Introduction
Acute lymphoblastic leukemia (ALL) is an aggressive
type of leukemia characterized by excessive lymphoblasts or
lymphocytes in the bone marrow and peripheral blood, which can
spread to the lymph nodes, spleen, liver, central nervous system
and other organs (1,2). While it predominantly affects
children, ALL represents 20% of leukemia cases in adults, of which
B cell lineage ALL constitutes 75% (3-5).
Over the past decades, treatment-related mortality rates during
induction chemotherapy in patients with ALL have decreased due to
improved supportive care (6). In
addition, the use of risk-directed therapy to optimize doses and
schedules of chemotherapy has been developed over the past 40 years
(7), which has increased the
survival probability to 80-90% (8).
Thus, most children with ALL can be cured; however, with increasing
age, adult patients have higher white blood cell counts, a lower
incidence of hyperdiploidy, an increased incidence of adverse
genetic abnormalities, and are at lower biological risk and reduced
tolerance to chemotherapy, which leads to poor prognosis in adult
patients with ALL (9-11).
Clinical studies have demonstrated that chemotherapy remains the
most common treatment option (12,13).
However, certain studies have reported that, in the treatment of
ALL, cell resistance is still an important problem in clinical
treatment and relapse (14,15).
Glucocorticoids (GCs), including dexamethasone
(DEX), are commonly used in ALL chemotherapy (16). GCs function by activating the GC
receptor (GR), a ligand-induced transcription factor, which in turn
regulates genes that induce leukemic cell apoptosis (17), indicating that GCs serve an
important role in the therapy and prognosis of ALL. However, it has
been reported that the long-term application of GCs, even at
therapeutic doses, can lead to adverse reactions and trigger
downregulation of GR, which results in resistance to GCs (18,19).
Therefore, inhibiting the downregulation of GR (thereby maintaining
the sensitivity of ALL to GCs) is essential for the improvement of
ALL treatment (19-22).
Thus, the exploration of a more effective combination therapy for
ALL has been of interest in recent years. Combination therapy can
reduce the dosage of GCs, lessen side effects and improve the
therapeutic effect, which is an effective method to reduce
chemotherapy resistance in ALL cells (23).
Andrographolide (AND) is the major bioactive
compound isolated from Andrographis paniculata, a medicinal
herb that is widely used in China and other parts of Asia for the
treatment of upper respiratory tract infections (24). It has been found that AND has a wide
range of therapeutic actions, including immunosuppressant (25), antithrombotic (26), anti-inflammatory (27), antineoplastic (28), antiviral (29,30),
antibacterial (31), antidiabetic
(30,32), antioxidative (33), antipyretic (34), anti-oedematogenic and
antinociceptive activities (35).
It has also been reported that AND is a common inhibitor of
autophagy (36,37). A previous study reported that AND
could inhibit the growth of the ALL cell line Jurkat (38), which provides some evidence for the
possible clinical application of AND in ALL.
Autophagy is an essential and conserved lysosomal
degradation pathway that controls the homeostasis of the cytoplasm
by bulk degradation of unnecessary or dysfunctional cellular
organelles and protein aggregates (39). In addition to the previously
described tumor-suppressive roles of autophagy, the pro-survival
function of autophagy under stress conditions, including nutrient
deprivation, hypoxia and therapeutic stress, has been found to
promote tumor growth and progression (40,41).
In the autophagy process, precursor microtubule-associated 1
protein light chain 3 (LC3) is processed by autophagy-related
protein (ATG) 4 into cytoplasmic soluble LC3Ⅰ, which is then
covalently linked to phosphatidylethanolamine (PE), by ATG7 and
ATG3, to become lipid-soluble LC3-PE (also known as LC3II), which
then participates in membrane elongation until the formation of the
autolysosome (42,43). Consequently, LC3II, an important
multi-signal transduction regulating protein located on the
autophagic vesicular membrane, is often used as a marker of
autophagy formation (44). Beclin1
was the first confirmed mammalian autophagy gene and the first gene
in the lysosomal degradation pathway of autophagy to be identified
as a tumor suppressor (45).
Studies have demonstrated that Beclin1 binds to proteins such as
vacuolated protein sorting associated protein (VPS) 15, VPS34, UV
radiation resistance associated gene product (UVRAG) and ATG14 to
form a class III PI3K complex, which controls autophagosome
formation and regulates autophagy activity (46,47).
In hematologic malignancies, autophagy can either act as a
chemoresistance mechanism or have tumor-suppressive functions
(21). When cells undergo lysosomal
autophagy, GRs are degraded by lysosomes (48), thereby reducing the sensitivity of
ALL cells to drugs. Therefore, we hypothesize that autophagy is
important in the resistance of ALL cells and the GR drug receptor
to GCs.
The PI3K/AKT/mTOR signaling pathway serves a crucial
role in cellular signaling and regulates various cellular
functions, including cell growth and death (49,50).
Upon entering the cell, PI3K catalyzes the generation of
phosphatidylinositol-3,4,5-triphosphate (PIP3), a second messenger,
alongside related active enzymes (such as tyrosine kinase and small
Ras-related GTPases). PIP3 subsequently activates AKT in
conjunction with phosphatidylinositol lipid-dependent protein
kinase 1. Activated AKT prevents tuberous sclerosis complex 1/2
complex degradation, promoting mTOR activation and inhibiting
cellular autophagy (51). It has
been demonstrated that activation of the PI3K/AKT/mTOR signaling
pathway can block cellular autophagy to enhance drug sensitivity in
tumor therapy (52). Consequently,
targeting the PI3K/AKT/mTOR signaling pathway to induce autophagy
may increase tumor cell susceptibility to drugs.
Based on the aforementioned studies, the present
study aimed to determine the synergistic antitumor effect of AND
and DEX on a DEX-resistant ALL cell line (CEM-C1). In terms of
autophagy and drug resistance, studying the inhibitory effect of
autophagy inhibitors combined with DEX on ALL has important
research value.
Materials and methods
Cell culture and drug treatment
The CEM-C1 cells used in the present study were
obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. CEM-C1 cells were maintained in
RPMI-1640 medium (cat. no. C11875500BT; Gibco; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (cat. no. S711-001S;
Lonsera; Shanghai Shuangru Biotechnology Co., Ltd.) and 1%
penicillin-streptomycin (cat. no. sv30010; HyClone; Cytiva). Cells
were kept in a humidified atmosphere of 5% CO2 at
37˚C.
AND (cat. no. 365645; Sigma-Aldrich; Merck KGaA) and
DEX (cat. no. A17590;Thermo Fisher Scientific, Inc.) were prepared
in DMSO and diluted in culture medium when needed. The final
concentration of DMSO was <0.5%, which did not affect cell
survival (53). Cells were treated
with different concentrations of the drugs: Control group
(RPMI-1640 medium without drugs), 5 µM AND, 50 µM DEX or 5 µM AND +
50 µM DEX, and then incubated for 24 h in a humidified atmosphere
of 5% CO2 at 37˚C.
Cell viability assay
CEM-C1 cells were cultured in 96-well plates and
seeded at 1x104 cells per well. After 24 h, different
concentrations of drugs were added and the cells were incubated for
a further 24 h in a humidified atmosphere of 5% CO2 at
37˚C. The cell treatment groups were as follows: i) Untreated
control group (control), RPMI-1640 medium without drugs; ii)
solvent control group (vehicle), 0.1% DMSO (the same concentration
used at the highest dose of AND); iii) AND only group, AND was
added at concentrations of 1.25, 2.5, 5, 10, 20 or 40 µM; iv) DEX
group, DEX was added at concentrations of 12.5, 25, 50, 100, 200 or
400 µM; and v) co-treatment group, AND and DEX were added at
various concentrations (1.25 µM AND + 12.5 µM DEX, 2.5 µM AND + 25
µM DEX, 5 µM AND + 50 µM DEX or 10 µM AND + 100 µM DEX). After
treatment, the cell viability was measured using a Cell Counting
Kit-8 (CCK-8; cat. no. CK04; Dojindo Laboratories, Inc.) assay:
CCK-8 (10 µl) was added to each well in the dark and incubated for
4 h. The optical density was measured with a microplate reader
(Bio-Rad Laboratories, Inc.) at a wavelength of 450 nm. The
survival rate of the drug-treated groups was compared with that of
the untreated control group.
Calculation of the combined effect of
AND and DEX
CompuSyn software (version 1.0; ComboSyn, Inc.) was
used to calculate the combination index (CI) (54). If the CI was <1, the combined
effect was considered synergistic; if CI=1, the effect of the two
drugs was considered to be additive; and if the CI was >1, the
combined effect was considered to be antagonistic.
