Introduction
Hepatocellular carcinoma (HCC) is the sixth most
common type of malignant tumor and the fourth leading cause of
cancer death worldwide. In 2018, global incidence of liver cancer
was 4.7% and the mortality rate was 8.2% (1). Incidence of HCC has been increasing
and the mortality rate has been growing faster than that of any
other malignant tumor (2).
Especially in developing countries, the prevalence of hepatitis B
virus (HBV)-associated HCC remains high and is associated with
local HBV epidemics (3).
HBV is a hepatotropic DNA virus infecting ~240
million people worldwide (4). HBV
DNA is detected in serum and tissue samples from patients with
HBV-related HCC (5). The mechanism
by which HBV promotes HCC is unclear, but it is hypothesized that
the pathogenesis of HCC is caused by chronic HB triggering
cirrhosis (6). The mortality
related to HBV-associated diseases, which account for ~600,000
deaths each year, is mainly caused by decompensated cirrhosis and
HCC (7). HBV mediates cell
proliferation and DNA damage by increasing oxidative stress and
inflammation, which are key links in the pathological changes of
HCC (8). A study also revealed that
the high expression of the HBV X gene leads to HCC in transgenic
mice (9). Therefore, it is
necessary to study the uncovered mechanism of HBV in pathological
changes in the liver.
MicroRNAs (miRNAs or miRs) have begun to receive
widespread attention in the field of virology (10-12).
miRNAs are non-coding RNAs with a length of ~22 nt that primarily
control gene expression via post-transcriptional regulation
patterns (13). Abnormally
expressed miRNAs are often detected in patients with HCC and HBV
infection and may be considered biomarkers for diagnosis and
prognosis assessment, such as miR-375, miR-92a, miR-25 and let-7f
(14,15). miRNAs serve a number of functions
in vivo to regulate cell invasion, migration, proliferation,
apoptosis and metabolism (16-19).
Additionally, miRNAs serve a critical role in the process of
oxidative stress, inflammation and autophagy (20,21).
HBV can influence the expression of miRNAs at multiple stages
following infection (22). However,
expression patterns of numerous miRNAs, such as miR-125b and
miR-210, remain controversial in patients with HBV-associated HCC:
Some researchers believe that miR-125b and miR-210 are highly
expressed in HCC, while others believe that the expression of the
two miRNAs is low in HCC (23-27).
Researchers have reported that miR-125b and miR-210
can be up- or downregulated in patients with HBV-associated HCC
(23-25).
Certain miRNAs have only been evaluated in a few studies of
HBV-related HCC, and their biological changes and mechanisms need
to be further elucidated, such as miR-374b (28,29).
Therefore, the regulatory mechanism of miRNAs in HCC needs to be
further studied.
Proteins serve a vital role in the occurrence and
development of tumors (30).
Previous studies have demonstrated that miRNAs in the cytoplasm
inhibit gene expression by binding to the 3' untranslated region
(UTR) of the gene to inhibit translation or direct degradation of
mRNAs (31,32). However, unlike the role of
cytoplasmic miRNA, miRNAs in the nucleus are primarily involved in
regulating transcription (33).
miRNAs, such as miR-373, miR-744 and miR-1186, are located in the
nucleus and can combine with promoter or enhancer regions to
activate gene expression (33). The
present study aimed to use bioinformatics methods using the Gene
Expression Omnibus (GEO) database and R programming language to
identify the HBV-associated miRNA-mRNA regulatory network.
Materials and methods
Analysis with R programming
language
The ‘limma’ package was obtained from Bioconductor
(bioconductor.org/biocLite) and was
employed to analyze the data via R software (version 4.0.0). RNA
sequencing (-seq) data was imported into the R environment and
converted into a data structure supported by ‘DESeq2’ package
(bioconductor.org/biocLite). Original
data was preprocessed by standardizing the data, removing
low-expression genes and batch effects. R language ‘limma’, ‘dplyr’
and ‘tidyr’ packages (bioconductor.org/biocLite) were used for differential
expression gene analysis. R package ‘ggplot2’ was applied to
visualize the results. Heatmap results of the miRNAs and
differential gene expression were produced with TBtools (version
1.09876) (34).
Non-coding RNA profiling by array
The miRNA expression database was established by
searching GEO Data Sets (ncbi.nlm.nih.gov). The inclusion criteria were samples
must be pathologically confirmed as HCC patients and miRNA
expression profiles are detected in tumor and adjacent normal
tissues. Through screening, two datasets containing samples from
tissues of patients with HCC were included: GSE67882 and GSE69580.
A total of 13 HCC and nine non-tumor tissues were included in the
present study. These patients with HCC had diagnosis confirmed
through pathological examination. The ‘limma’ R package was applied
to compare expression levels of miRNAs between HBV-related HCC and
the corresponding non-tumor tissues from the two series of matrix
files. After data normalization, expression levels of the miRNAs in
the two datasets were analyzed.
Analysis of prognostic value of
differentially expressed miRNAs (DE-miRNAs)
The prognostic values of DE-miRNAs in HCC were
analyzed using the Kaplan-Meier Plotter database (kmplot.com/analysis/index.php?p=service&cancer=liver_mirna).
Through comparing the relationship between miRNA expression levels
and the survival rate of HCC patients, the DE-miRNAs with an impact
on the prognosis of patients with HBV-related HCC were selected for
further analysis.
DE-miRNA target gene prediction
miRNet (mirnet.ca/miRNet/home.xhtml), an easy-to-use analysis
tool based on an online network, was used to predict the miRNA
target genes (35). miRNet
integrates data from 11 high-quality miRNA-target interaction
databases and displays the results through a network visualization
system. Gephi software (version 0.9.2, gephi.org/users/), a network visualization software
based on the JAVA working environment, displayed the regulatory
association between miRNAs and their target genes.
miRNA-hub gene prediction
Cytoscape software (version 3.7.1, https://github.com/cytoscape/cytoscape)
was used to evaluate the miRNA hub genes. For all DE-miRNA target
genes, Molecular Complex Detection plug-in (version 3.3.10,
http://apps.cytoscape.org/) was used for
scoring according to the interaction between the genes. The higher
the score, the more key the role the hub genes play in the entire
gene network.
Functional annotation and signal
pathway enrichment analysis
Database for Annotation, Visualization and
Integrated Discovery (DAVID; david.ncifcrf.gov/home.jsp) was applied for Gene
Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
analysis of DE-miRNA target genes. For GSE101728 data, containing
the expression profiling of lncRNA and mRNA in HCC (36), the enrichment of KEGG pathway
according to the top 25% of the highest and lowest GRB2 expression
patients' genes expression profiling was assessed.
HBV-associated HCC target gene
prediction
HBV-associated experimental data were screened
through the GEO database. In the GSE100400 dataset, the researchers
combined circularized chromosome conformation capture (4C) with
RNA-seq and chromatin immunoprecipitation (ChIP)-seq to illuminate
the nuclear landscape associated with HBV episomes and HBx
(37). To the best of our
knowledge, the aforementioned study is the first to use 4C to
identify regional virus-host genome interactions. By using R
programming language tool, expression matrix of RNA-seq data in
GSE100400 was obtained and the difference in gene expression was
calculated.
The Cancer Genome Atlas (TCGA)
database analysis
UALCAN database (ualcan.path.uab.edu/) is an easy-to-use interactive
web portal that for in-depth analysis of TCGA gene expression data.
UALCAN includes TCGA RNA-seq and clinical data from 31 types of
cancer (38). This was used to
select genes with significant differences in expression and
prognosis between tumors and corresponding adjacent tissues.
Immune infiltrate analysis
The Tumor Immune Estimation Resource (TIMER)
database (cistrome.shinyapps.io/timer/) is designed to
investigate the molecular characteristics of tumor-immune
interactions and analyze the correlation between gene expression
and the abundance of immune infiltrates (39). This was used to screen genes related
to immune infiltration.
