1. Introduction
Melatonin (N-acetyl-5-methoxy-tryptamine) is an
amphiphilic tryptophan-derived indoleamine highly conserved among
vertebrates (1). Although melatonin
is primarily synthesized in the pineal gland, smaller amounts are
detected in peripheral tissues, including the gastrointestinal
tract, skin, retina and bone marrow (2,3). The
release of melatonin from the pineal gland is rhythmically
regulated by retinal photoreceptors, with low levels during the day
and high levels at night (4).
Melatonin cannot be stored in tissues and is immediately released
into the blood to modulate physiological conditions in peripheral
tissues (5). Therefore, melatonin
has a short half-life of 30-60 min in the body (6). Melatonin regulates several
physiological functions, including circadian rhythm regulation,
acting as an antioxidant and free radical scavenger, possessing
anti-inflammatory properties, modulating mitochondrial homeostasis,
and enhancing nitric oxide bioavailability (7).
A total of four enzymes involved in the melatonin
synthesis pathway, tryptophan hydroxylase (TPH), aromatic amino
acid decarboxylase (DDC), aryl-alkyl-amine N-acetyl-transferase
(AANAT) and hydroxy-indole-O-methyl-transferase (HIOMT), are
primarily expressed with a circadian rhythm in the suprachiasmatic
nucleus (SCN), pineal gland and retina (Fig. 1) (8). TPH and DDC sequentially catalyze the
conversion of tryptophan to serotonin, which is a precursor to
melatonin (9). AANAT converts
serotonin to N-acetyl-serotonin, which is then converted to
melatonin by HIOMT (10). Melatonin
modulates diverse physiological conditions via a combination of
both melatonin receptor-mediated and receptor-independent
mechanisms (11). Specifically,
melatonin receptor activation plays a pivotal role in regulating
circadian gene expression, thereby influencing the sleep-wake
cycle, body temperature, blood pressure, metabolism, urine
production and hormone secretion (12). Conversely, the receptor-independent
actions of melatonin are largely attributed to its potent
free-radical-scavenging and antioxidant properties. The
electron-rich aromatic indole ring in melatonin makes it a potent