Wright-Giemsa staining
The CEM-C1 cells were cultured in 6-well plates at
1x106 cells per well. After the aforementioned drug
treatment, cells were washed with PBS three times and suspended in
0.5 ml PBS per well. The smear slides were constructed slowly with
15 µl cell suspension, then fixed with 4% paraformaldehyde (cat.
no. P1110; Beijing Solarbio Science & Technology Co., Ltd.) at
room temperature for 30 min. Following this, the cells were washed
three times with PBS. The slides were then dried and stained by
Wright-Giemsa (cat. no. G1020; Beijing Solarbio Science &
Technology Co., Ltd.) staining at room temperature, according to
the manufacturer's instructions. The slides were then washed with
PBS, dried and observed under a light microscope (Olympus
Corporation). The quantification of stained cells was performed
using ImageJ software (version 4.1; National Institutes of
Health).
Reverse transcription-quantitative PCR
(RT-qPCR)
After the aforementioned drug treatments in a 6-well
plate, total RNA in the treated cells was isolated using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.).
RNA was reverse transcribed into cDNA using the RevertAid First
Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Inc.). The
temperature protocol for the RT was as follows: 25˚C for 5 min,
42˚C for 60 min and 70˚C for 5 min. GR, LC3 and Beclin1 were
detected by qPCR using the SYBR Green Kit (Shanghai Yeasen
Biotechnology Co., Ltd.). The amplification parameters were as
follows: 95˚C for 10 min, followed by 40 cycles of 95˚C for 10 sec,
60˚C for 20 sec and 72˚C for 20 sec. The primer sequences used were
as follows: GR forward, 5'-AGGACCACCTCCCAAACTCT-3' and reverse,
5'-TGTTTTTCGAGCTTCCAGGT-3'; LC3 forward,
5'-CCGACTTATTCGAGAGCAGCATCC-3' and reverse,
5'-GTCCGTTCACCAACAGGAAGAAGG-3'; Beclin1 forward,
5'-ATCTAAGGAGCTGCCGTTATAC-3' and reverse,
5'-CTCCTCAGAGTTAAACTGGGTT-3'; and GAPDH forward,
5'-CAGGAGGCATTGCTGATGAT-3' and reverse, 5'-GAAGGCTGGGGCTCATTT-3'.
The primers were designed and synthetized by Sangon Biotech Co.,
Ltd. Expression was analyzed using the 2-ΔΔCq method
(55) with GAPDH as the reference
gene. All experiments were conducted in triplicate.
Western blotting
Western blotting experiments were conducted as
previously described (53). The
drug-treated cells were lysed at 4˚C in RIPA buffer (Thermo Fisher
Scientific, Inc.), which contained protease and phosphatase
inhibitors (Roche Diagnostics GmbH). The supernatant was collected
for protein determination using a BCA protein assay kit (Epizyme,
Inc.; Ipsen Biopharmaceuticals, Inc.). An equal amount of protein
(30 µg/lane) was resolved via 8, 10 or 15% SDS-PAGE, and separated
proteins were then transferred to a PVDF membrane. The membranes
were blocked with 5% non-fat milk (Beijing Solarbio Science &
Technology Co., Ltd.) in tris-buffered saline containing 0.1%
Tween-20 for 2 h at room temperature, and then incubated with
primary antibodies (1:1,000) overnight at 4˚C. The antibodies used
in the present study were: GR (cat. no. 3660S), LC3Ⅱ/LC3Ⅰ (cat. no.
12741), Beclin1 (cat. no. 3495S), PI3K (cat. no. 4257S),
phosphorylated (p-)PI3K (cat. no. 4228S), AKT (cat. no. 4691S),
p-AKT (cat. no. 4060S), mTOR (cat. no. 2983S), p-mTOR (cat. no.
5536S) (all purchased from Cell Signaling Technology, Inc.), GAPDH
(cat. no. AF0006) and β-actin (cat. no. AF0003) (all purchased from
Beyotime Institute of Biotechnology). The membranes were then
incubated with horseradish peroxidase-labeled secondary antibodies
[anti-mouse IgG (1:2,000; cat. no. 7076; Cell Signaling Technology,
Inc.) and anti-rabbit IgG (1:2,000; cat. no. 7074; Cell Signaling
Technology, Inc.)] for 2 h at room temperature. The immunoreactive
bands were visualized using an ECL western blot detection kit
(Epizyme, Inc.; Ipsen Biopharmaceuticals, Inc.) and a Tanon 5200
Imaging System (Tanon Science and Technology Co., Ltd.). The
intensity of the immunoblotting bands was measured using ImageJ
software.
LysoTracker Red staining
After the aforementioned drug treatment in 6-well
plates, cells were stained with LysoTracker Red (50 nM), a specific
red fluorescent dye for lysosomes, for 45 min at 37˚C, followed by
counterstaining with Hoechst 33342 for 15 min at room temperature
in the dark. The cells were then observed and imaged using an
inverted fluorescence microscope (Olympus Corporation).
Small interfering RNA (siRNA) and
transfection
The siRNA experiments were divided into five groups:
The control (untreated control group, RPMI-1640 medium only), 50 nM
negative control (NC; non-targeting sequence; cat. no.
siN0000001-1-5), 50 nM positive control (PC; h-ACTB;
siP0000002-1-5), si-LC3-1, si-LC3-2 and si-LC3-3 (all 50 nM)
groups. The si-LC3 and NC sequences were designed and synthesized
by Guangzhou RiboBio Co., Ltd. The si-LC3 nucleotide sequences used
were as follows: si-LC3-1, 5'-ATTCCTGTACATGGTCTAT-3'; si-LC3-2,
5'-TATGCCTCCCAGGAGACGT-3'; and si-LC3-3, 5'-GATTCCTGTACATGGTCTA-3'.
For transfection, cells were seeded into a 6-well plate at a
density of 5x105 cells per well. When the cell density
reached 70-80%, 250 µl transfection complex solution was added,
which was prepared following the instructions of the siRNA reagent
kit (Guangzhou RiboBio Co., Ltd.) and Lipofectamine®
3000 reagent (Thermo Fisher Scientific, Inc.). In brief, solution A
(5 µl 20 µM siRNA stock solution + 120 µl RPMI-1640) was mixed with
solution B (3.5 µl Lipofectamine 3000 + 125 µl RPMI-1640) and
incubated for 20 min at room temperature to form the transfection
complex solution. Cells were kept in a humidified atmosphere of 5%
CO2 at 37˚C for 24 h. After 24 h of transfection, total
cellular proteins were extracted and the transfection efficiency
was analyzed by western blotting as aforementioned.
Statistical analysis
Statistical analyses were conducted using SPSS 25.0
(IBM Corp.). All data are presented as the mean ± standard
deviation and all experiments were repeated three times.
Differences among groups were analyzed using one-way ANOVA,
followed by Tukey's post hoc test for multiple comparisons.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Co-treatment with AND + DEX inhibits
the viability of CEM-C1 cells
As shown in Fig.
1A-C, compared with the control group, the viability of the
cells incubated with AND, DEX or AND + DEX decreased with
increasing drug concentrations. According to Fig. 1D, the CI values were all <1,
indicating that the combination of the two drugs had a synergistic
effect.
