; data
not shown). Other interactions between age, DBP and Alu methylation
level, the percentages of mCuC and
uCmC patterns were not statistically
significant (P>0.05).
Discussion
Samples were classified into two groups based on age
at the time of sampling. Results showed that the Alu methylation
levels in the young and old age groups were statistically different
(P≤0.05), with the Alu methylation levels in the old age group
being statistically lower than those in the young group.
Several studies have reported an inverse correlation
between advanced chronological age and global hypomethylation in
various tissues, including fibroblasts from the lung (9,39,40).
The methylation level of the Alu element, with ~1.4 million copies
interspersed in the human genome, has long been regarded to reflect
the global methylation level (10).
Cho et al (41) showed that
Alu hypomethylation is present in cancerous samples from older
patients. A similar association between age and Alu hypomethylation
was observed in normal cells, such as white blood cells (13). However, the correlation between Alu
methylation patterns and age in human dermal fibroblasts is yet to
be determined.
COBRA was selected to measure the methylation levels
based on the comparability of the results with those of
pyrosequencing and high-throughput sequencing methods (9,37).
COBRA-IRS can also illustrate methylation patterns that are
potential markers in the diagnosis of several diseases, including
cancer, autism spectrum disorders and schizophrenia (42-44).
The results of the present study are similar to
those of other studies in different tissues; the Alu methylation
levels in human dermal fibroblasts decrease with age (10,45).
There are two possible explanations for such a correlation: First,
in agreement with multiple studies on human diploid fibroblasts,
the change in methylation patterns may be the result of age-related
chromatin remodeling, which is thought to be the basis of altered
epigenetic control of retrotransposable elements in aging (45,46).
Second, according to a study by Patchsung et al (37), the manipulation of Alu methylation
levels could result in the reversal of aging phenotypes and
increase cell resistance to DNA damage. These results led to the
hypothesis that, rather than being a downstream event of other
aging mechanisms, Alu hypomethylation may play an active role in
the aging process or both. Alu hypomethylation, an effect of
chronological aging, may reactivate retrotransposable elements,
causing genomic instability and making cells more susceptible to
DNA damage accumulation. Another possible mechanism by which Alu
methylation stabilizes DNA is by relieving DNA tension, which makes
the DNA less susceptible to pathological endogenous double-strand
breaks. This was illustrated in a study by Patchsung et al
(37), who found that increasing
Alu methylation reduced endogenous double-strand breaks and made
cells less susceptible to DNA-damaging agents (37,47).
The current methylation pattern analysis also suggested that the
age-related methylation change of Alu may be a dynamic process
similar to uCuC and
mCuC loci percentage, which was found to be
at the bottom and peak in the middle-aged group compared with the
expected trend of continuous change from young to old age. It was
hypothesized that this phenomenon may be explained by the dynamic
transfer of methyl groups between loci, resulting in a distinct
pattern for each age range while retaining the overarching trend of
overall methylation loss. Further studies are required to examine
this hypothesis by increasing the sample size and age
stratification to study the molecular events underlying the process
in depth. It was also shown that no notable changes in the
uCmC loci were reported in another study by
our team in burned skin (35).
Based on this contradiction, the study of methylation patterns
points to multiple mechanisms underlying the Alu methylation
change. Further studies should confirm these hypotheses and
determine whether the proposed mechanisms apply to human dermal
fibroblasts.
Subgroup analyses based on sex and surgical site
found no differences in the Alu methylation levels. The observation
regarding Alu methylation and sex is consistent with a study
conducted in another cell type, the peripheral blood mononuclear
cell, by Jintaridth and Mutirangura (9). The correlation between Alu methylation
and age found in the current study, although statistically
significant, was presented with a small margin between the age
groups. This is in concordance with other studies, one of which
found that the difference in Alu methylation between young and old
age groups was only 1% (48).
Furthermore, high intragroup variability was observed, which
suggests that the human dermal fibroblast Alu methylation levels
may depend on several factors other than age, such as DBP. Future
studies should include other demographic traits as control
variables to avoid confounding effects. Intragroup variability may
also occur because the Alu methylation status is more strongly
correlated with physiological aging than with chronological aging
(45). To uncover this, further
studies should consider classifying samples using an aging
phenotype based on criteria such as the activity of
β-galactosidase, and the proliferation profile and expression of
apoptotic proteins. Studies should also collect information on
lifestyle factors including occupation, diet, exercise and outdoor
activities, a number of which are important in aging and Alu
methylation. It is noteworthy that the present study was conducted
using skin from the eyelids and other facial areas. Skins in these
regions are most prone to aging due to their thinness and greater
exposure to sunlight (21).