Cell culture and transfection
Human hepatoblastoma cell lines (HepG2 and
HepG2.2.15) were purchased from Guangzhou Cellcook Biotech Co.,
Ltd. and were authenticated via STR profiling. The hepatoblastoma
cells were grown in Minimal Essential Medium supplemented with
non-essential amino acids and 10% fetal bovine serum (all WISENT,
Inc.). Cells were placed in a cell incubator (Thermo Fisher
Scientific, Inc.) at 37˚C with 5% carbon dioxide. miRNA-93 mimics
and inhibitors and negative controls (Guangzhou RiboBio Co., Ltd.)
were prepared for cell transfection. The confluence of the cells
was maintained at ~60% before transfection. Cells were starved in
serum-free Minimal Essential Medium (WISENT, Inc.) at 37˚C with 5%
carbon dioxide for 2 h and transfected with 1 µg miRNA-93 mimics
and inhibitors and their respective blank vectors using
Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) for 36 h at 37˚C. Subsequent experiment was conducted within
72 h. The sequences of the miRNA-93 mimics/inhibitors are displayed
in Table SI. The sequence of
mature miRNA-93 was 5'-CAAAGUGCUGUUCGUGCAGGUAG-3'. The sequence of
mimics negative control was 5'-UUGUACUACACAAAAGUACUG-3' and
inhibitor negative control was 5'-CAGUACUUUUGUGUAGUACAA-3'.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA of the cells was extracted using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.). RT was performed
using a PrimeScript™ RT reagent Kit (Takara Bio, Inc.,
No RR047A) according to the manufacturer's instructions. A SYBR
Premix Ex Taq II kit (Takara Bio, Inc.) was used for RT-qPCR with
thermocycling conditions as follows: initial denaturation 95˚C, 30
sec; 40 cycles of denaturation 95˚C, 30 sec, annealing at 60˚C 30
sec and extension 72˚C, 30 sec. StepOne Plus system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was employed. Target
gene expression was normalized using the
2-ΔΔCq method (40). The sequences of mature miRNAs are
presented in Table SII; RT-qPCR
primers (Shanghai GenePharma Co., Ltd.) are displayed in Table SIII.
Statistical analysis
All experimental data with at least 3 times are
presented as the mean ± standard deviation using GraphPad Prism
(version 5.0; Dotmatics) and the statistical analysis was performed
using SPSS 20.0 (IBM Corp.). Differences between the two groups was
analyzed using paired t tests. For >2 groups in TCGA database,
One-way ANOVA was used followed by Fisher's least significant
difference post hoc test. The correlation analysis between two
genes was evaluated using the Spearman rank correlation
coefficient. All plots were drawn with GraphPad Prism software. For
the results of miRNA microarray, P<0.05 and
|log2fold-change (FC)|> 1 were considered to define
the DE-miRNAs. The association between expression of DE-miRNAs and
the prognostic value was analyzed using log-rank test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Screening of potential DE-miRNAs in
HBV-associated HCC
The original data of miRNA arrays GSE67882 and
GSE69580 were obtained and normalized (Fig. S1). A total of 13 HCC and nine
non-tumor tissues were included for further analysis. The ‘limma’ R
package was used to generate volcano plots of miRNA expression
(Fig. S2). The top 10 up- and
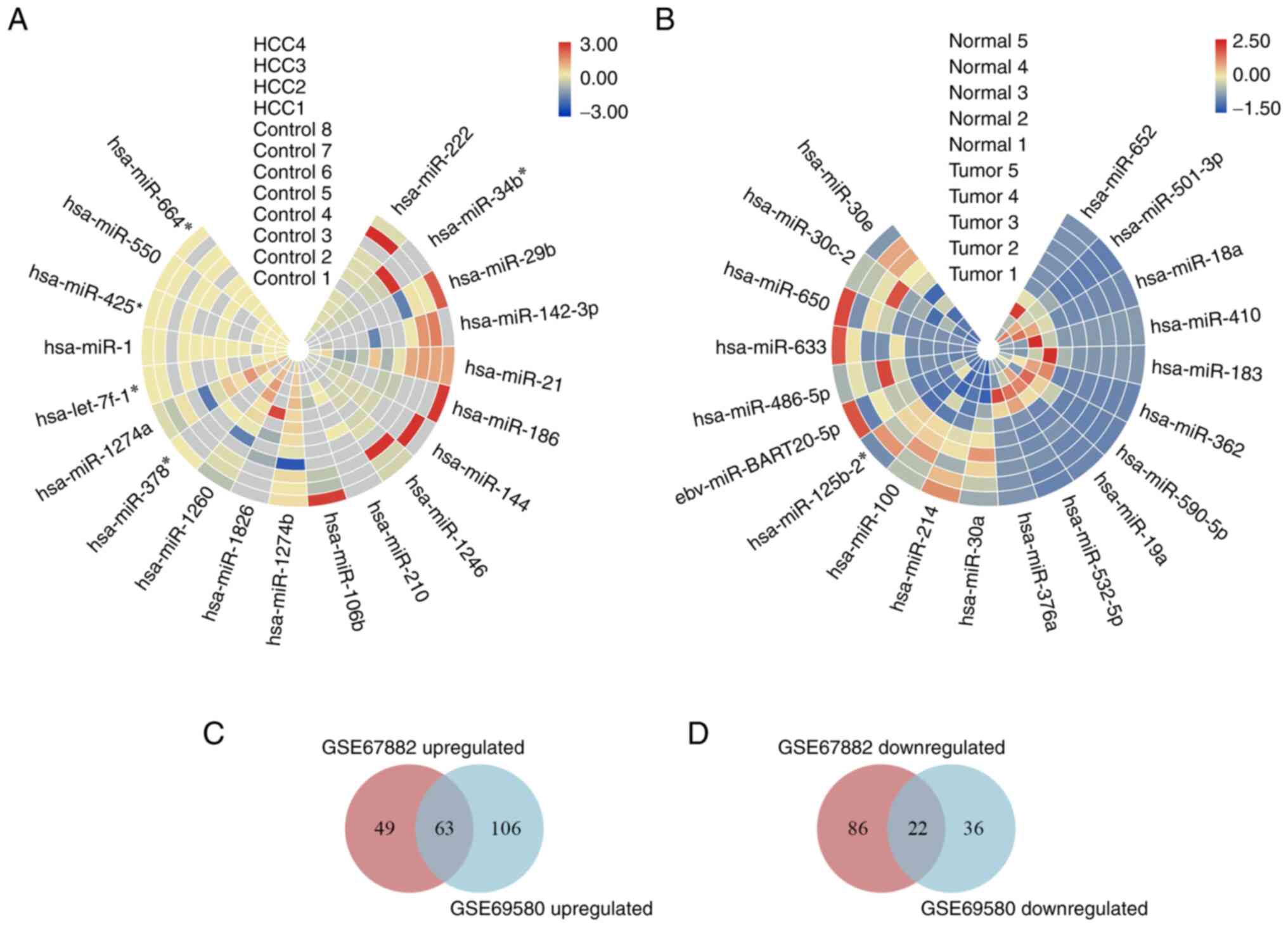
downregulated genes are displayed as heatmaps (Fig. 1A and B). Intersection analysis showed that there
were 63 up- and 22 downregulated genes (Fig. 1C and D). The candidate DE-miRNAs were defined
according to two indicators |log2FC|>1 and P<0.05.
Following screening, nine up- and one downregulated DE-miRNA were
obtained (Table I).
 | Table IDE-miRNAs in hepatitis B
virus-associated hepatocellular carcinoma. |
Table I
DE-miRNAs in hepatitis B
virus-associated hepatocellular carcinoma.