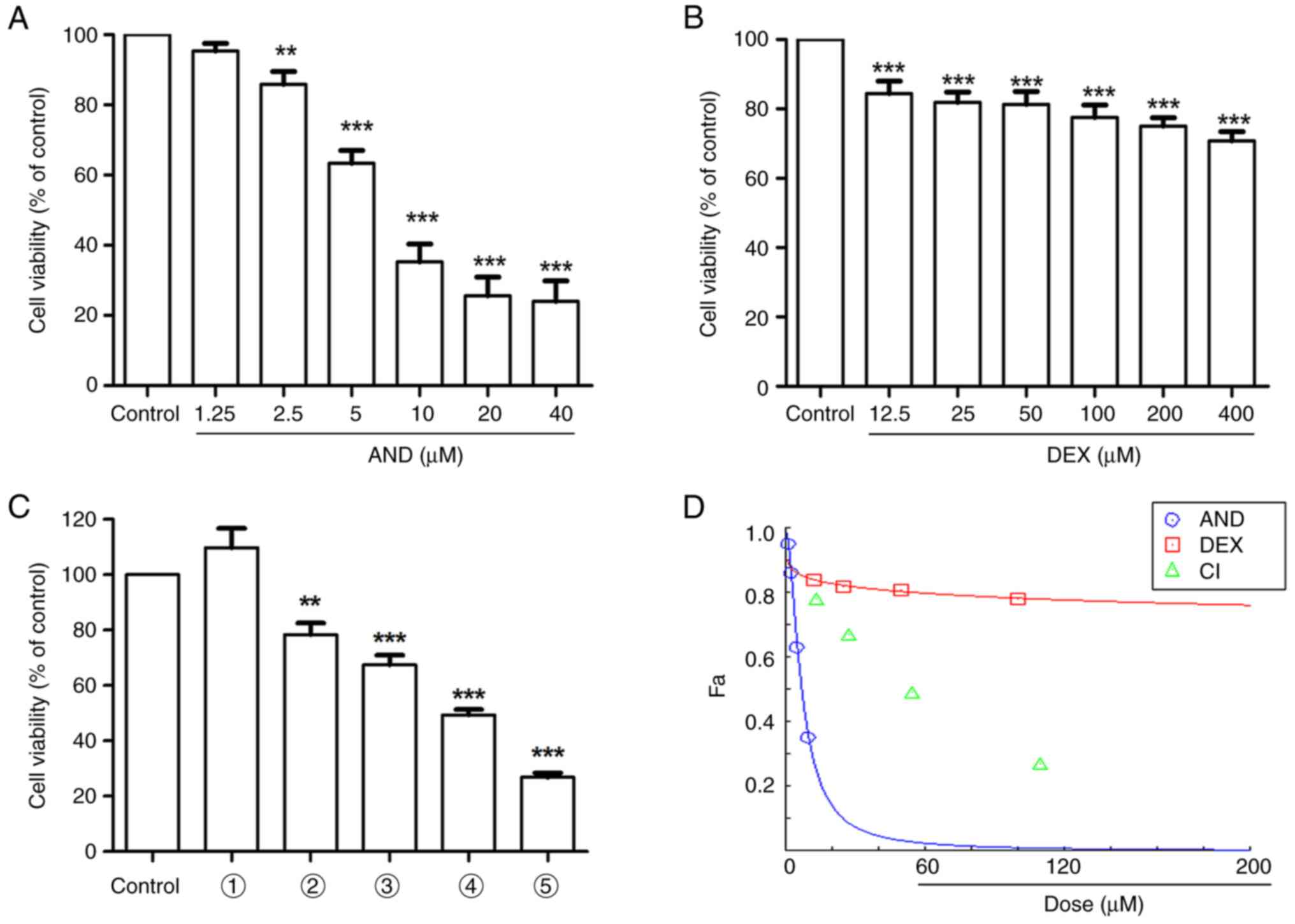 | Figure 1Effect of AND, DEX or AND + DEX on
the viability of CEM-C1 cells. Viability of CEM-C1 cells following
(A) AND or (B) DEX treatment, measured using a CCK-8 assay. (C)
Effect of AND + DEX on CEM-C1 cell viability, measured using a
CCK-8 assay. 1, vehicle; 2, 1.25 µM AND + 12.5 µM DEX; 3, 2.5 µM
AND + 25 µM DEX; 4, 5 µM AND + 50 µM DEX; and 5, 10 µM AND + 100 µM
DEX. (D) CompuSyn software was used to analyze the CI presented in
(C). n=3; **P<0.01, ***P<0.001 vs.
control. AND, andrographolide; CCK-8, Cell Counting Kit-8; CI,
combination index; DEX, dexamethasone; Fa, fraction affected. |
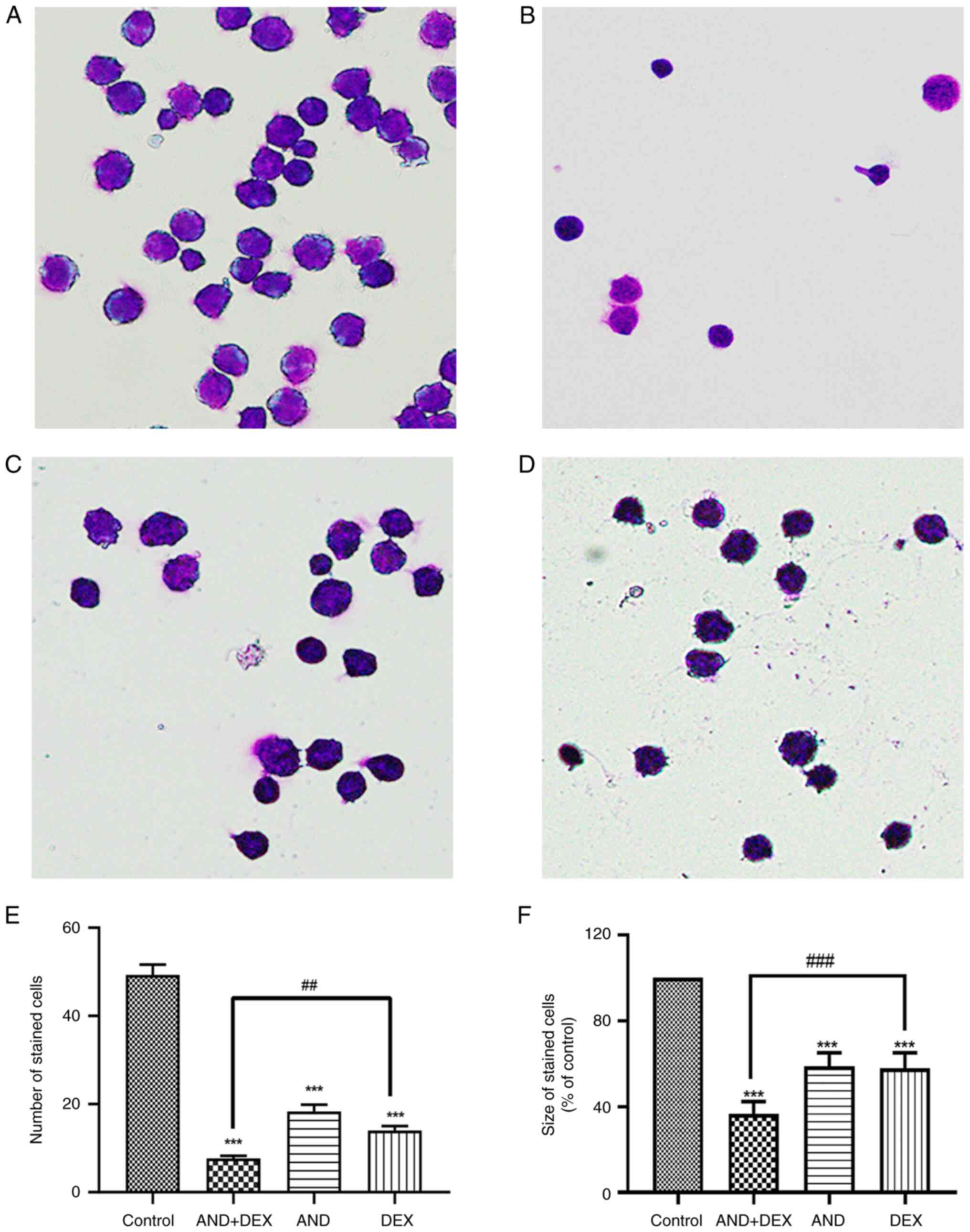
Wright-Giemsa staining indicates
alterations of the cell morphology
According to the results of the Wright-Giemsa
staining (Fig. 2), compared with
the administration of 5 µM AND or 50 µM DEX groups, the CEM-C1
cells were smaller in the AND + DEX group (Fig. 2F). In addition, the cells were
uniformly stained to purple and the number of stained cells was
reduced in the AND + DEX group (Fig.
2E). These results indicated that, compared with the AND or DEX
groups, co-treatment with 5 µM AND + 50 µM DEX had a greater
inhibitory effect on the growth of CEM-C1 cells.