electron donor, significantly reducing oxidative stress (13). This direct interaction with reactive
oxygen species allows melatonin to protect cellular components from
oxidative damage, contributing to its broad protective effects
(14). In general, melatonin binds
with high affinity and specificity to membrane receptors in the
brain and periphery to trigger physiological responses by altering
cell signaling pathways (15).
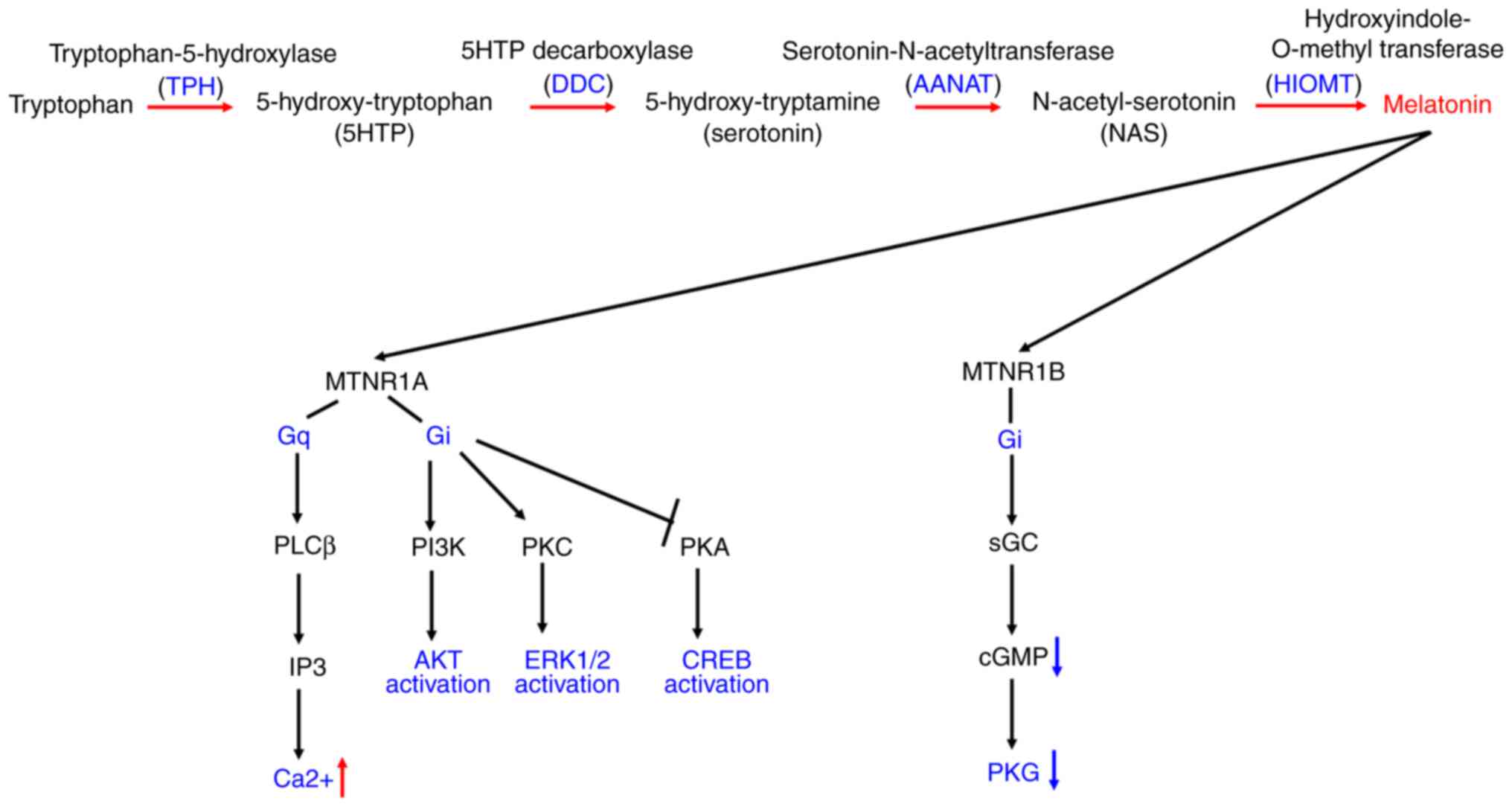 | Figure 1Diagrammatic representation of the
melatonin synthesis pathway and melatonin receptor-mediated cell
signaling. The metabolic pathway converting tryptophan to melatonin
involves the enzymes TPH, DDC, AANAT and HIOMT (also known as
ASMT). Melatonin activation of MTNR1A receptors triggers Gq
activation, leading to increased levels of calcium and IP3.
Additionally, it induces Gi-dependent activation of the PI3K/AKT
and PKC/ERK pathways, while causing Gi-dependent inactivation of
the PKA/CREB axis. MTNR1B coupling to Gi results in PKG
inactivation and a decrease in intracellular cGMP levels. TPH,
tryptophan hydroxylase; DDC, aromatic amino acid decarboxylase;
AANAT, aryl-alkyl-amine N-acetyltransferase; HIOMT,
hydroxy-indole-O-methyl-transferase; MTNR1A, melatonin receptor 1A;
MTNR1B, melatonin receptor 1B; PKC, protein kinase C; cGMP,
3'-5'-cyclic guanosine monophosphate; PKG, protein kinase G; IP3,
inositol 1,4,5-triphosphate; PLCb, phospholipase C beta. |
The melatonin signaling pathway consists of
melatonin synthesis enzymes and melatonin-mediated cellular
responses, which are evolutionarily conserved in both brain and
peripheral tissues. However, most studies focus on the role of the
melatonin signaling pathway in brain tissue. In the present review,
the expression of melatonin signaling components and their mediated
roles in renal tissue under various biological conditions are
discussed, and evidence regarding the benefits of targeting
membranous nephropathy (MN) with this approach is presented. The
present review offers new insights into the melatonin signaling
pathway in the pathology, treatment, and prevention of
nephropathy.