| A, Upregulated |
|---|
| | GSE67882 | GSE69580 |
|---|
| DE-miRNA | P-value |
log2FC | P-value |
log2FC |
|---|
| hsa-miR-15a | 0.007808345 | 1.611184956 | 0.019320684 | 1.29726588 |
| hsa-miR-29b |
1.82x10-6 | 3.659264924 | 0.027230532 | 2.09606417 |
| hsa-miR-93 | 0.030902439 | 1.22769598 | 0.005843196 | 1.842708148 |
| hsa-miR-106b | 0.002537174 | 2.156166989 | 0.000566846 | 2.676683779 |
| hsa-miR-146b | 0.003197486 | 2.058917809 | 0.004808375 | 2.831128268 |
| hsa-miR-210 | 0.000261703 | 2.486264664 | 0.003623182 | 3.288727003 |
| hsa-miR-374b | 0.021895978 | 1.863305503 | 0.045327223 | 3.113135194 |
| hsa-miR-660 | 0.017308666 | 1.21052014 | 0.029714338 | 5.260854 |
| hsa-miR-886 | 0.032721809 | 1.547302991 | 0.013391202 | 1.871457118 |
| B,
Downregulated |
| | GSE67882 | GSE69580 |
| DE-miRNA | P-value |
log2FC | P-value |
log2FC |
| hsa-miR-125b | 0.03390546 | -1.098388512 | 0.012086138 | -4.843165124 |
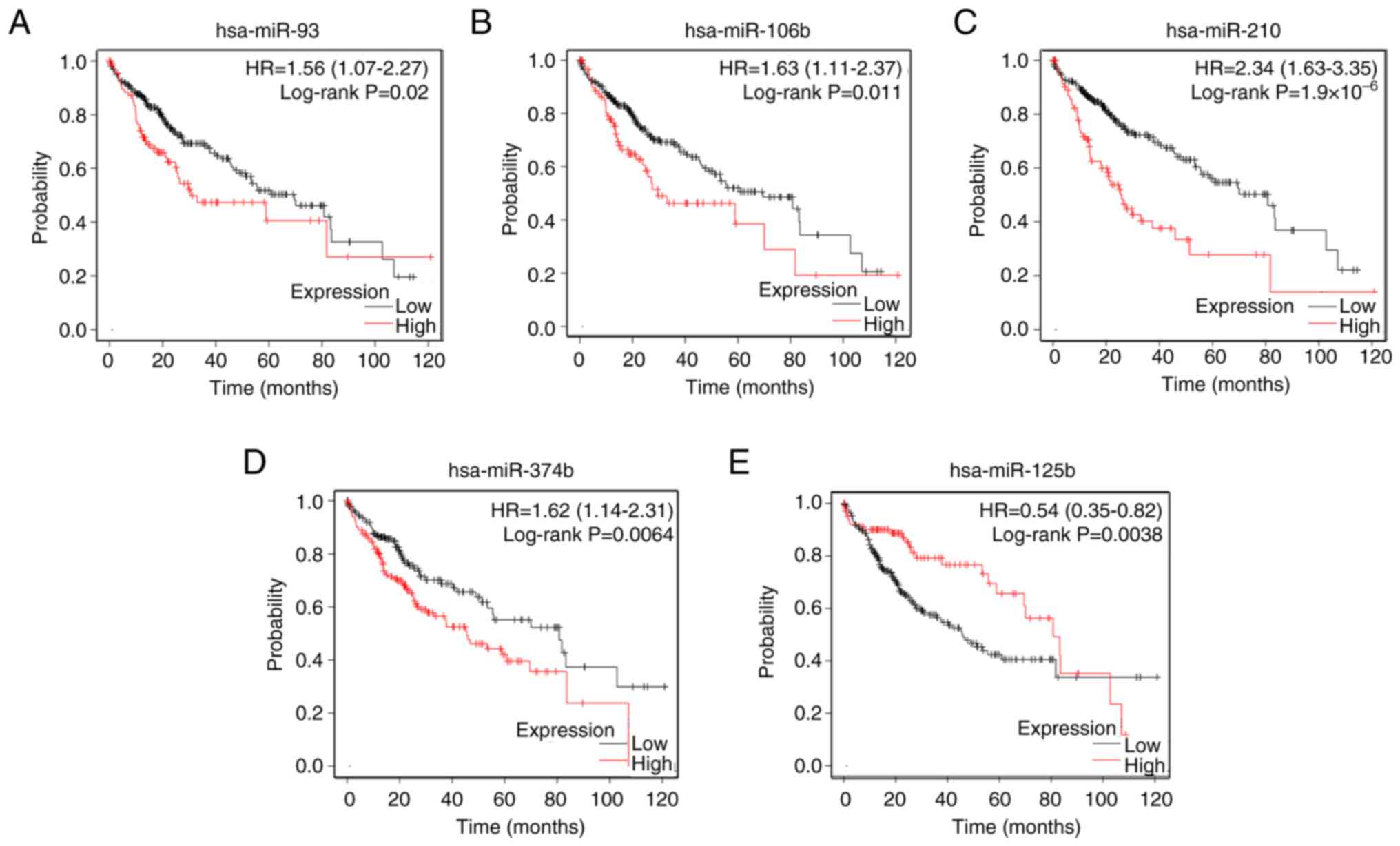
Prognostic value of DE-miRNAs
To evaluate candidate DE-miRNAs, their prognostic
value was analyzed. Kaplan-Meier Plotter database showed that among
nine upregulated miRNAs, only four miRNAs, namely miRNA-93,
miRNA-106b, miRNA-210 and miRNA-374b, had a significant effect on
prognosis of patients with HCC (Fig.
2A-D). The prognostic analysis of miRNA-125b showed that the
lower the expression of miRNA-125b, the worse the prognosis
(Fig. 2E). Finally, five miRNAs
whose expression levels and prognosis were significantly different
between HCC and normal tissue were identified as candidate
DE-miRNAs for further study.
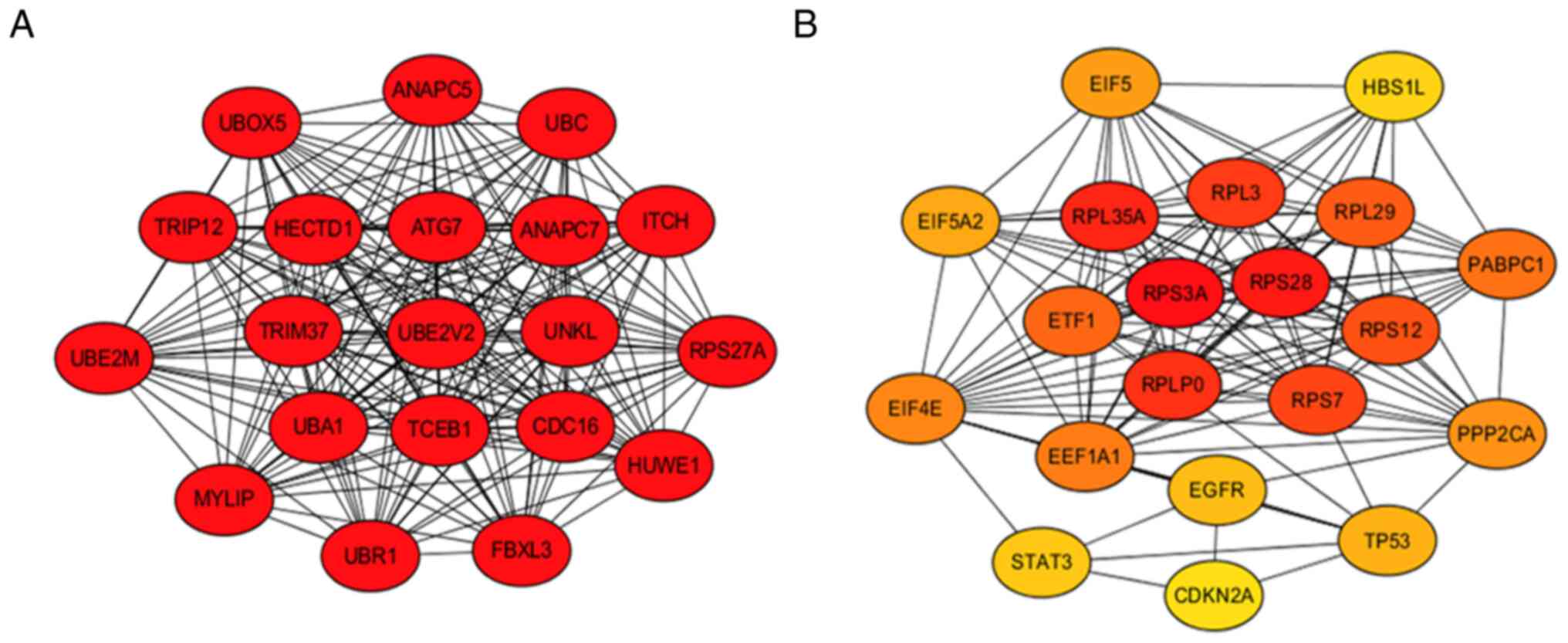
Prediction of DE-miRNAs target and hub
genes
The target genes of DE-miRNAs were predicted in
miRNet database. miRNA-93 had 1,220, miRNA-106b had 1,091,
miRNA-210 had 124, miRNA-374b had 234 and miRNA-125b had 432
potential target genes (Fig. S3A
and B; Table II). Protein-protein interaction
network analysis was performed on predicted target genes of
DE-miRNA using the STRING database and Cytoscape software (version
3.7.1; Fig. 3A and B). The top 20 resulting hub genes are
displayed, as well as their scores (Table SIV).