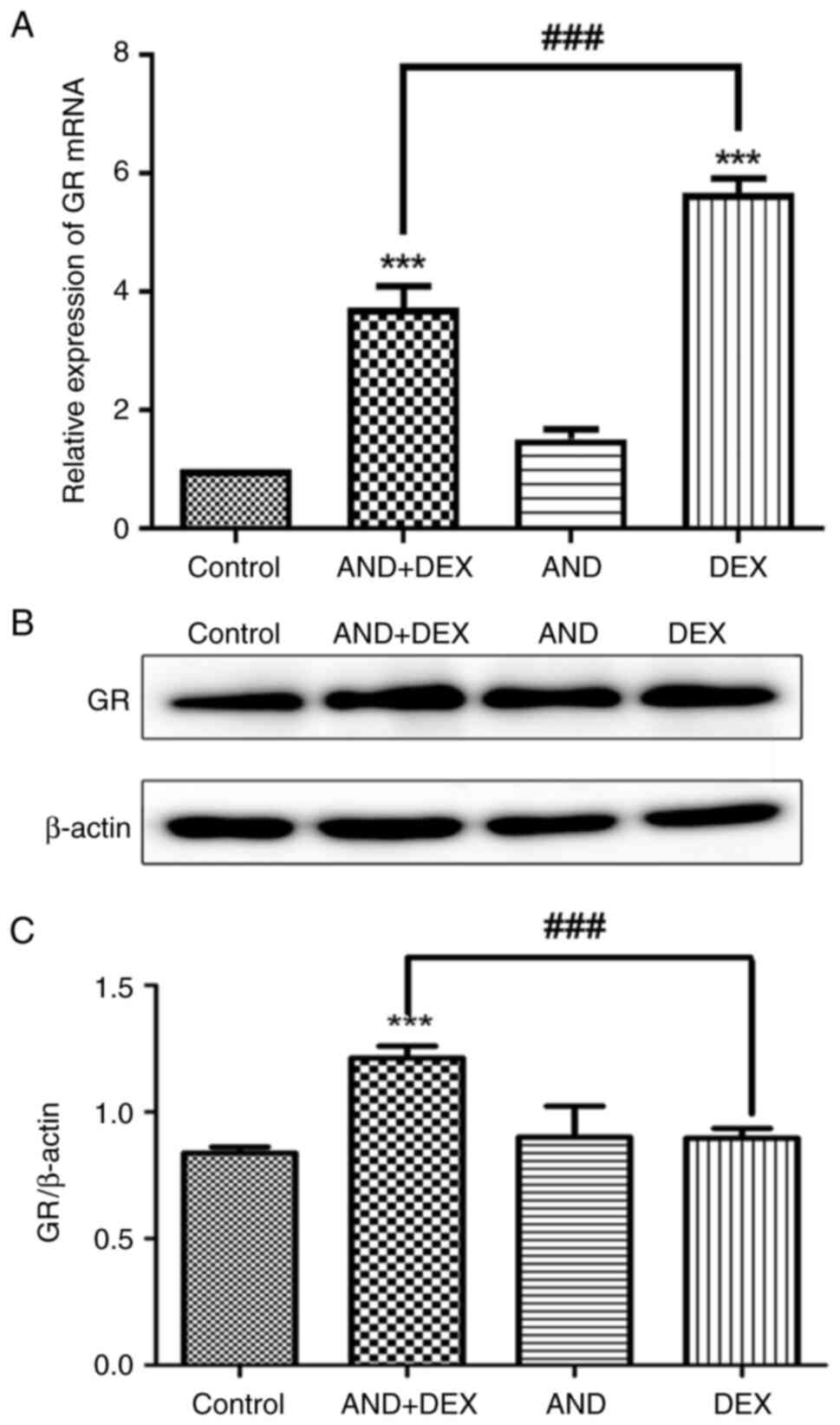
Co-treatment with AND + DEX
upregulates GR expression
GR mRNA expression in CEM-C1 cells following drug
treatment was detected via RT-qPCR. As shown in Fig. 3A, compared with that in the control
group, GR expression was significantly increased in the DEX and AND
+ DEX groups, and GR expression in the AND + DEX group was higher
than that in the DEX group. Subsequently, changes in GR protein
expression were analyzed by western blotting, which further
confirmed that GR expression in CEM-C1 cells of the AND + DEX group
was significantly upregulated compared with that in the control and
DEX groups (Fig. 3B and C). The aforementioned results suggested
that the combination of the two drugs upregulated the transcription
and post-transcription levels of GR.
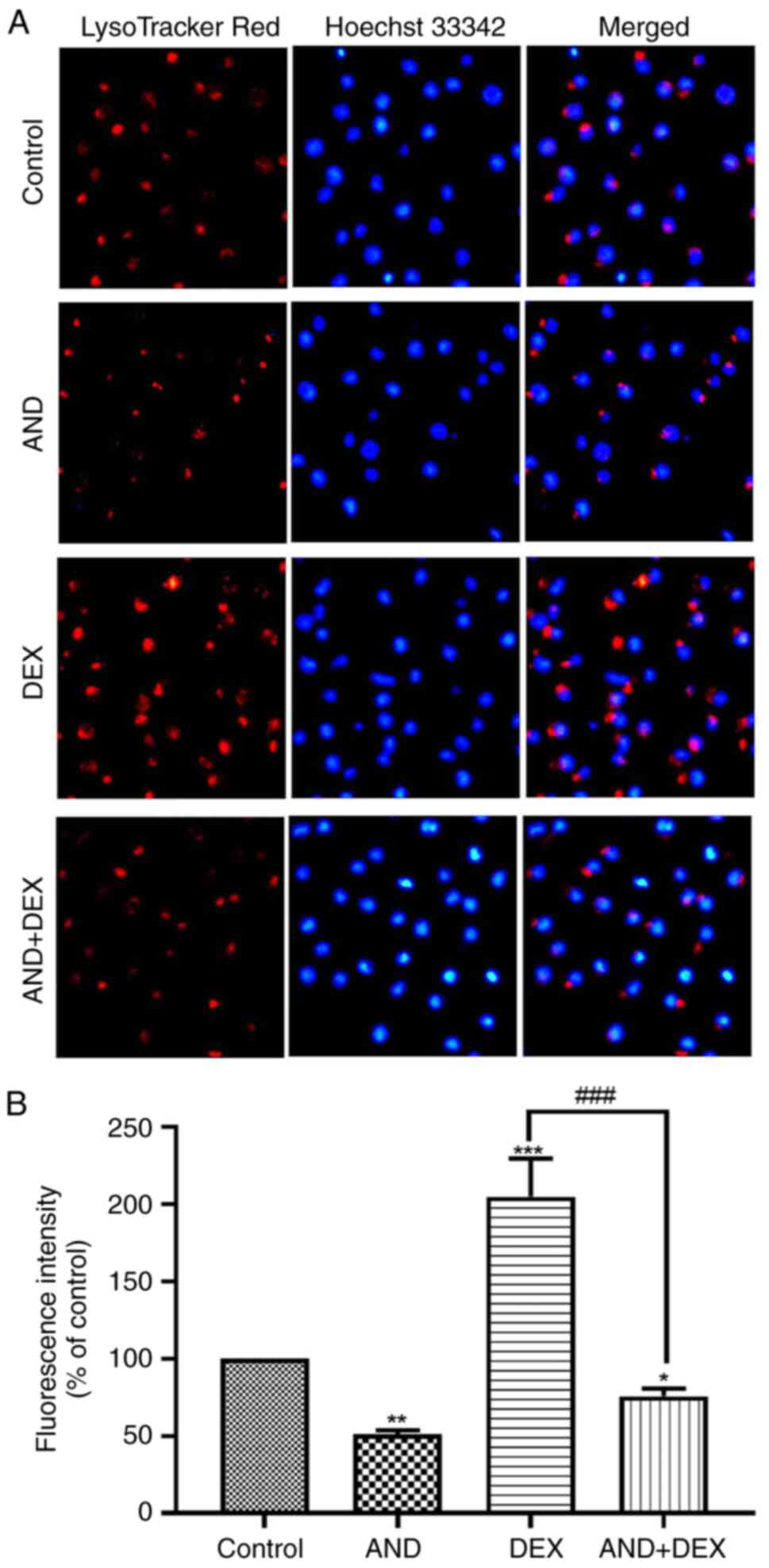
Co-treatment with AND + DEX alkalizes
the lysosomal lumen
LysoTracker Red is sensitive to pH alteration and
can be used to label and track acidic organelles (such as
autolysosomes) in live cells. The fluorescence intensity of
LysoTracker Red has a negative correlation with the pH of the
lysosome (56). As shown in
Fig. 4A, the nucleus of CEM-C1
cells exhibited blue fluorescence following incubation with Hoechst
33342 and the cells exhibited red fluorescence following staining
with LysoTracker Red. Compared with that of the 50 µM DEX group,
the LysoTracker Red fluorescence intensity of the AND + DEX group
was significantly reduced (Fig.