2. Melatonin synthesis enzyme in kidney
Melatonin, a small amphipathic indolamine, is
ubiquitous across nearly all organisms, from bacteria to humans,
and plays a crucial role in regulating circadian rhythms and
sleep-wake cycles (16). The
circadian production of melatonin is regulated by endogenous
oscillators within the body and synchronized by daily and seasonal
variations in the environmental light-dark cycle (17). The pineal gland and SCN are the
major sources of melatonin in vertebrates, and they appear to
regulate target cells via an exocrine mechanism (18). Most of the research on the gene
expression of melatonin-synthesizing enzymes focuses on retinal
cells and pinealocytes, with limited research specifically focused
on the kidney (19). In the present
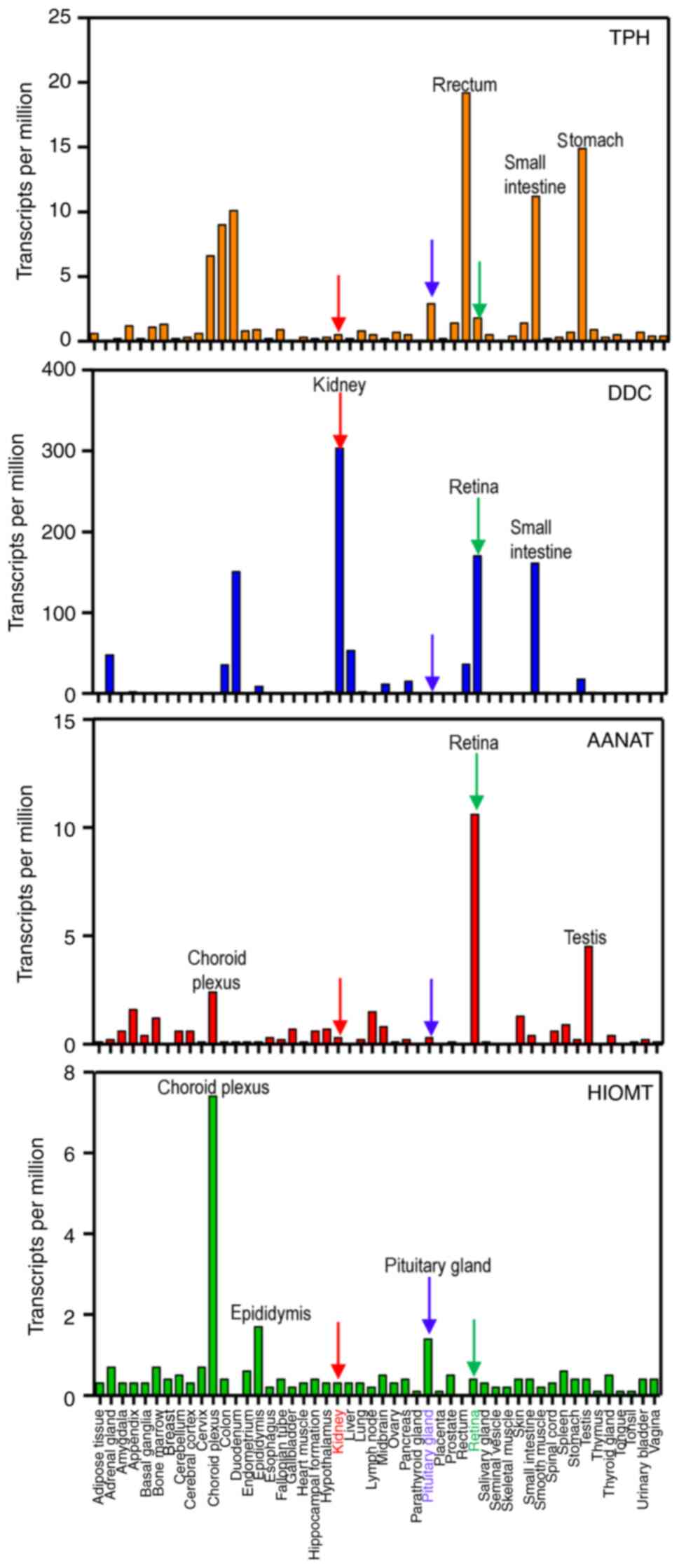
review, focus was addressed on the expression levels of the four
enzymes of the melatoninergic pathway (TPH, DDC, AANAT and HIOMT)
in central and peripheral locations of human tissue using data from
The Human Protein Atlas (20,21).