 | Table IITarget genes of DE-miRNAs in
hepatocellular carcinoma. |
Table II
Target genes of DE-miRNAs in
hepatocellular carcinoma.
| A, Upregulated |
|---|
| DE-miRNA | Number of
genes |
|---|
| hsa-miRNA-93 | 1,220 |
| hsa-miRNA-106b | 1,091 |
| hsa-miRNA-210 | 124 |
| hsa-miRNA-374b | 234 |
| B,
Downregulated |
| DE-miRNA | Number of
genes |
| hsa-miRNA-125b | 432 |
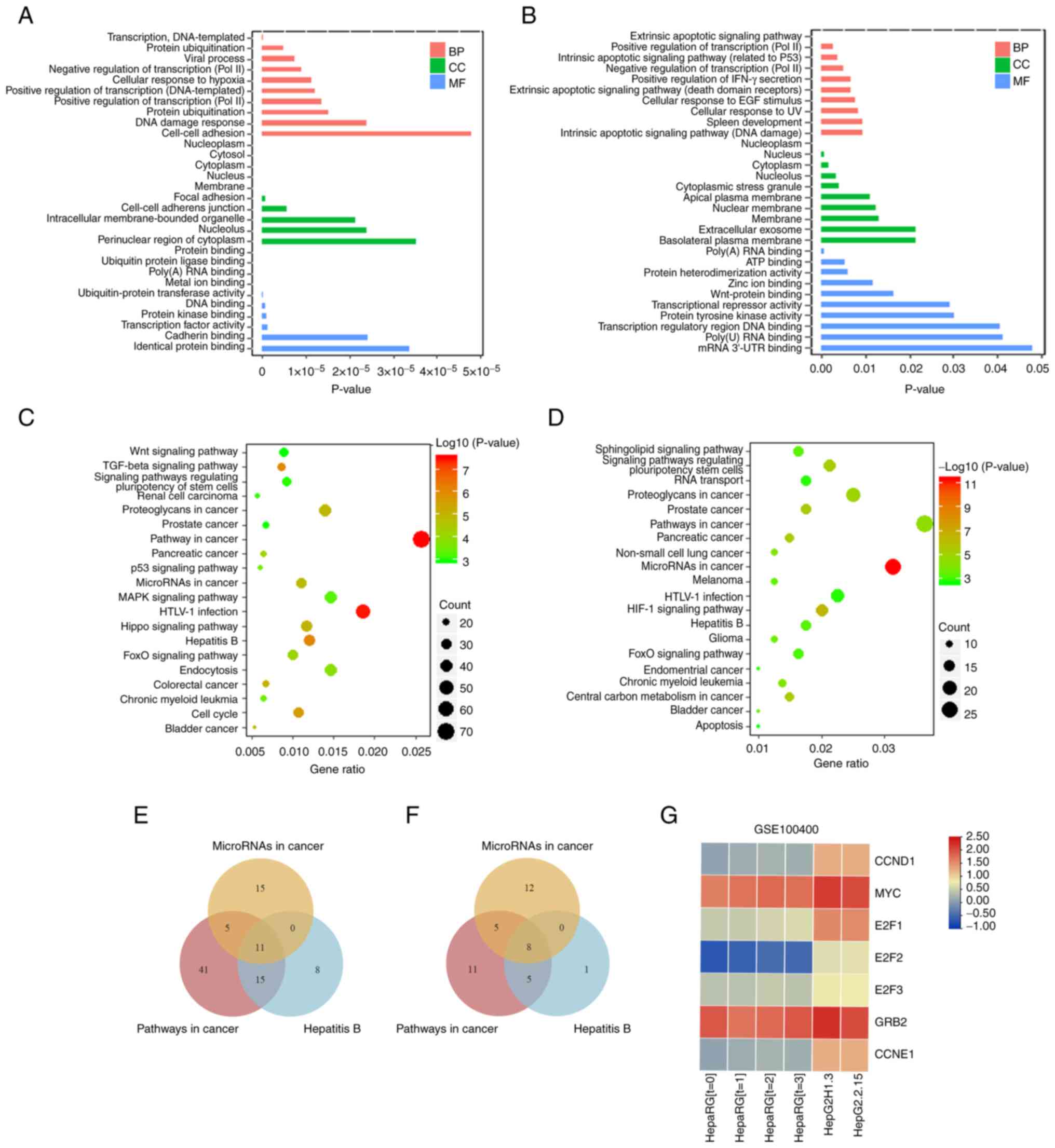
Functional annotation and pathway
enrichment analysis
The present study evaluated the terms involved in
these target genes through GO analysis. DAVID (version 6.8)
database was used to perform GO enrichment analysis of 2,669
predicted target genes of up- and 432 predicted target genes of
downregulated DE-miRNAs. The results were divided into three
categories: Biological process, cellular component and molecular
function. The top ten terms are listed according to P-value
(Fig. 4A and B). KEGG pathway enrichment analysis of
target genes was performed using the DAVID database to evaluate the
signaling pathways. The top 20 terms are displayed according to
P-value (Fig. 4C and D). Notably, three signal pathways
(‘pathway in cancer’, ‘hepatitis B’ and ‘microRNAs in cancer’) were
co-enriched, which were associated with HBV-related HCC. To
identify potential target genes regulated by HBV, these signaling
pathways were selected for intersection analysis, resulting in 11
and 8 sets of intersecting genes for up- and downregulated genes,
respectively (Fig. 4E and F). For these predicted target genes,
experimental data from GEO database were further investigated. In
GSE100400 dataset, RNA-seq, ChIP-seq and 4C methods were combined
to identify target genes regulated by HBV (37). Since this dataset provided a
reliable and well-analyzed differential gene expression profile, it
was cross-analyzed with the present findings to predict the
potential miRNA-mRNA regulatory relationship network in HBV-related
HCC. An intersection analysis of 11 up- and 8 downregulated target
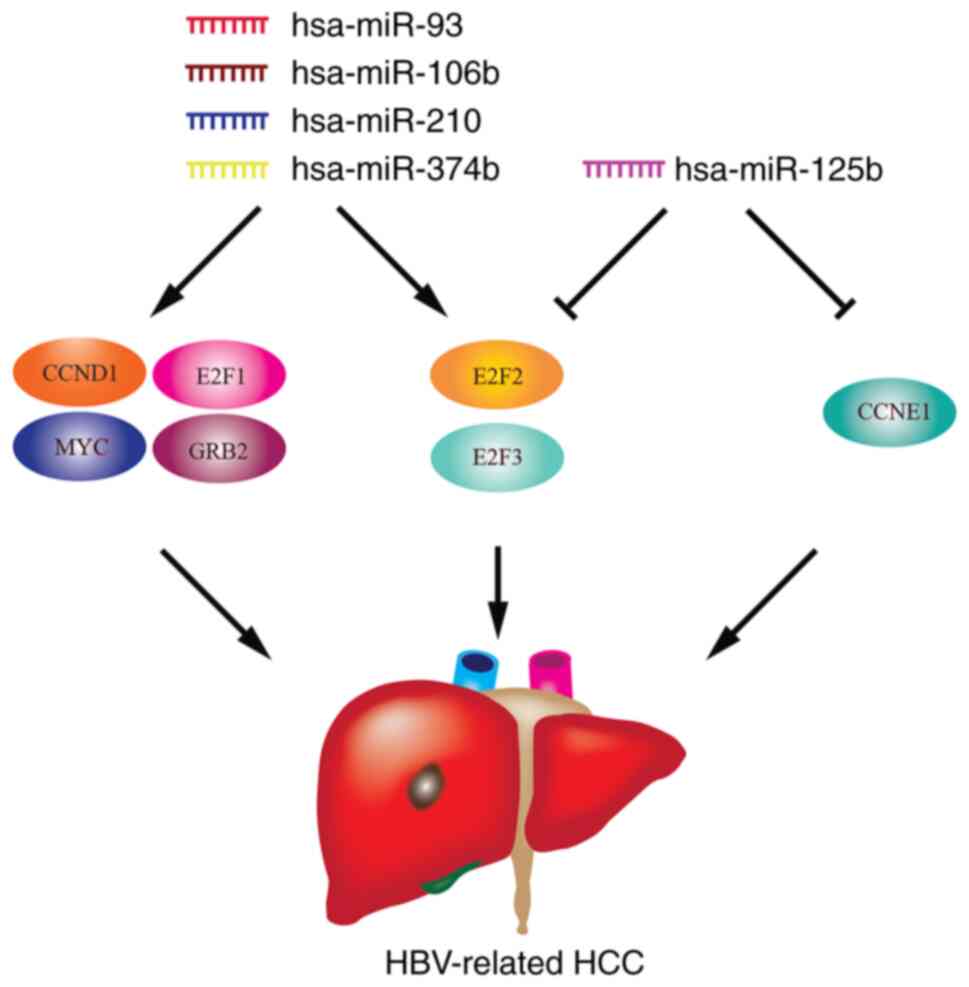
genes with the results of GSE100400 was conducted. A total of seven
HBV-associated intersecting genes was obtained: CCND1, MYC, E2F1,
E2F2, E2F3, GRB2 and CCNE1. The sequencing expression profiles of
these genes in the GSE100400 dataset is shown in Fig. 4G. The expression levels of the
DE-miRNA target genes were analyzed via UALCAN cancer database
(Fig. S4). The present study
summarized the potential miRNA-mRNA regulatory association network
model in HBV-associated HCC (Fig.