4B), which suggested that the lysosomal lumen of CEM-C1 cells
was alkalized and the lysosomal pH was increased by co-treatment
with AND + DEX.
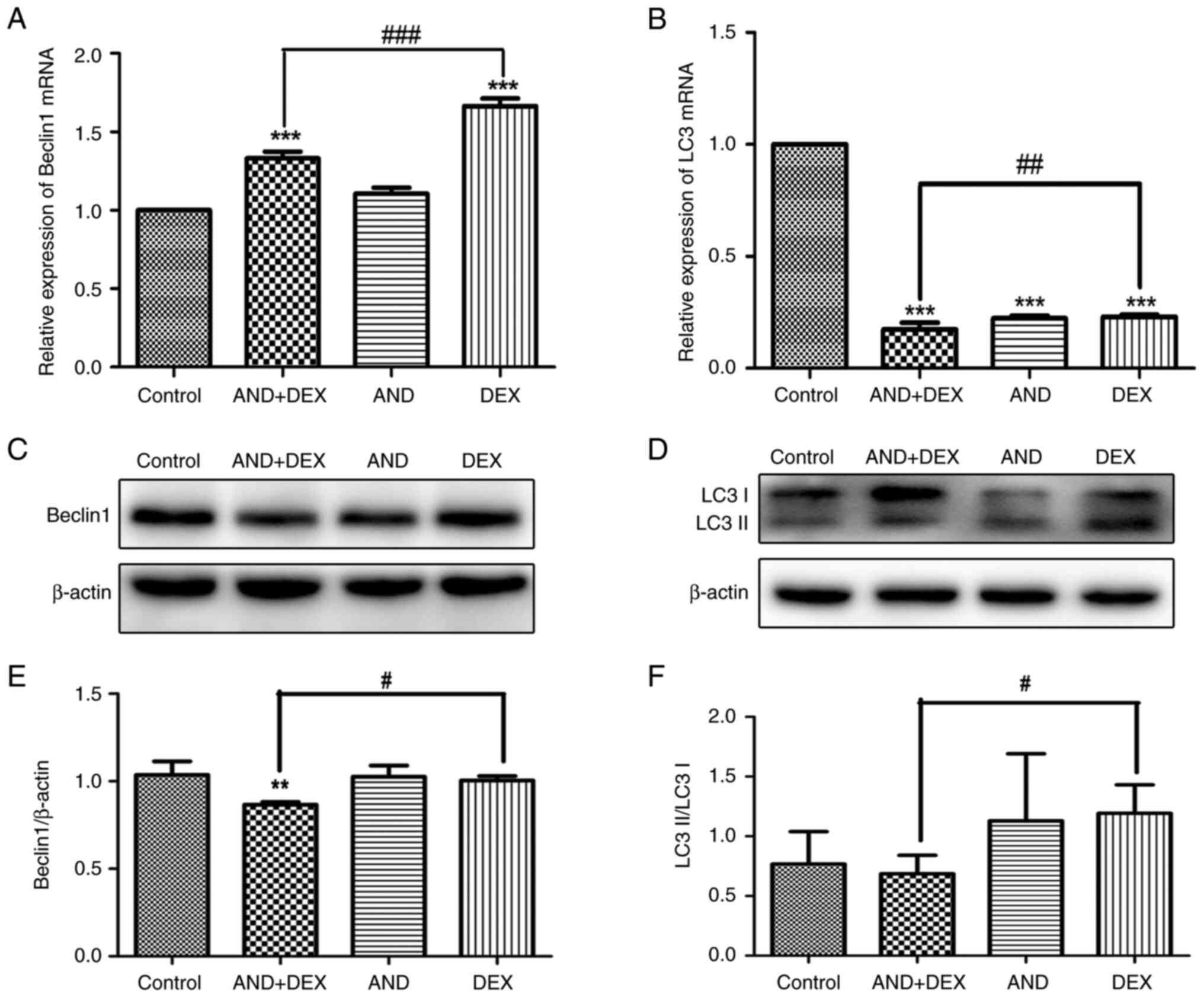
Co-treatment with AND + DEX inhibits
the expression of autophagy-related genes
The mRNA expression levels of Beclin1 and LC3 in
CEM-C1 cells following treatment were determined by RT-qPCR. As
shown in Fig. 5A, compared with
that in the control group, Beclin1 mRNA expression was
significantly increased in the DEX and AND + DEX groups, and the
level in the combination group was lower than that in the DEX
group. Similarly, LC3 mRNA expression was significantly decreased
in the drug-treated groups, and the LC3 mRNA expression in the AND
+ DEX group was lower than that in the DEX group (Fig. 5B). The changes in Beclin1 and LC3
protein expression were subsequently examined via western blotting.
As shown in Fig. 5C and E, the protein expression levels of Beclin1
in the AND + DEX group were reduced compared with those in the
control and 50 µM DEX groups. Additionally, as shown in Fig. 5D and F, compared with the 50 µM DEX group, the
conversion of LC3I to LC3II was reduced and the protein expression
levels of LC3II/LC3Ⅰ were downregulated in the AND + DEX group.
These results indicated that the combination of these two drugs
regulated the transcription and post-transcriptional levels of
autophagy-related genes.
Co-treatment with AND + DEX alters the
PI3K/AKT/mTOR signaling pathway in CEM-C1 cells
Western blotting was conducted to detect the
expression levels of key proteins in the PI3K/AKT/mTOR signaling
pathway. As shown in Fig. 6A-D, the
levels of p-PI3K, p-AKT and p-mTOR were significantly upregulated
in the AND + DEX group compared with the control group. However,
the expression levels of total PI3K, AKT and mTOR were not affected
(Fig. 6A), indicating that AND acts
with DEX to alter the activation of the aforementioned proteins,
thus inhibiting autophagy.
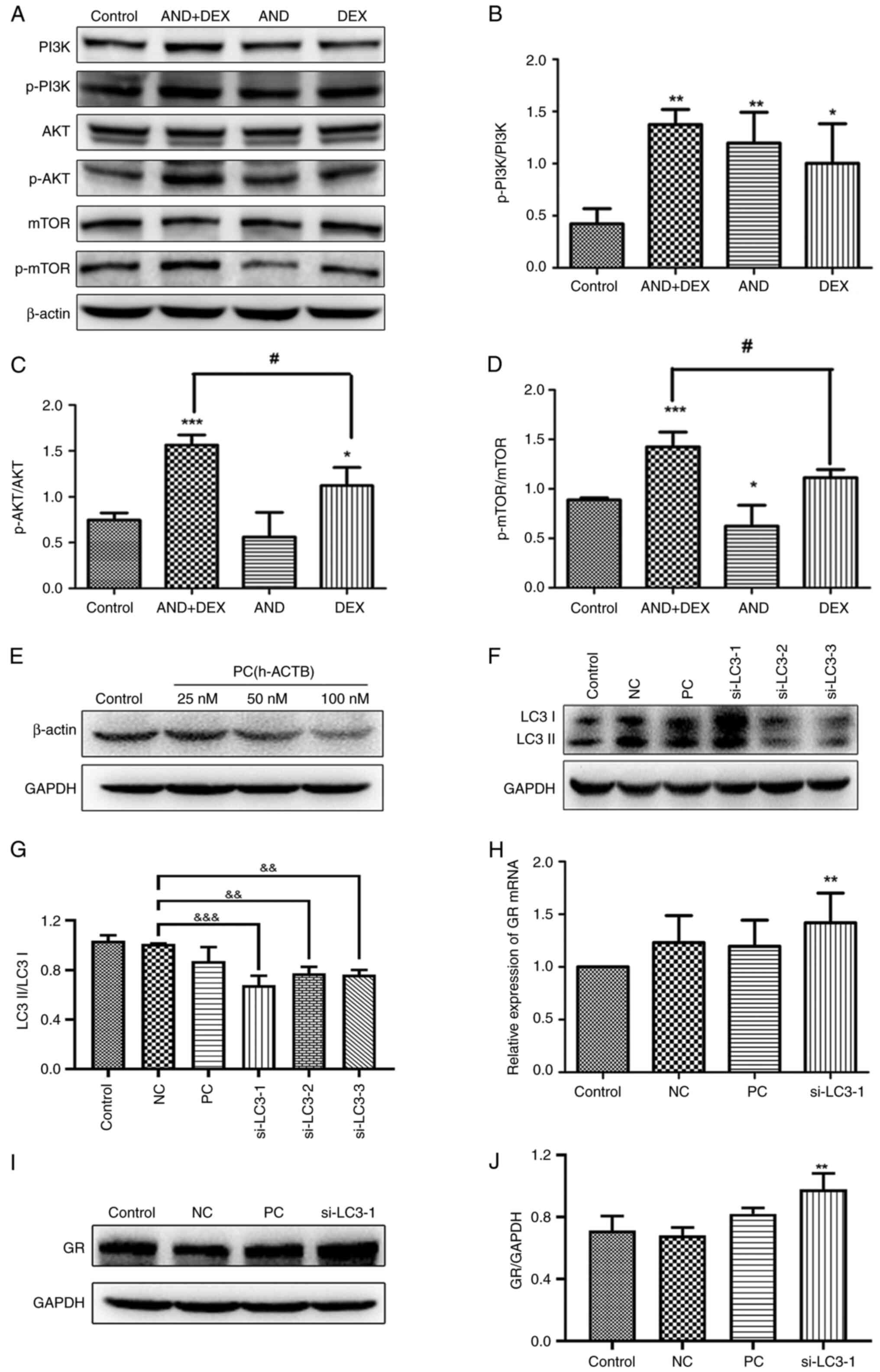 | Figure 6Effect of AND (5 µM), DEX (50 µM) or
AND (5 µM) + DEX (50 µM) on the PI3K/AKT/mTOR signaling pathway,
and GR expression after knockdown of the autophagy-related gene,
LC3. (A) Western blot images of proteins from the PI3K-AKT-mTOR
signaling pathway after drug treatment of CEM-C1 cells. The mean
ratio of (B) p-PI3K/PI3K, (C) p-AKT/AKT and (D) p-mTOR/mTOR
following the densitometric semi-quantification of bands. (E)
Transfection efficiency of the PC was determined by western
blotting. (F) Transfection efficiency of si-LC3 was determined by
western blotting, followed by the (G) semi-quantitation of the
ratio of LC3II to LC3I. (H) Reverse transcription-quantitative PCR
was used to detect GR mRNA expression following knockdown of LC3.