Although TPH and DDC contribute to melatonin synthesis, they also
play roles in other physiological processes independent of
melatonin production. The results indicated that the top three
tissues with the highest expression of TPH are the rectum, stomach
and small intestine; the highest expression of DDC is found in the
kidney, retina and small intestine; the highest expression of AANAT
is found in the retina, choroid plexus and testis; and the highest
expression of HIOMT is found in the choroid plexus, epididymis and
pituitary gland (Fig. 2).
Consistent with previous findings, melatonin biosynthesis enzymes
are identified in the retina, pituitary gland, and gastrointestinal
tract (22,23). Notably, the kidney exhibited low
expression levels of TPH, AANAT, and HIOMT, and the highest levels
of DDC (Fig. 2). These results
suggest that melatonin-synthesizing enzymes are expressed in renal
tissue. However, there are a few concerns regarding these results
from The Human Protein Atlas (20,21).
First, the kidney consists of different cell types organized into
sub-anatomical tissue structures. These melatonin synthesis genes
could be expressed in immune cells or other cell types within renal
tissue. Moreover, RNA expression does not necessarily represent
protein production in the kidney. Although the highest levels of
DDC have been noticed in the kidney, it is important to note that
DDC is not solely responsible for melatonin production. DDC protein
also catalyzes the decarboxylation of L-3,4-dihydroxyphenylalanine
to dopamine, L-5-hydroxytryptophan to serotonin, and L-tryptophan
to tryptamine (24). Collectively,
whether these melatonin synthesis enzymes produce melatonin in the
kidney needs to be further validated.
Previous studies showed that AANAT transcripts and
melatonin can be detected in renal tubular epithelial cells (TECs)
in both human cells and mouse kidneys (25,26).
The molecular mechanism indicated that cAMP responsive element
binding protein (CREB) increases AANAT transcriptional activity in
TECs, while it is decreased by c-Fos. The c-Fos family forms the
activator protein-1, functioning as transcriptional activators or
repressors depending on the diverse collection of interacting
proteins (27). Functional results
revealed that AANAT expression is decreased by c-Fos, leading to
enhanced cell damage in an albumin-injury cell model. Notably,
melatonin levels increased from 10 to 100 pg/ml following c-Fos
knockdown in the TEC cell line HK-2(25). Although these results suggest that
TECs produce melatonin in vitro, the precise niche within
renal tissue still needs to be confirmed. Melatonin metabolism is
mostly metabolized in the liver and kidneys by several enzymes of
the cytochrome P450 system (28,29).
Although only a small percentage (<5%) of blood melatonin is
unmetabolized and excreted into the urine (30), these unmetabolized melatonin could
be identify in renal tissue. Therefore, the specific knock-out of
AANAT or HIOMT in glomerular or TEC mice should be used to
determine endogenous melatonin production.