5).
Verification of candidate miRNAs and
potential target genes and exploration of the regulatory
association
To validate the results of the bioinformatics
analysis, the expression levels of DE-miRNAs and target genes in
HepG2 and HepG2.2.1.5 (a cell line that stably expresses HBV)
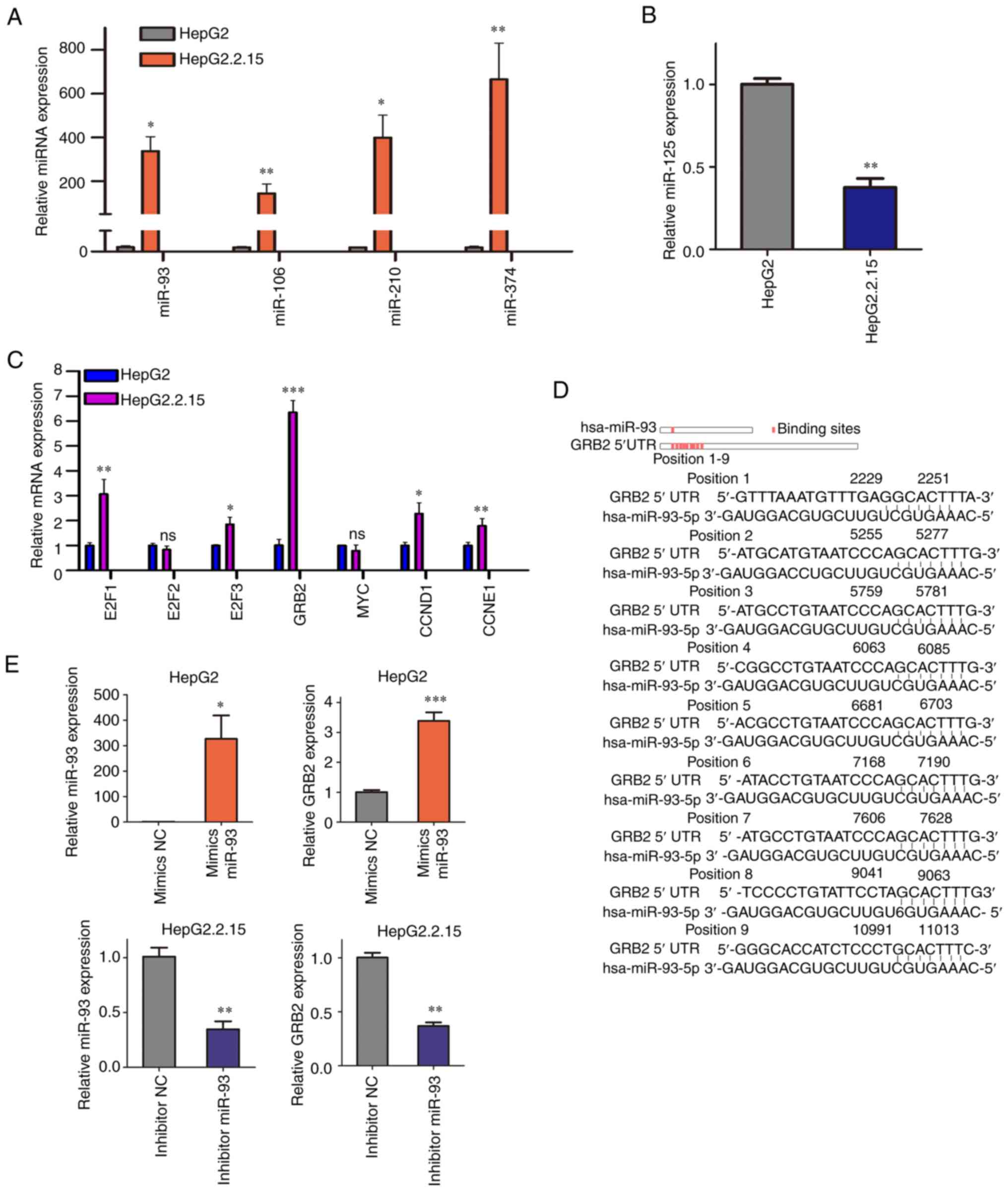
(41) cells were detected. Compared
with those in HepG2 cells, miRNA-93, miRNA-106b, miRNA-210 and
miRNA-374b showed significantly higher expression levels in
HepG2.2.1.5 cells, while miRNA-125b showed significantly lower
expression (Fig. 6A and B). Similarly, compared with HepG2 cells,
expression levels of E1F1, E2F3, GRB2, CCND1 and CCNE1 were higher
than those in HepG2.2.15 cells, while expression levels of E2F2 and
MYC were not significantly different (Fig. 6C).
Base pairing analysis demonstrated that miRNA-93 had
binding sites not only in the 3' but also the 5' UTR of GRB2. A
total of nine potential binding sites was predicted between
miRNA-93 and 5' UTR of GRB2 (Fig.
6D). As aforementioned, in HepG2.2.15 cells with HBV
overexpression, the expression levels of miRNA-93 and GRB2 were
upregulated. According to the predicted binding site, miRNA-93 has
a regulatory effect of activation by targeting the GRB2 5' UTR.
Therefore, more attention was paid to the active regulation between
miRNA-93 and GRB2.
Transfection was performed to detect the effect of
miRNA-93 on GRB2 expression levels in vitro. Mimics of
miRNA-93 were transfected into HepG2 cells and changes in GRB2 were
detected. With the increase in miRNA-93, the expression of GRB2
also increased (Fig. 6E). In
addition, transfection of miRNA-93 inhibitors in HepG2.2.15 cells
confirmed that the expression of GRB2 decreased with the knockdown
of miRNA-93 (Fig. 6E). Thus, GRB2
was a target gene of miRNA-93 and was positively regulated by
miRNA-93.