GAPDH was used as the internal reference. (I) Western blot images
and (J) semi-quantification of GR protein expression following
knockdown of LC3. n=3. *P<0.05,
**P<0.01, ***P<0.001 vs. control;
#P<0.05 vs. DEX only; &&P<0.01,
&&&P<0.001 vs. NC. AND, andrographolide;
DEX, dexamethasone; GR, glucocorticoid receptor; LC3,
microtubule-associated 1 protein light chain 3; NC, negative
control; p-, phosphorylated; PC, positive control; si, small
interfering RNA. |
GR expression increases following
knockdown of the autophagy-related gene, LC3
As shown in Fig. 6E,
compared with the control group, β-actin expression was
downregulated by PC siRNA transfection at 50 and 100 nM, which did
not affect the expression of GAPDH, indicating that PC siRNA
successfully knocked down β-actin expression and that the
transfection conditions tested were suitable for subsequent
experiments. Moreover, since the initial transfection concentration
recommended by the manufacturer was also 50 nM, 50 nM was selected
for subsequent experiments. In Fig.
6F and G, compared with those
in the NC group, the expression levels of LC3 in CEM-C1 cells
transfected with si-LC3-1, si-LC3-2 and si-LC3-3 were decreased,
indicating that LC3 knockdown was successful. It was also
demonstrated that the knockdown effect of si-LC3-1 was the
greatest. Therefore, si-LC3-1 was selected for subsequent
experiments. GR expression after knockdown of LC3 expression was
subsequently detected by RT-qPCR and western blotting. As shown in
Fig. 6H-J, compared with that in
the control group, GR mRNA and protein expression was increased
following knockdown of LC3 expression, and the difference was
statistically significant.
Discussion
The issue of determining prognosis in relapsed
patients with ALL has been a persistent concern, with
drug-resistant ALL phenotypes being the most prominent biological
feature of relapse (57).
Combination therapy has been proposed as a potential strategy to
overcome this issue (22,23,58,59).
In current clinical practice, combination therapy for cancer can
reduce drug toxicity and avoid the occurrence of rapid drug
resistance (60-62).
A previous study has shown that single-agent chemotherapy can
trigger autophagy, and the presence of autophagy may induce the
multi-drug resistance mechanism of cancer cells (63). It has been reported that AND is a
common autophagy inhibitor, and the present study demonstrated that
the intensity of the LysoTracker Red fluorescence was attenuated in
the AND group compared with the DEX group, suggesting that AND can
inhibit autophagy by alkalizing the lysosomal lumen (36,37).
In the present study, the combination of 5 µM AND + 50 µM DEX was
used to inhibit ALL cell viability. The results of the CCK-8 assay
demonstrated that the inhibition rate was as high as 51% in the 5
µM AND + 50 µM DEX group, and the CI indicated that the combination
of the two drugs had a synergistic effect. The results of the
present study also demonstrated that GR expression was upregulated
at the mRNA and protein levels in the AND + DEX group compared with
the 50 µM DEX group, which indicated that the combination of the
two drugs could increase the transcription of GR and inhibit the
degradation of GR protein. These findings, in combination with the
aforementioned findings, demonstrated that the mechanism behind GR
upregulation by the combination of these two drugs warrants further
study.
Autophagy is an evolutionarily conserved process
that serves an important role in tumor cell resistance to
chemotherapy, and changes in autophagy-related gene expression may
contribute to GC resistance in ALL (64). Beclin1, an essential protein for
autophagy, transitions from its metastable homodimeric state to
interact with key modulators, such as ATG14L or UVRAG, and form
functionally distinct ATG14L or UVRAG-containing Beclin1-VPS34
subcomplexes. The Beclin1-VPS34 complex serves essential roles in
membrane-mediated transport processes, including autophagy and
endosomal trafficking (65,66). In coordination with the Unc-51-like
autophagy activating kinase 1 complex, the Beclin1 complex
regulates early events in the initiation of autophagosome formation
(67,68). Another crucial autophagy protein,
LC3B (also known as ATG8F), facilitates autophagosome elongation
and maturation, leading to the sequestration of autophagic cargoes
(69). The results of the present
study demonstrated that the expression levels of LC3II/LC3I and
Beclin1 were significantly reduced following the co-treatment with
AND + DEX, compared with DEX alone. In addition, GR expression was
upregulated following AND + DEX treatment and following knockdown
of LC3 expression by siRNA transfection, indicating that treatment
with AND + DEX reduced the degradation of GR by inhibiting
autophagy. Therefore, AND + DEX combination therapy may serve an
anticancer role. In tumor cells, autophagy is regulated by multiple
signaling pathways (70), including
the PI3K/AKT/mTOR signaling pathway. The PI3K/AKT/mTOR signaling
pathway is widely recognized as a fundamental intracellular
signaling pathway that plays a role in cell physiology, cancer cell
metastasis and tumorigenesis, and inhibits autophagy when activated
(71,72). The present study demonstrated that,
compared with the control group, the p-PI3K, p-AKT and p-mTOR
protein levels were significantly upregulated in the AND + DEX
group, further indicating that this treatment combination could
inhibit autophagy by activating the PI3K/AKT/mTOR signaling
pathway.
In summary, in the present study, AND was found to
upregulate transcriptional and post-transcriptional GR expression
by inhibiting autophagy and the autophagy-dependent PI3K/AKT/mTOR
signaling pathway, thus increasing the sensitivity of
drug-resistant cells and serving an anti-ALL role. Therefore, the
present study provided a theoretical basis for a novel treatment
for ALL, and the presented results warrant further study.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by The National Natural
Science Foundation of China (grant no. 81660031) and The Innovation
and Entrepreneurship Training Program for College Students of
Guilin Medical University (grant no. 202010601013).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ and XQ designed the study. XL, TW, WC and JZ
conducted experiments. JD, YJ and WL contributed new reagents and
analytic tools. XL, JZ, YJ and WL contributed to acquisition of
data. XL, TW, JD and XQ performed data analysis. YZ, XL, TW and WC
wrote or contributed to the writing of the manuscript. XL and TW
confirm the authenticity of all the raw data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors are named inventors on Chinese patent,
ZL202010550895.1, entitled ‘The application of the combination of
andrographolide and dexamethasone in the preparation of compound
anti-acute lymphoblastic leukemia drugs’ held by Guilin Medical
University (Guilin, China) for the combination of andrographolide
and dexamethasone as a treatment for acute lymphoblastic leukemia,
which was approved on April 13, 2021.