Therefore, the effect of aging may be exacerbated, and intrinsic
chronological aging may also be supplemented by photoaging.
Although these two modes of aging are similar epigenetically,
further research should be carried out involving the use of
fibroblasts from other areas of the body to confirm the phenomenon
observed in the current study is generally applicable to dermal
fibroblasts (49).
The current study classified the patients into three
groups according to their blood pressure levels. SBP and DBP were
considered separately. The results showed a statistically
significant increase in Alu methylation in the high-normal DBP
group compared with that in the normal DBP group. No differences
were found between groups classified according to SBP. This is in
line with most studies showing that changes in DNA methylation,
including Alu methylation, usually occur in conjunction with
elevated DBP (49,50). SBP and Alu methylation were reported
only by one study and are considered to be more susceptible to
transient changes such as anxiety, exertion and other disrupting
factors; they are therefore not reflective of the baseline patient
status (49,51). A debatable aspect of the results of
the present study was that it showed a positive correlation between
DBP and Alu methylation, a result supported by a study by Alexeeff
et al (51) but
contradicting others (48-50).
This contradiction may be due to the different tissue types used in
the analysis. All previous studies have used peripheral leukocytes
as a model, whereas the present study used dermal fibroblasts.
Therefore, the alteration of methylation levels might be regulated
by a different process.
Results also confirm the findings of several studies
showing that blood pressure can affect dermal fibroblasts. A study
by Delva et al (52)
illustrated the link between hypertension and alterations in
collagen synthesis and the proliferative activity of dermal
fibroblasts. Another study by Kosugi et al (29) reported an increase in the activity
of phospholipase C, a target of angiotensin II, vasopressin and
thromboxane A2 in dermal fibroblasts obtained from patients with
hypertension. Both studies suggested that dermal fibroblasts may
play a role in the pathogenesis of hypertension.
Similar to that in the aging study, patterns of Alu
methylation were identified using COBRA analysis. Overall,
methylation was higher in the high-normal DBP group than that in
the normal DBP group. The unmethylated allele, the
uCuC methylation pattern, was significantly
lower in the high-normal DBP group than in the normal and high DBP
groups. Therefore, it was hypothesized that similar to that in
aging, these alterations are dynamic processes. Two-way ANOVA
analysis of the uCuC methylation pattern
revealed a statistically significant interaction between aging and
DBP, suggesting that both processes share an underlying epigenetic
event or complement each other in the progression of the
pathogenetic process. This is a likely possibility, given that
hypertension is known to be an age-related disease. Given the
connection between dermal fibroblasts and hypertension in a
previous study and the pattern of our results, it is also possible
that rather than being the victim of damage by hypertension, dermal
fibroblasts may play an active role. Further studies could answer
this question as this may serve as a basis for a better
understanding of the pathogenesis of hypertension. Furthermore, the
result of two-way ANOVA, which revealed a statistically significant
difference in the uCuC percentage but not
overall Alu methylation, indicating a correlation between
uCuC percentage and diastolic hypertension.
Therefore, further validation is suggested to investigate the
potential utility of the uCuC percentage as a
more efficient marker than the Alu methylation level.
In conclusion, Alu hypomethylation is correlated
with chronological age in human dermal fibroblasts. The Alu
methylation levels were significantly lower in the young age group
than those in the old age group. Alu methylation was also higher in
fibroblasts in patients with high-normal DBP than in those with
normal DBP. Dynamic alteration in the methylation of Alu element
was also observed in both aging and diastolic hypertension. Taken
together, Alu methylation may play an active role in the aging of
skin fibroblasts and the pathogenesis of hypertension. The present
study could serve as a foundation for further investigations of
skin aging, its role in the pathogenesis of hypertension and the
development of therapeutics to reverse this process.
Supplementary Material
Representative agarose gel
electrophoresis of the combined bisulfite restriction analysis
method of Alu repetitive sequence.