3. Melatonin receptor and its mediated
signaling
In higher vertebrates, the melatonin receptor family
consists of two members, melatonin receptor 1A (MTNR1A) and
melatonin receptor 1B (MTNR1B), both exhibiting high affinity for
the natural ligand melatonin (Fig.
1) (31). The amino acid
homology between human MTNR1A and MTNR1B is ~60% overall, with a
higher similarity of ~73% within the transmembrane domains
(31). Interestingly, the human
MTNR1A receptor exhibits greater resemblance to the rodent MTNR1A
than to the MTNR1B receptors found in bovine, ovine and porcine
species (32). Other nuclear
receptors with melatonin at low affinity include the retinoic acid
receptor-related orphan receptor alpha (RORα) and the vitamin D
receptor (VDR) (33,34). However, these results need
replication to confirm the interaction between melatonin and RORAα
and VDR in future studies. Moreover, the quinone reductase 2 (QR2)
enzyme likely corresponds to the melatonin receptor binding site,
binding melatonin in the µM range (35). Further studies are needed to
identify the subcellular localization of QR2.
These high-affinity melatonin receptors are
expressed in multiple tissues, including the heart and arteries,
adrenal gland, kidney, lung, liver, gallbladder, small intestine,
adipocytes, ovaries, uterus, breast, prostate, skin, lymphocytes
and central nervous system (36).
Melatonin receptors are derived from various tissues, with the
highest density expressed in the SCN, retina and anterior pituitary
(37). The coupling of high
affinity melatonin receptors MTNR1A and MTNR1B varies in
distribution throughout the body (38).
Both melatonin receptors couple to heterotrimeric G
proteins of the Gi subfamily, resulting in decreased adenylyl
cyclase activity and diminished cyclic adenosine monophosphate
(cAMP) production (39). Therefore,
melatonin signaling leads to the downregulation of genes controlled
by the CREB protein (40).
Melatonin receptors may also interact with other G proteins in a
context-specific manner (41).
Since the interaction between MTNR1A and Gi coupling exhibits
higher affinity compared with MTNR1A-Gq coupling, MTNR1A activation
primarily triggers the Gi/PKA/CREB, Gi/phosphatidylinositol
3'-kinase (PI3K)/AKT, Gi/protein kinase C (PKC)/ERK1/2, and
Gq/phospholipase C beta (PLCβ)/inositol 1,4,5-triphosphate
(IP3)/Ca2+ pathways (Fig.
1) (42). The binding of
melatonin to MTNR1A stimulates PLC activity, leading to the
conversion of phosphatidylinositol 4,5-biphosphate to
diacylglycerol and IP3(44).
Elevated levels of these secondary messengers activate several
kinases, including PKC, calmodulin kinases and mitogen-activated
protein kinases (43). Activation
of melatonin receptors leads to alterations in the ERK kinases'
signaling pathway (44). However,
MTNR1B activation decreases 3'-5'-cyclic guanosine monophosphate
levels, leading to reduced protein kinase G activity (Fig. 1) (45). Moreover, the Gi subfamily exhibits
slightly higher affinity in coupling to MTNR1A than to MTNR1B
(46). In a physiological context,
melatonin signaling through MTNR1A and MTNR1B may differ. Notably,
cells and tissues expressing receptors are not the only targets of
melatonin's physiological actions, as melatonin also exerts
non-receptor-dependent mechanisms of action, such as direct
antioxidant effects through the chelation of oxygen radical
species.
4. Melatonin receptor in kidney with MN
Both melatonin receptors are expressed in various
types of organs and tissues (47).
However, there is a different expression pattern in different cells
within the same organ. For example, in adult human islet cells,
MTNR1B receptor is expressed higher than MTNR1A in both α and β
cells; the expression levels of MTNR1B is similar in α and β cells
(48). The expression levels of
melatonin receptors in the renal tissue were previously determined
by the authors; the results revealed that MTNR1A is more highly and
abundantly expressed in glomerular and TECs than MTNR1B (49). Notably, MTNR1A is expressed
predominantly in TECs rather than in glomeruli at both the RNA and
protein levels (49).