References
|
1
|
Olivas-Aguirre M, Torres-López L, Pottosin
I and Dobrovinskaya O: Overcoming glucocorticoid resistance in
acute lymphoblastic leukemia: repurposed drugs can improve the
protocol. Front Oncol. 11(617937)2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Gregory S: Adult acute lymphoblastic
leukemia: treatment and management updates. Semin Oncol Nurs.
35(150951)2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Koh WJ, Greer BE, Abu-Rustum NR, Campos
SM, Cho KR, Chon HS, Chu C, Cohn D, Crispens MA, Dizon DS, et al:
Vulvar cancer, version 1.2017, NCCN clinical practice guidelines in
oncology. J Natl Compr Canc Netw. 15:92–120. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Huang FL, Yu SJ and Li CL: Role of
autophagy and apoptosis in acute lymphoblastic leukemia. Cancer
Control. 28(10732748211019138)2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Chennamadhavuni A, Lyengar V, Mukkamalla
SKR and Shimanovsky A: Leukemia. In: StatPearls [Internet].
Treasure Island (FL): StatPearls Publishing, 2023.
|
|
6
|
Park H, Youk J, Shin DY, Hong J, Kim I,
Kim NJ, Lee JO, Bang SM, Yoon SS, Park WB and Koh Y: Micafungin
prophylaxis for acute leukemia patients undergoing induction
chemotherapy. BMC Cancer. 19(358)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rafei H, Kantarjian HM and Jabbour EJ:
Recent advances in the treatment of acute lymphoblastic leukemia.
Leuk Lymphoma. 60:2606–2621. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Kato M and Manabe A: Treatment and biology
of pediatric acute lymphoblastic leukemia. Pediatr Int. 60:4–12.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Martinelli G, Papayannidis C, Piciocchi A,
Robustelli V, Soverini S, Terragna C, Marconi G, Lemoli RM, Guolo
F, Fornaro A, et al: INCB84344-201: Ponatinib and steroids in
frontline therapy for unfit patients with Ph+ acute lymphoblastic
leukemia. Blood Adv. 6:1742–1753. 2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Imai K: Acute lymphoblastic leukemia:
Pathophysiology and current therapy. Rinsho Ketsueki. 58:460–470.
2017.PubMed/NCBI View Article : Google Scholar : (In Japanese).
|
|
11
|
Aureli A, Marziani B, Venditti A,
Sconocchia T and Sconocchia G: Acute lymphoblastic leukemia
immunotherapy treatment: Now, next, and beyond. Cancers (Basel).
15(3346)2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lato MW, Przysucha A, Grosman S,
Zawitkowska J and Lejman M: The new therapeutic strategies in
pediatric T-cell acute lymphoblastic leukemia. Int J Mol Sci.
22(4502)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Guo XM, Fang YJ, Lv CL, Wang YR and Sun
XY: Changes of peripheral blood marrow-derived suppressor cell
level after chemotherapy induction remission by VDLP regimen and
their relationship with immune system in B-ALL children. Zhongguo
Shi Yan Xue Ye Xue Za Zhi. 25:1611–1614. 2017.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
14
|
Follini E, Marchesini M and Roti G:
Strategies to overcome resistance mechanisms in T-cell acute
lymphoblastic leukemia. Int J Mol Sci. 20(3021)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Samii B, Jafarian A, Rabbani M, Zolfaghari
B, Rahgozar S and Pouraboutaleb E: The effects of Astragalus
polysaccharides, tragacanthin, and bassorin on
methotrexate-resistant acute lymphoblastic leukemia. Res Pharm Sci.
18:381–391. 2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Inaba H and Pui CH: Glucocorticoid use in
acute lymphoblastic leukaemia. Lancet Oncol. 11:1096–1106.
2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kruth KA, Fang M, Shelton DN, Abu-Halawa
O, Mahling R, Yang H, Weissman JS, Loh ML, Müschen M, Tasian SK, et
al: Suppression of B-cell development genes is key to
glucocorticoid efficacy in treatment of acute lymphoblastic
leukemia. Blood. 129:3000–3008. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bedewy AM, El-Maghraby SM, Kandil NS and
El-Bendary WR: The prognostic value of glucocorticoid receptors for
adult acute lymphoblastic leukemia. Blood Res. 50:235–241.
2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xu JY and Luo JM: Association between BIM
gene and glucocorticoid resistance in children with acute
lymphoblastic leukemia. Zhongguo Dang Dai Er Ke Za Zhi. 19:945–949.
2017.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
20
|
Gong H, Liu L, Cui L, Ma H and Shen L:
ALKBH5-mediated m6A-demethylation of USP1 regulated T-cell acute
lymphoblastic leukemia cell glucocorticoid resistance by Aurora B.
Mol Carcinog. 60:644–657. 2021.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Yuan N, Song L, Zhang S, Lin W, Cao Y, Xu
F, Fang Y, Wang Z, Zhang H, Li X, et al: Bafilomycin A1 targets
both autophagy and apoptosis pathways in pediatric B-cell acute
lymphoblastic leukemia. Haematologica. 100:345–356. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wandler AM, Huang BJ, Craig JW, Hayes K,
Yan H, Meyer LK, Scacchetti A, Monsalve G, Dail M, Li Q, et al:
Loss of glucocorticoid receptor expression mediates in vivo
dexamethasone resistance in T-cell acute lymphoblastic leukemia.
Leukemia. 34:2025–2037. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Kaveh K, Takahashi Y, Farrar MA, Storme G,
Guido M, Piepenburg J, Penning J, Foo J, Leder KZ and Hui SK:
Combination therapeutics of Nilotinib and radiation in acute
lymphoblastic leukemia as an effective method against
drug-resistance. PLoS Comput Biol. 13(e1005482)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Burgos RA, Alarcón P, Quiroga J, Manosalva
C and Hancke J: Andrographolide, an anti-inflammatory multitarget
drug: All roads lead to cellular metabolism. Molecules.
26(5)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Guo BJ, Liu Z, Ding MY, Li F, Jing M, Xu
LP, Wang YQ, Zhang ZJ, Wang Y, Wang D, et al: Andrographolide
derivative ameliorates dextran sulfate sodium-induced experimental
colitis in mice. Biochem Pharmacol. 163:416–424. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li X, Yuan K, Zhu Q, Lu Q, Jiang H, Zhu M,
Huang G and Xu A: Andrographolide ameliorates rheumatoid arthritis
by regulating the apoptosis-NETosis balance of neutrophils. Int J
Mol Sci. 20(5035)2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ahmed S, Kwatra M, Ranjan Panda S, Murty
USN and Naidu VGM: Andrographolide suppresses NLRP3 inflammasome
activation in microglia through induction of parkin-mediated
mitophagy in in-vitro and in-vivo models of Parkinson disease.
Brain Behav Immun. 91:142–158. 2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Doi H, Matsui T, Dijkstra JM, Ogasawara A,
Higashimoto Y, Imamura S, Ohye T, Takematsu H, Katsuda I and
Akiyama H: Andrographolide, isolated from Andrographis paniculata,
induces apoptosis in monocytic leukemia and multiple myeloma cells
via augmentation of reactive oxygen species production. F1000Res.
10(542)2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Latif R and Wang CY: Andrographolide as a
potent and promising antiviral agent. Chin J Nat Med. 18:760–769.
2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Adiguna SP, Panggabean JA, Atikana A,
Untari F, Izzati F, Bayu A, Rosyidah A, Rahmawati SI and Putra MY:
Antiviral activities of andrographolide and its derivatives:
Mechanism of action and delivery system. Pharmaceuticals (Basel).
14(1102)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang L, Bao M, Liu B, Zhao H, Zhang Y, Ji
X, Zhao N, Zhang C, He X, Yi J, et al: Effect of andrographolide
and its analogs on bacterial infection: A review. Pharmacology.
105:123–134. 2020.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Kumar G, Singh D, Tali JA, Dheer D and
Shankar R: Andrographolide: Chemical modification and its effect on
biological activities. Bioorg Chem. 95(103511)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mussard E, Cesaro A, Lespessailles E,
Legrain B, Berteina-Raboin S and Toumi H: Andrographolide, a
natural antioxidant: An update. Antioxidants (Basel).
8(571)2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Jadhav AK and Karuppayil SM: Andrographis
paniculata (Burm. F) wall ex nees: Antiviral properties. Phytother
Res. 35:5365–5373. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Tandoh A, Danquah CA, Benneh CK, Adongo
DW, Boakye-Gyasi E and Woode E: Effect of diclofenac and
andrographolide combination on carrageenan-induced paw edema and
hyperalgesia in rats. Dose Response.