Patient and control subject
demographic and Alu methylation data.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by the Thailand Science
Research and Innovation Fund Chulalongkorn University.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SJ, NK, AM and JM conceptualized the study. SJ, PH,
SK, NB, TS and JM carried out formal analysis. NK, AM and JM
acquired funding. SJ, PH, SK, NB, TS, NK, AM and JM carried out the
investigation. SJ, PH, SK, NB, TS, NK, AM and JM developed the
methodology used. JM completed project administration and provided
resources. JM and NK used software. AM and JM supervised the study.
NK, AM and JM visualized and validated the data, and wrote the
original draft. SJ, PH, SK, NB,TS, NK, AM and JM carried out
investigation. SJ, PH, SK, NB, TS, NK, AM and JM reviewed and
edited the draft. NK, AM and JM confirm the authenticity of all the
raw data. All authors have read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
The authors state that they have obtained ethics
approval from the institutional review board of the Faculty of
Medicine, Chulalongkorn University, Bangkok, Thailand (IRB No.
353/63), for the research described in the present study. In
addition, written informed consent was obtained from all
participants before skin sample collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Campisi J, Andersen JK, Kapahi P and Melov
S: Cellular senescence: A link between cancer and age-related
degenerative disease? Semin Cancer Biol. 21:354–359.
2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Sesti F, Liu S and Cai SQ: Oxidation of
potassium channels by ROS: A general mechanism of aging and
neurodegeneration? Trends Cell Biol. 20:45–51. 2010.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Levy MZ, Allsopp RC, Futcher AB, Greider
CW and Harley CB: Telomere end-replication problem and cell aging.
J Mol Biol. 225:951–960. 1992.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Calvanese V, Lara E, Kahn A and Fraga MF:
The role of epigenetics in aging and age-related diseases. Ageing
Res Rev. 8:268–276. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Shames DS, Minna JD and Gazdar AF: DNA
methylation in health, disease, and cancer. Curr Mol Med. 7:85–102.
2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lim DH and Maher ER: DNA methylation: A
form of epigenetic control of gene expression. Obstet Gynaecol.
12:37–42. 2010.
|
|
7
|
Luo Y, Lu X and Xie H: Dynamic Alu
methylation during normal development, aging, and tumorigenesis.
Biomed Res Int. 2014(784706)2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Erichsen L, Beermann A, Arauzo-Bravo MJ,
Hassan M, Dkhil MA, Al-Quraishy S, Hafiz TA, Fischer JC and
Santourlidis S: Genome-wide hypomethylation of LINE-1 and Alu
retroelements in cell-free DNA of blood is an epigenetic biomarker
of human aging. Saudi J iol Sci. 25:1220–1226. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Jintaridth P and Mutirangura A:
Distinctive patterns of age-dependent hypomethylation in
interspersed repetitive sequences. Physiol Genomics. 41:194–200.
2010.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bollati V, Schwartz J, Wright R, Litonjua
A, Tarantini L, Suh H, Sparrow D, Vokonas P and Baccarelli A:
Decline in genomic DNA methylation through aging in a cohort of
elderly subjects. Mech Ageing Dev. 130:234–239. 2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jintaridth P, Tungtrongchitr R,
Preutthipan S and Mutirangura A: Hypomethylation of Alu elements in
post-menopausal women with osteoporosis. PLoS One.
8(e70386)2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Thongsroy J, Patchsung M and Mutirangura
A: The association between Alu hypomethylation and severity of type
2 diabetes mellitus. Clin Epigenetics. 9(93)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Bollati V, Galimberti D, Pergoli L, Dalla
Valle E, Barretta F, Cortini F, Scarpini E, Bertazzi PA and
Baccarelli A: DNA methylation in repetitive elements and Alzheimer
disease. Brain Behav Immun. 25:1078–1083. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tigges J, Krutmann J, Fritsche E,
Haendeler J, Schaal H, Fischer JW, Kalfalah F, Reinke H,
Reifenberger G, Stühler K, et al: The hallmarks of fibroblast
ageing. Mech Ageing Dev. 138:26–44. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Varani J, Dame MK, Rittie L, Fligiel SE,
Kang S, Fisher GJ and Voorhees JJ: Decreased collagen production in
chronologically aged skin: Roles of age-dependent alteration in
fibroblast function and defective mechanical stimulation. Am J
Pathol. 168:1861–1868. 2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Quan T, Qin Z, Xia W, Shao Y, Voorhees JJ
and Fisher GJ: Matrix-degrading metalloproteinases in photoaging. J
Investig Dermatol Symp Proc. 14:20–24. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Talwar HS, Griffiths CE, Fisher GJ,
Hamilton TA and Voorhees JJ: Reduced type I and type III
procollagens in photodamaged adult human skin. J Invest Dermatol.