Prediction of the mechanism of GRB2
promotion of HCC
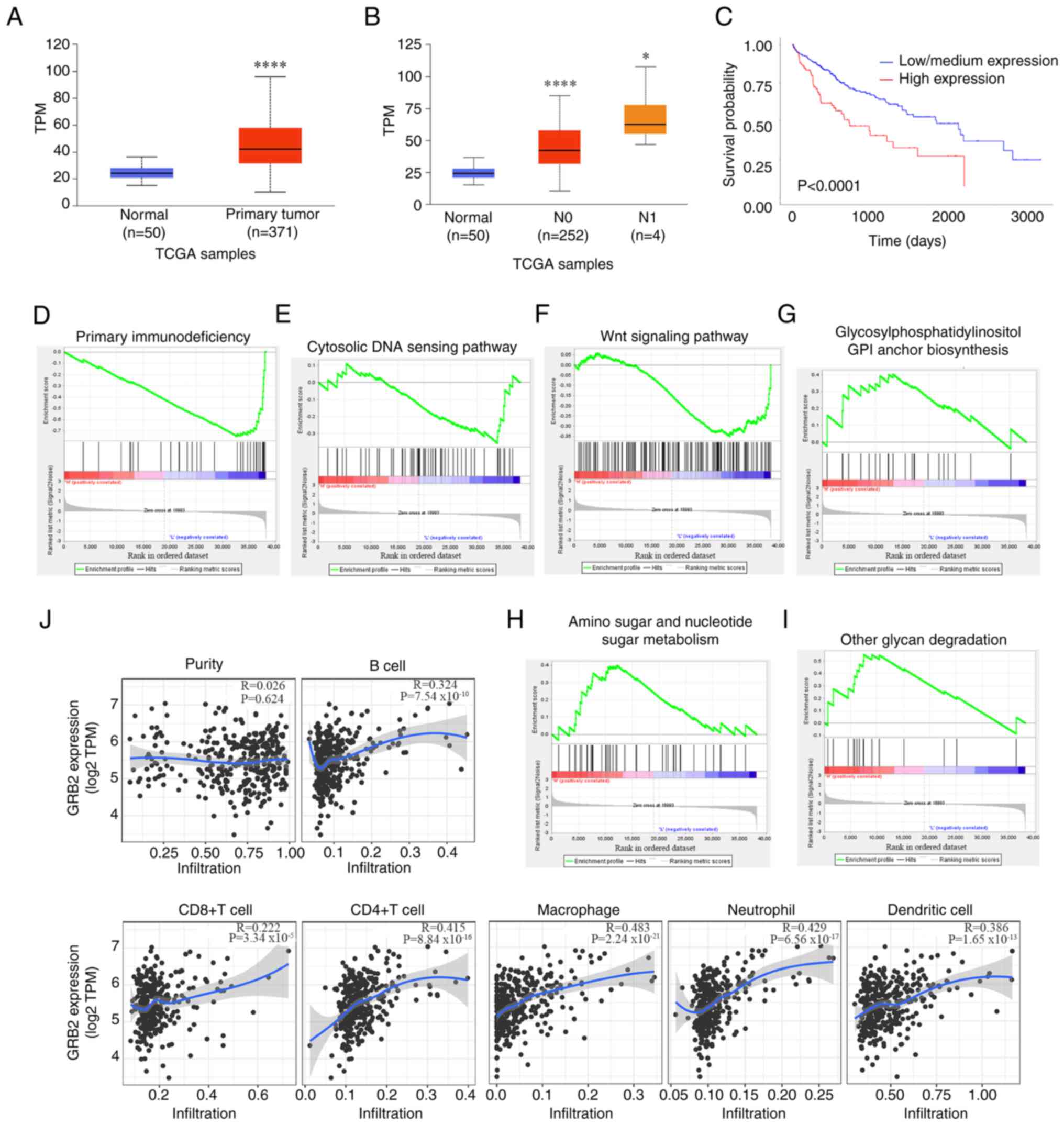
TCGA database demonstrated that expression levels of
GRB2 in HCC were significantly higher than those in non-tumor
tissues (Fig. 7A). Expression of
GRB2 was associated with lymph node metastasis in patients with HCC
(Fig. 7B). The higher the GRB2
expression, the lower the overall survival rate of patients with
HCC (Fig. 7C).
The present study analyzed the role of GRB2 in
occurrence and development of HCC. After GSE101728 data
normalization, the present study assessed enrichment of the KEGG
pathway between the top 25% of the highest and lowest GRB2
expression (Fig. 7D-I). Primary
immunodeficiency signaling pathway had the smallest P-value in the
low GRB2 expression group.
The association between GRB2 expression and immune
infiltration levels in HCC were assessed using the TIMER database.
Spearman's correlation was assessed based on microarray expression
levels. High GRB2 expression had positive correlations with the
infiltration levels of B (R=0.324) and CD4+ T cells
(R=0.415), macrophages (R=0.483), neutrophils (R=0.429) and
dendritic cells (R=0.386) in HCC (Fig.
7J). GRB2 expression had a weakly positive correlation with the
infiltration of CD8+ T cells (R=0.222) and no
significant correlation with tumor purity (R=0.026). These findings
suggest that GRB2 may serve a specific role in immune infiltration
in HCC, especially for CD4+ T cells, macrophages and
neutrophils.
Discussion
Although HBV-related miRNA-mRNA regulatory networks
have been considered in previous studies (42,43),
the miRNA-mRNA network regulated by HBV needs to be further
explored due to continuous updating of databases with different
patient samples and technologies. Advanced studies have implied
that these factors can predict bioinformatic data more accurately
(44,45).
HBV is widely integrated into the host genome and
has four open reading frames, primarily encoding four viral
proteins: Polymerase, HBx, envelope and core protein. HBx) serves a
key role in regulating the replication of HBV and is involved in
carcinogenesis associated with HBV (46). HBx can be detected in the nucleus,
cytoplasm and plasma (47,48). In the nucleus, HBx directly
regulates downstream target genes through gene transcription, while
in the cytoplasm, HBx regulates downstream target genes via
multiple signaling pathways (49).
Similarly, miRNAs are widely distributed in human cells and can be
detected in the cytoplasm and nucleus (33). HBx and miRNAs are colocalized in the
nucleus, which suggests that HBx is a key molecule that
participates in HBV regulation of miRNAs (50,51).
miRNAs are important gene expression regulators that
can bind to complementary target mRNAs and inhibit their expression
(52,53). They mainly play a role in
translation inhibition by targeting the 3' UTR of mRNA and causing
its cleavage and degradation (54).
Analysis of the interaction sites between miRNAs and target genes
demonstrated that most miRNAs have binding sites in the 3' UTR of
the gene and a small number of predicted binding sites are located
in the coding sequence region of the gene (54). However, the present study
demonstrated that miRNA-93 upregulates the expression of GRB2,
while most miRNAs inhibit gene expression (55). A previous study showed that miRNAs
located in the nucleus change the chromatin state by binding to
enhancers to activate gene transcription (56). Subsequent research confirmed the
phenomenon of miRNA activation (57). miRNAs can suppress gene expression
in the cytoplasm and activate gene transcription in the nucleus. In
breast cancer, miRNA-93 is localized in the nucleus (58). The present study did not clarify the
miRNA-93 localization or chromatin state of the GRB2 enhancer
region. This would provide understanding of how miRNA-93 is
involved in transcriptional regulation in the nucleus.
GRB2, an oncogenic functional gene in HCC, mainly
regulates cell proliferation and invasion via the ERK1/2/AKT axis
(59). A previous article
identified that GRB2-associated binding proteins serve important
roles in the regulation of immune response and cancer cell
signaling (60). In addition,
miRNA-93 regulates FAT atypical cadherin 4 expression, which is
associated with prognosis and immune cell infiltration in HCC
(61). The aforementioned report
suggested that miRNA-93 was indirectly involved in tumor immune
regulation. Several studies have reported that tumor-infiltrating
lymphocytes are an independent predictor of lymph node status and
survival in cancer (62,63). The present bioinformatics analysis
indicated that GRB2 expression can be treated as a biomarker to
evaluate the prognosis of HCC patients and regulate the tumor
immune microenvironment. There was moderate correlation between
GRB2 and immune cells, such as CD4+ T cells, macrophages
and neutrophils. The high expression of GRB2 in HCC is accompanied
by aggregation of these immune cells. GRB2 may lead to progression
of HCC by affecting the tumor immune microenvironment (64). HBV-positive patients have long-term
chronic viral infection (65).
Liver macrophages are stimulated by HBV-associated inflammatory
cytokines and chemokines and can adjust their phenotype based on
different signals from the liver microenvironment, such as danger
signals, fatty acids, and phagocytosis of cell debris, thereby
affecting tumor progression (66).
In previous studies, macrophages were not considered to have
antigen-specific immune memory (67,68).
However, macrophages, as a class of immune cells, can also form
antigen-specific immune memory (69). When macrophages encounter a specific
antigen again, they stimulate a stronger immune response. This
indicates a function of macrophages in HBV-related HCC.
Collectively, the present study summarized a
potential miRNA-mRNA regulatory network in HBV-associated HCC and
demonstrated miRNA-93 positively regulated immune
infiltration-related GRB2 gene expression through the 5'UTR region.
This study reports for the first time the correlation between GRB2
expression and immune cell infiltration. Restoring GRB2 levels may
be a novel treatment strategy for HCC in future.
Supplementary Material
Normalized data of GSE67882 and
GSE69580. GSE67882 data (A) before and (B) following normalization.
GSE69580 data (C) before and (D) following normalization.