20(15593258221103846)2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yuwen D, Mi S, Ma Y, Guo W, Xu Q, Shen Y
and Shu Y: Andrographolide enhances cisplatin-mediated anticancer
effects in lung cancer cells through blockade of autophagy.
Anticancer Drugs. 28:967–976. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Mi S, Xiang G, Yuwen D, Gao J, Guo W, Wu
X, Wu X, Sun Y, Su Y, Shen Y and Xu Q: Inhibition of autophagy by
andrographolide resensitizes cisplatin-resistant non-small cell
lung carcinoma cells via activation of the Akt/mTOR pathway.
Toxicol Appl Pharmacol. 310:78–86. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yang T, Yao S, Zhang X and Guo Y:
Andrographolide inhibits growth of human T-cell acute lymphoblastic
leukemia Jurkat cells by downregulation of PI3K/AKT and
upregulation of p38 MAPK pathways. Drug Des Devel Ther.
10:1389–1397. 2016.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kocak M, Ezazi Erdi S, Jorba G, Maestro I,
Farrés J, Kirkin V, Martinez A and Pless O: Targeting autophagy in
disease: Established and new strategies. Autophagy. 18:473–495.
2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chen Y and Gibson SB: Three dimensions of
autophagy in regulating tumor growth: Cell survival/death, cell
proliferation, and tumor dormancy. Biochim Biophys Acta Mol Basis
Dis. 1867(166265)2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wei H, Wang C, Croce CM and Guan JL:
p62/SQSTM1 synergizes with autophagy for tumor growth in vivo.
Genes Dev. 28:1204–1216. 2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer.
19(12)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Amaravadi RK, Kimmelman AC and Debnath J:
Targeting autophagy in cancer: Recent advances and future
directions. Cancer Discov. 9:1167–1181. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Pugsley HR: Quantifying autophagy:
Measuring LC3 puncta and autolysosome formation in cells using
multispectral imaging flow cytometry. Methods. 112:147–156.
2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Shi B, Ma M, Zheng Y, Pan Y and Lin X:
mTOR and Beclin1: Two key autophagy-related molecules and their
roles in myocardial ischemia/reperfusion injury. J Cell Physiol.
234:12562–12568. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Nishimura T and Tooze SA: Emerging roles
of ATG proteins and membrane lipids in autophagosome formation.
Cell Discov. 6(32)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Tran S, Fairlie WD and Lee EF: BECLIN1:
Protein structure, function and regulation. Cells.
10(1522)2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Dunn WA Jr: Autophagy and related
mechanisms of lysosome-mediated protein degradation. Trends Cell
Biol. 4:139–143. 1994.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Gu X, Guo W, Zhao Y, Liu G, Wu J and Chang
C: Deoxynivalenol-induced cytotoxicity and apoptosis in IPEC-J2
cells through the activation of autophagy by inhibiting
PI3K-AKT-mTOR signaling pathway. ACS Omega. 4:18478–18486.
2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Song G, Lu H, Chen F, Wang Y, Fan W, Shao
W, Lu H and Lin B: Tetrahydrocurcumin-induced autophagy via
suppression of PI3K/Akt/mTOR in non-small cell lung carcinoma
cells. Mol Med Rep. 17:5964–5969. 2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Fattahi S, Amjadi-Moheb F, Tabaripour R,
Ashrafi GH and Akhavan-Niaki H: PI3K/AKT/mTOR signaling in gastric
cancer: Epigenetics and beyond. Life Sci.
262(118513)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Xu Z, Han X, Ou D, Liu T, Li Z, Jiang G,
Liu J and Zhang J: Targeting PI3K/AKT/mTOR-mediated autophagy for
tumor therapy. Appl Microbiol Biotechnol. 104:575–587.
2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Li X, Zhang W, Liang L, Duan X, Deng J and
Zhou Y: Natural product-derived icaritin exerts anti-glioblastoma
effects by positively modulating estrogen receptor β. Exp Ther Med.
19:2841–2850. 2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Duarte D, Falcão SI, El Mehdi I,
Vilas-Boas M and Vale N: Honeybee venom synergistically enhances
the cytotoxic effect of CNS drugs in HT-29 colon and MCF-7 breast
cancer cell lines. Pharmaceutics. 14(511)2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Feng L, Lu CK, Wu J, Chan LL and Yue J:
Identification of anhydrodebromoaplysiatoxin as a dichotomic
autophagy inhibitor. Mar Drugs. 21(46)2023.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Pierro J, Hogan LE, Bhatla T and Carroll
WL: New targeted therapies for relapsed pediatric acute
lymphoblastic leukemia. Expert Rev Anticancer Ther. 17:725–736.
2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Habiel DM, Krepostman N, Lilly M,
Cavassani K, Coelho AL, Shibata T, Elenitoba-Johnson K and Hogaboam
CM: Senescent stromal cell-induced divergence and therapeutic
resistance in T cell acute lymphoblastic leukemia/lymphoma.
Oncotarget. 7:83514–83529. 2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Rose-James A, Shiji R, Kusumakumary P,
Nair M, George SK and Sreelekha TT: Profiling gene mutations,
translocations, and multidrug resistance in pediatric acute
lymphoblastic leukemia: A step forward to personalizing medicine.
Med Oncol. 33(98)2016.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Palmer AC, Chidley C and Sorger PK: A
curative combination cancer therapy achieves high fractional cell
killing through low cross-resistance and drug additivity. Elife.
8(e50036)2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Jaaks P, Coker EA, Vis DJ, Edwards O,
Carpenter EF, Leto SM, Dwane L, Sassi F, Lightfoot H, Barthorpe S,
et al: Effective drug combinations in breast, colon and pancreatic
cancer cells. Nature. 603:166–173. 2022.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Pemovska T, Bigenzahn JW and Superti-Furga
G: Recent advances in combinatorial drug screening and synergy
scoring. Curr Opin Pharmacol. 42:102–110. 2018.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye
WC, Zhang DM and Chen ZS: Autophagy and multidrug resistance in
cancer. Chin J Cancer. 36(52)2017.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Sarang Z, Gyurina K, Scholtz B, Kiss C and
Szegedi I: Altered expression of autophagy-related genes might
contribute to glucocorticoid resistance in precursor B-cell-type
acute lymphoblastic leukemia. Eur J Haematol. 97:453–460.
2016.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Li Y, Qu M, Xing F, Li H, Cheng D, Xing N
and Zhang W: The protective mechanism of dexmedetomidine in
regulating Atg14L-Beclin1-Vps34 complex against myocardial
ischemia-reperfusion injury. J Cardiovasc Transl Res. 14:1063–1074.
2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Wu S, He Y, Qiu X, Yang W, Liu W, Li X, Li
Y, Shen HM, Wang R, Yue Z and Zhao Y: Targeting the potent Beclin
1-UVRAG coiled-coil interaction with designed peptides enhances
autophagy and endolysosomal trafficking. Proc Natl Acad Sci USA.
115:E5669–E5678. 2018.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Wu W, Wang X, Sun Y, Berleth N, Deitersen
J, Schlutermann D, Stuhldreier F, Wallot-Hieke N, José Mendiburo M,
Cox J, et al: TNF-induced necroptosis initiates early autophagy
events via RIPK3-dependent AMPK activation, but inhibits late
autophagy. Autophagy. 17:3992–4009. 2021.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Nakahira K, Pabon Porras MA and Choi AM:
Autophagy in pulmonary diseases. Am J Respir Crit Care Med.
194:1196–1207. 2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Wesch N, Kirkin V and Rogov VV:
Atg8-family proteins-structural features and molecular interactions
in autophagy and beyond. Cells. 9(2008)2020.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Prieto-Domínguez N, Ordóñez R, Fernández
A, García-Palomo A, Muntané J, González-Gallego J and Mauriz JL:
Modulation of autophagy by sorafenib: Effects on treatment
response. Front Pharmacol. 7(151)2016.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Yang G, Li Z, Dong L and Zhou F: lncRNA
ADAMTS9-AS1 promotes bladder cancer cell invasion, migration, and
inhibits apoptosis and autophagy through PI3K/AKT/mTOR signaling
pathway. Int J Biochem Cell Biol. 140(106069)2021.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Petrulea MS, Plantinga TS, Smit JW,
Georgescu CE and Netea-Maier RT: PI3K/Akt/mTOR: A promising
therapeutic target for non-medullary thyroid carcinoma. Cancer
Treat Rev. 41:707–713. 2015.PubMed/NCBI View Article : Google Scholar
|