105:285–290. 1995.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Shin JW, Kwon SH, Choi JY, Na JI, Huh CH,
Choi HR and Park KC: Molecular mechanisms of dermal aging and
antiaging approaches. Int J Mol Sci. 20(2126)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Langton AK, Sherratt MJ, Griffiths CE and
Watson RE: Differential expression of elastic fibre components in
intrinsically aged skin. Biogerontology. 13:37–48. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Boukamp P: Skin aging: A role for
telomerase and telomere dynamics? Curr Mol Med. 5:171–177.
2005.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zou Z, Long X, Zhao Q, Zheng Y, Song M, Ma
S, Jing Y, Wang S, He Y, Esteban CR, et al: A single-cell
transcriptomic atlas of human skin aging. Dev Cell. 56:383–397.e8.
2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Williams B, Mancia G, Spiering W, Agabiti
Rosei E, Azizi M, Burnier M, Clement DL, Coca A, de Simone G,
Dominiczak A, et al: 2018 ESC/ESH Guidelines for the management of
arterial hypertension: The Task Force for the management of
arterial hypertension of the European Society of Cardiology (ESC)
and the European Society of Hypertension (ESH). G Ital Cardiol
(Rome). 19 (11 Suppl 1):3S–73S. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Huang C and Ogawa R: The link between
hypertension and pathological scarring: Does hypertension cause or
promote keloid and hypertrophic scar pathogenesis? Wound Repair
Regen. 22:462–466. 2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Stanley AC, Fernandez NN, Lounsbury KM,
Corrow K, Osler T, Healey C, Forgione P, Shackford SR and Ricci MA:
Pressure-induced cellular senescence: A mechanism linking venous
hypertension to venous ulcers. J Surg Res. 124:112–117.
2005.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Healey C, Forgione P, Lounsbury KM, Corrow
K, Osler T, Ricci MA and Stanley A: A new in vitro model of venous
hypertension: The effect of pressure on dermal fibroblasts. J Vasc
Surg. 38:1099–1105. 2003.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Etufugh CN and Phillips TJ: Venous ulcers.
Clin Dermatol. 25:121–130. 2007.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Díez J: Mechanisms of cardiac fibrosis in
hypertension. J Clin Hypertens (Greenwich). 9:546–550.
2007.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wall SJ, Sampson MJ, Levell N and Murphy
G: Elevated matrix metalloproteinase-2 and-3 production from human
diabetic dermal fibroblasts. Br J Dermatol. 149:13–16.
2003.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kosugi T, Osanai T, Kamada T, Nakano T and
Okumura K: Phospholipase C activity is enhanced in skin fibroblasts
obtained from patients with essential hypertension. J Hypertens.
21:583–590. 2003.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ansurudeen I, Sunkari VG, Grünler J,
Peters V, Schmitt CP, Catrina SB, Brismar K and Forsberg EA:
Carnosine enhances diabetic wound healing in the db/db mouse model
of type 2 diabetes. Amino Acids. 43:127–134. 2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Moulin L, Cenizo V, Antu AN, André V, Pain
S, Sommer P and Debret R: Methylation of LOXL1 Promoter by DNMT3A
in aged human skin fibroblasts. Rejuvenation Res. 20:103–110.
2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Koch CM, Suschek CV, Lin Q, Bork S,
Goergens M, Joussen S, Pallua N, Ho AD, Zenke M and Wagner W:
Specific age-associated DNA methylation changes in human dermal
fibroblasts. PLoS One. 6(e16679)2011.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Casillas MA Jr, Lopatina N, Andrews LG and
Tollefsbol TO: Transcriptional control of the DNA
methyltransferases is altered in aging and
neoplastically-transformed human fibroblasts. Mol Cell Biochem.