Volcano plots of GSE67882 and GSE69580
data. Volcano plot of (A) GSE67882 and (B) GSE69580 data. The
candidate differentially expressed microRNAs were defined as
fold-change >2 and P<0.05.
miRNet prediction of potential target
genes of DE-miRNAs in HBV-related HCC. (A) Four up- and (B) one
downregulated DE-miRNAs and corresponding predicted target genes.
Certain genes were simultaneously regulated by several miRNAs.
Expression levels of seven
differentially expressed microRNA target genes were dysregulated in
UALCAN database. Differential expression levels of (A) Cyclin D1),
(B) MYC, (C) E2F1 (E2F Transcription Factor 1), (D) E2F2 (E2F
Transcription Factor 2), (E) E2F3 (E2F Transcription Factor 3), (F)
GRB2 (Growth Factor Receptor Bound Protein 2) and (G) CCNE1 (Cyclin
E1) in tumor and normal tissue according to ethnicity. ns, not
significant.
Sequences of the miRNA-93
mimics/inhibitors used for transfection.
Sequence of mature miRs.
Reverse transcription-quantitative PCR
primers.
Hub genes identified in the cytoHubba
network.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81972664).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
CZ and HF conceived the study. CZ, HS, ML and YQ
designed the methodology, performed experiments and analyzed data.
CZ wrote the manuscript. CZ, HS, ML and HF edited the manuscript.
HF supervised the study. CZ and HS confirm the authenticity of all
the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Younes R and Bugianesi E: Should we
undertake surveillance for HCC in patients with NAFLD? J Hepatol.
68:326–334. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Sartorius K, Makarova J, Sartorius B, An
P, Winkler C, Chuturgoon A and Kramvis A: The regulatory role of
MicroRNA in hepatitis-B virus-associated hepatocellular carcinoma
(HBV-HCC) pathogenesis. Cells. 8(1504)2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
El-Serag HB: Epidemiology of viral
hepatitis and hepatocellular carcinoma. Gastroenterology.
142:1264–1273.e1. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sagnelli C, Macera M, Pisaturo M, Zampino
R, Coppola M and Sagnelli E: Occult HBV infection in the
oncohematological setting. Infection. 44:575–582. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ringelhan M, O'Connor T, Protzer U and
Heikenwalder M: The direct and indirect roles of HBV in liver
cancer: Prospective markers for HCC screening and potential
therapeutic targets. J Pathol. 235:355–367. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Szpakowski JL and Tucker LY: Causes of
death in patients with hepatitis B: A natural history cohort study
in the United States. Hepatology. 58:21–30. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Choi YM, Lee SY and Kim BJ: Naturally
occurring hepatitis B virus mutations leading to endoplasmic
reticulum stress and their contribution to the progression of
hepatocellular carcinoma. Int J Mol Sci. 20(597)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kim CM, Koike K, Saito I, Miyamura T and
Jay G: HBx gene of hepatitis B virus induces liver cancer in
transgenic mice. Nature. 351:317–320. 1991.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Sadri Nahand J, Bokharaei-Salim F,
Karimzadeh M, Moghoofei M, Karampoor S, Mirzaei HR, Tabibzadeh A,
Jafari A, Ghaderi A, Asemi Z, et al: MicroRNAs and exosomes: Key
players in HIV pathogenesis. HIV Med. 21:246–278. 2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Petrini E, Caviglia GP, Abate ML, Fagoonee
S, Smedile A and Pellicano R: MicroRNAs in HBV-related
hepatocellular carcinoma: Functions and potential clinical
applications. Panminerva Med. 57:201–209. 2015.PubMed/NCBI
|
|
12
|
Pu C, Wang J, Wei Q and Han P: Commentary
on ‘Asymptomatic prostatic inflammation in men with clinical BPH
and erectile dysfunction affects the positive predictive value of
prostate-specific antigen’. Agnihotri S, Mittal RD, Kapoor R,
Mandhani A, Department of Urology & Renal Transplantation,
Sanjay Gandhi Post Graduate Institute of Medical Sciences, Lucknow,
India: Urol Oncol 2014; [Epub ahead of print]. doi:
10.1016/j.urolonc.2014.03.004. Urol Oncol. 33:103–104.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kelly EJ and Russell SJ: MicroRNAs and the
regulation of vector tropism. Mol Ther. 17:409–416. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY,
Zhang JF, Shen HB, Zhang CY and Zen K: Serum microRNA profiles
serve as novel biomarkers for HBV infection and diagnosis of
HBV-positive hepatocarcinoma. Cancer Res. 70:9798–9807.
2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bertoli G, Cava C and Castiglioni I:
MicroRNAs: New biomarkers for diagnosis, prognosis, therapy
prediction and therapeutic tools for breast cancer. Theranostics.
5:1122–1143. 2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ma X, Feng J, Lu M, Tang W, Han J, Luo X,
Zhao Q and Yang L: microRNA-501-5p promotes cell proliferation and
migration in gastric cancer by downregulating LPAR1. J Cell
Biochem. 121:1911–1922. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Vautrot V, Chanteloup G, Elmallah M,
Cordonnier M, Aubin F, Garrido C and Gobbo J: Exosomal miRNA: Small
molecules, big impact in colorectal cancer. J Oncol.
2019(8585276)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Sur S, Steele R, Shi X and Ray RB:
miRNA-29b inhibits prostate tumor growth and induces apoptosis by
increasing bim expression. Cells. 8(1455)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Vienberg S, Geiger J, Madsen S and
Dalgaard LT: MicroRNAs in metabolism. Acta Physiol (Oxf).
219:346–361. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Portal-Núñez S, Esbrit P, Alcaraz MJ and
Largo R: Oxidative stress, autophagy, epigenetic changes and
regulation by miRNAs as potential therapeutic targets in
osteoarthritis. Biochem Pharmacol. 108:1–10. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Guo D, Liu J, Wang W, Hao F, Sun X, Wu X,
Bu P, Zhang Y, Liu Y, Liu F, et al: Alteration in abundance and
compartmentalization of inflammation-related miRNAs in plasma after
intracerebral hemorrhage. Stroke. 44:1739–1742. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ura S, Honda M, Yamashita T, Ueda T,
Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K and Kaneko
S: Differential microRNA expression between hepatitis B and
hepatitis C leading disease progression to hepatocellular
carcinoma. Hepatology. 49:1098–1112. 2009.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Giray BG, Emekdas G, Tezcan S, Ulger M,
Serin MS, Sezgin O, Altintas E and Tiftik EN: Profiles of serum
microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for
HBV-positive hepatocellular carcinoma. Mol Biol Rep. 41:4513–4519.
2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ninomiya M, Kondo Y, Kimura O, Funayama R,
Nagashima T, Kogure T, Morosawa T, Tanaka Y, Nakayama K and
Shimosegawa T: The expression of miR-125b-5p is increased in the
serum of patients with chronic hepatitis B infection and inhibits
the detection of hepatitis B virus surface antigen. J Viral Hepat.
23:330–339. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Xu J, An P, Winkler CA and Yu Y:
Dysregulated microRNAs in hepatitis B virus-related hepatocellular
carcinoma: Potential as biomarkers and therapeutic targets. Front
Oncol. 10(1271)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Morishita A, Fujita K, Iwama H, Chiyo T,
Fujihara S, Oura K, Tadokoro T, Mimura S, Nomura T, Tani J, et al:
Role of microRNA-210-3p in hepatitis B virus-related hepatocellular
carcinoma. Am J Physiol Gastrointest Liver Physiol. 318:G401–G409.
2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Meng FL, Wang W and Jia WD: Diagnostic and
prognostic significance of serum miR-24-3p in HBV-related
hepatocellular carcinoma. Med Oncol. 31(177)2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Xie KL, Zhang YG, Liu J, Zeng Y and Wu H:
MicroRNAs associated with HBV infection and HBV-related HCC.
Theranostics. 4:1176–1192. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang C, Su K, Lin H, Cen B, Zheng S and Xu
X: Identification and verification of a novel
MAGI2-AS3/miRNA-374-5p/FOXO1 network associated with HBV-related
HCC. Cells. 11(3466)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ranjpour M, Katare DP, Wajid S and Jain
SK: HCC specific protein network involving interactions of EGFR
with A-Raf and transthyretin: experimental analysis and
computational biology correlates. Anticancer Agents Med Chem.