252:33–43. 2003.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Tirado-Magallanes R, Rebbani K, Lim R,
Pradhan S and Benoukraf T: Whole genome DNA methylation: Beyond
genes silencing. Oncotarget. 8:5629–5637. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Meevassana J, Serirodom S, Prabsattru P,
Boonsongserm P, Kamolratanakul S, Siritientong T, Mutirangura A and
Angspatt A: Alu repetitive sequence CpG methylation changes in burn
scars. Burns. 48:1417–1424. 2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Dagan T, Sorek R, Sharon E, Ast G and
Graur D: AluGene: A database of Alu elements incorporated within
protein-coding genes. Nucleic Acids Res. 32(Database
issue):D489–D492. 2004.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Patchsung M, Settayanon S, Pongpanich M,
Mutirangura D, Jintarith P and Mutirangura A: Alu siRNA to increase
Alu element methylation and prevent DNA damage. Epigenomics.
10:175–185. 2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Thongsroy J and Mutirangura A: The
association between Alu hypomethylation and the severity of
hypertension. PLoS One. 17(e0270004)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Unnikrishnan A, Hadad N, Masser DR,
Jackson J, Freeman WM and Richardson A: Revisiting the genomic
hypomethylation hypothesis of aging. Ann N Y Acad Sci. 1418:69–79.
2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ciccarone F, Tagliatesta S, Caiafa P and
Zampieri M: DNA methylation dynamics in aging: how far are we from
understanding the mechanisms? Mech Ageing Dev. 174:3–17.
2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Cho NY, Kim BH, Choi M, Yoo EJ, Moon KC,
Cho YM, Kim D and Kang GH: Hypermethylation of CpG island loci and
hypomethylation of LINE-1 and Alu repeats in prostate
adenocarcinoma and their relationship to clinicopathological
features. J Pathol. 211:269–727. 2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Pobsook T, Subbalekha K, Sannikorn P and
Mutirangura A: Improved measurement of LINE-1 sequence methylation
for cancer detection. Clin Chim Acta. 412:314–321. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Saeliw T, Tangsuwansri C, Thongkorn S,
Chonchaiya W, Suphapeetiporn K, Mutirangura A, Tencomnao T, Hu VW
and Sarachana T: Integrated genome-wide Alu methylation and
transcriptome profiling analyses reveal novel epigenetic regulatory
networks associated with autism spectrum disorder. Mol Autism.
9(27)2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kalayasiri R, Kraijak K, Mutirangura A and
Maes M: Paranoid schizophrenia and methamphetamine-induced paranoia
are both characterized by a similar LINE-1 partial methylation
profile, which is more pronounced in paranoid schizophrenia.
Schizophr Res. 208:221–227. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Cardelli M: The epigenetic alterations of
endogenous retroelements in aging. Mech Ageing Dev. 174:30–46.
2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Gentilini D, Mari D, Castaldi D, Remondini
D, Ogliari G, Ostan R, Bucci L, Sirchia SM, Tabano S, Cavagnini F,
et al: Role of epigenetics in human aging and longevity:
Genome-wide DNA methylation profile in centenarians and
centenarians' offspring. Age (Dordr). 35:1961–1973. 2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Mutirangura A: Is global hypomethylation a
nidus for molecular pathogenesis of age-related noncommunicable
diseases? Epigenomics. 11:577–579. 2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Xiao FH, Kong QP, Perry B and He YH:
Progress on the role of DNA methylation in aging and longevity.
Brief Funct Genomics. 15:454–459. 2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Orioli D and Dellambra E: Epigenetic
regulation of skin cells in natural aging and premature aging
diseases. Cells. 7(268)2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Bellavia A, Urch B, Speck M, Brook RD,
Scott JA, Albetti B, Behbod B, North M, Valeri L, Bertazzi PA, et
al: DNA hypomethylation, ambient particulate matter, and increased
blood pressure: Findings from controlled human exposure
experiments. J Am Heart Assoc. 2(e000212)2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Alexeeff SE, Baccarelli AA, Halonen J,
Coull BA, Wright RO, Tarantini L, Bollati V, Sparrow D, Vokonas P
and Schwartz J: Association between blood pressure and DNA
methylation of retrotransposons and pro-inflammatory genes. Int J
Epidemiol. 42:270–280. 2013.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Delva P, Lechi A, Pastori C, Degan M,
Sheiban I, Montesi G, Pea M, Meneguzzi A and Menegazzi M: Collagen
I and III mRNA gene expression and cell growth potential of skin
fibroblasts in patients with essential hypertension. J Hypertens.
20:1393–1399. 2002.PubMed/NCBI View Article : Google Scholar
|