18:1163–1176. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Liu H, Lei C, He Q, Pan Z, Xiao D and Tao
Y: Nuclear functions of mammalian MicroRNAs in gene regulation,
immunity and cancer. Mol Cancer. 17(64)2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Huang V and Li LC: miRNA goes nuclear. RNA
Biol. 9:269–273. 2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Chen C, Chen H, Zhang Y, Thomas HR, Frank
MH, He Y and Xia R: TBtools: An integrative toolkit developed for
interactive analyses of big biological data. Mol Plant.
13:1194–1202. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Fan Y, Siklenka K, Arora SK, Ribeiro P,
Kimmins S and Xia J: miRNet-dissecting miRNA-target interactions
and functional associations through network-based visual analysis.
Nucleic Acids Res. 44 (W1):W135–W141. 2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhu HR, Yu XN, Zhang GC, Shi X,
Bilegsaikhan E, Guo HY, Liu LL, Cai Y, Song GQ, Liu TT, et al:
Comprehensive analysis of long non-coding RNA-messenger
RNA-microRNA co-expression network identifies cell cycle-related
lncRNA in hepatocellular carcinoma. Int J Mol Med. 44:1844–1854.
2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Hensel KO, Cantner F, Bangert F, Wirth S
and Postberg J: Episomal HBV persistence within transcribed host
nuclear chromatin compartments involves HBx. Epigenetics Chromatin.
11(34)2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658.
2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110.
2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang X, Hu H, Hu B, Xia H, Cheng X, Zheng
J, Zhang Z and Liu H: Dihydromyricetin inhibits Hepatitis B virus
replication by activating NF-κB, MAPKs, and autophagy in HepG2.2.15
cells. Mol Biol Rep. 50:1403–1414. 2023.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lou W, Liu J, Ding B, Chen D, Xu L, Ding
J, Jiang D, Zhou L, Zheng S and Fan W: Identification of potential
miRNA-mRNA regulatory network contributing to pathogenesis of
HBV-related HCC. J Transl Med. 17(7)2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Ren M, Qin D, Li K, Qu J, Wang L, Wang Z,
Huang A and Tang H: Correlation between hepatitis B virus protein
and microRNA processor Drosha in cells expressing HBV. Antiviral
Res. 94:225–231. 2012.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhang M, Bai X, Zeng X, Liu J, Liu F and
Zhang Z: circRNA-miRNA-mRNA in breast cancer. Clin Chim Acta.
523:120–130. 2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Ma J, Wang P, Huang L, Qiao J and Li J:
Bioinformatic analysis reveals an exosomal miRNA-mRNA network in
colorectal cancer. BMC Med Genomics. 14(60)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Guerrieri F, Belloni L, D'Andrea D,
Pediconi N, Le Pera L, Testoni B, Scisciani C, Floriot O, Zoulim F,
Tramontano A and Levrero M: Genome-wide identification of direct
HBx genomic targets. BMC Genomics. 18(184)2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Su Q, Schröder CH, Hofmann WJ, Otto G,
Pichlmayr R and Bannasch P: Expression of hepatitis B virus X
protein in HBV-infected human livers and hepatocellular carcinomas.
Hepatology. 27:1109–1120. 1998.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Liang XH, Stemler M, Will H, Braun R, Tang
ZY and Schröder CH: Low incidence and high titers of antibodies to
hepatitis B virus X-protein in sera of Chinese patients with
hepatocellular carcinoma. J Med Virol. 25:329–337. 1988.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Levrero M and Zucman-Rossi J: Mechanisms
of HBV-induced hepatocellular carcinoma. J Hepatol. 64 (Suppl
1):S84–S101. 2016.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Peng F, Xiao X, Jiang Y, Luo K, Tian Y,
Peng M, Zhang M, Xu Y and Gong G: HBx down-regulated Gld2 plays a
critical role in HBV-related dysregulation of miR-122. PLoS One.
9(e92998)2014.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zhou SJ, Deng YL, Liang HF, Jaoude JC and
Liu FY: Hepatitis B virus X protein promotes CREB-mediated
activation of miR-3188 and Notch signaling in hepatocellular
carcinoma. Cell Death Differ. 24:1577–1587. 2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Michlewski G and Cáceres JF:
Post-transcriptional control of miRNA biogenesis. RNA. 25:1–16.
2019.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Bernardo BC, Ooi JYY, Lin RCY and McMullen
JR: miRNA therapeutics: A new class of drugs with potential
therapeutic applications in the heart. Future Med Chem.
7:1771–1792. 2015.PubMed/NCBI View Article : Google Scholar
|
|
54
|
lyu Z, Mao Z, Wang H, Fang Y, Chen T, Wan
Q, Wang M, Wang N, Xiao J, Wei H, et al: MiR-181b targets Six2 and
inhibits the proliferation of metanephric mesenchymal cells in
vitro. Biochem Biophys Res Commun. 440:495–501. 2013.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Schmiedel JM, Klemm SL, Zheng Y, Sahay A,
Blüthgen N, Marks DS and van Oudenaarden A: Gene expression.
MicroRNA control of protein expression noise. Science. 348:128–132.
2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Xiao M, Li J, Li W, Wang Y, Wu F, Xi Y,
Zhang L, Ding C, Luo H, Li Y, et al: MicroRNAs activate gene
transcription epigenetically as an enhancer trigger. RNA Biol.
14:1326–1334. 2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Suzuki HI, Young RA and Sharp PA:
Super-enhancer-mediated RNA processing revealed by integrative
MicroRNA network analysis. Cell. 168:1000–1014.e15. 2017.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Li N, Miao Y, Shan Y, Liu B, Li Y, Zhao L
and Jia L: MiR-106b and miR-93 regulate cell progression by
suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell
Death Dis. 8(e2796)2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Liang C, Xu Y, Ge H, Xing B, Li G, Li G
and Wu J: miR-564 inhibits hepatocellular carcinoma cell
proliferation and invasion by targeting the GRB2-ERK1/2-AKT axis.
Oncotarget. 8:107543–107557. 2017.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Vaughan TY, Verma S and Bunting KD:
Grb2-associated binding (Gab) proteins in hematopoietic and immune
cell biology. Am J Blood Res. 1:130–134. 2011.PubMed/NCBI
|
|
61
|
Li J, Lv M, Huang Q, Hu R, Zhong X, Sun X,
Feng W, Han Z, Ma M, Zhang W and Zhou X: FAT4 expression in
peripheral blood mononuclear cells is associated with prognosis and
immune cell infiltration in hepatocellular carcinoma. Sci Rep.
13(15735)2023.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Dunn GP, Dunn IF and Curry WT: Focus on
TILs: Prognostic significance of tumor infiltrating lymphocytes in
human glioma. Cancer Immun. 7(12)2007.PubMed/NCBI
|
|
63
|
Azimi F, Scolyer RA, Rumcheva P, Moncrieff
M, Murali R, McCarthy SW, Saw RP and Thompson JF:
Tumor-infiltrating lymphocyte grade is an independent predictor of
sentinel lymph node status and survival in patients with cutaneous
melanoma. J Clin Oncol. 30:2678–2683. 2012.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Zhang X and Zhang J, Gao F, Fan S, Dai L
and Zhang J: KPNA2-associated immune analyses highlight the
dysregulation and prognostic effects of GRB2, NRAS, and their
RNA-binding proteins in hepatocellular carcinoma. Front Genet.
11(593273)2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Lee CC, Li IJ, Chen YC, Cheng JW, Wu HH,
Weng CH, Fang JT and Tian YC: Comparable ten-year outcome in
hemodialysis patients with hepatitis C virus and hepatitis B virus
coinfection and single hepatitis B virus infection. Blood Purif.
32:89–95. 2011.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Tacke F: Targeting hepatic macrophages to
treat liver diseases. J Hepatol. 66:1300–1312. 2017.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Maheshwari A: Innate immune memory in
macrophages. Newborn (Clarksville). 2:60–79. 2023.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Kawasaki T, Ikegawa M, Yunoki K, Otani H,
Ori D, Ishii KJ, Kuroda E, Takamura S, Kitabatake M, Ito T, et al:
Alveolar macrophages instruct CD8+ T cell expansion by
antigen cross-presentation in lung. Cell Rep.
41(111828)2022.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Dai H, Lan P, Zhao D, Abou-Daya K, Liu W,
Chen W, Friday AJ, Williams AL, Sun T, Chen J, et al: PIRs mediate
innate myeloid cell memory to nonself MHC molecules. Science.
368:1122–1127. 2020.PubMed/NCBI View Article : Google Scholar
|