Membranous glomerulonephritis is an
autoimmune-mediated glomerular nephritis characterized by
subepithelial immune complex deposits. The majority of primary MN
cases are caused by circulating antibodies that target the M-type
phospholipase A2 receptor located on the podocyte membrane,
triggering glomerular injury through a complement-dependent process
(50). Since TEC injury occurs
during the progression of MN, it often lacks a definitive cure and
may progress to chronic kidney disease (CKD) and end-stage renal
disease (ESRD), thereby posing a substantial public health threat
(51). Collectively, averting the
advancement of CDK or ESRD necessitates a more comprehensive
mechanistic understanding of MN.
Few previous studies have documented that MTNR1A
expression is decreased in various types of diseases, including the
substantia nigra and amygdala in Parkinson's disease, as well as in
human ductal breast cancer (52,53).
Using both clinical and experimental MN kidneys, the expression of
MTNR1A in TECs was validated. The downregulation of MTNR1A suggests
a protective role against MN progression. To assess the role of
reduced MTNR1A expression in MN progression, the biological effects
of luzindole (a competitive antagonist of MTNR1A/MTNR1B) were
evaluated in the MN mouse model. Consistent with previous findings,
MN mice exhibited symptoms of proteinuria, hypercholesterolemia and
hypoalbuminemia (54-59).
Remarkably, the inhibition of the MTNR1A receptor using luzindole
in MN mice exacerbated renal dysfunction, which was concomitant
with the upregulation of the core clock gene period 2 (PER2)
(49). The activating transcription
factor 4 and CREB heterodimer is essential for the transactivation
of the PER2 gene depending on cAMP stimulation (60), suggesting that luzindole blocks the
MTNR1A-mediated cell signaling. However, luzindole may impact blood
melatonin synthesis or other hormone production in different
tissues, subsequently affecting cAMP production in the kidney.
Therefore, the specific knockout of MTNR1A in mice TECs should be
employed to elucidate the MTNR1A-mediated physiological condition
and signaling pathway during the progression of MN.
5. Regulation of MTNR1A expression in renal
TECs
Considering the documented downregulation of MTNR1A
in MN, gaining a deeper understanding of its regulation will be
crucial in delineating the pathogenesis of this condition and
improving both diagnostic and therapeutic approaches. Indeed,
molecular mechanisms indicate that cytoplasmic heterogeneous
nuclear ribonucleoprotein L (hnRNPL) exerts a stabilizing effect on
the MTNR1A transcript through its interaction with MTNR1A RNA,
serving to protect it from degradation by the exosome component 10
(EXOSC10) protein (Fig. 3)
(26,61). EXOSC10 is an evolutionarily
conserved nuclear protein that interacts with the RNA exosome
complex and exhibits 3'-5'exoribonuclease activity (62). Furthermore, the activation of MTNR1A
by pituitary homeobox-1 (PITX1) is upregulated at the
transcriptional level (Fig. 3)
(49,63,64).
The predicted PITX1-binding region on the MTNR1A promoter, located
between positions -545 and -426, has been examined. Chromatin
immunoprecipitation-quantitative PCR results demonstrated
significant PITX1 recruitment to this region (49). These findings indicate that PITX1
directly transactivates MTNR1A in renal TECs. TECs depleted of
MTNR1A, PITX1 and CREB exhibit increased PER2 expression at both
the RNA and protein levels compared with control cells, suggesting
that the PITX1/MTNR1A/CREB axis regulates PER2 expression (Fig. 3). PER2 levels can affect peripheral
organs by modulating clock-controlled genes, as it is one of the
key components of the circadian clock gene (65). Taken together, MTNR1A expression is
upregulated by PITX1 and hnRNPL at the transcriptional and
post-transcriptional levels, respectively.
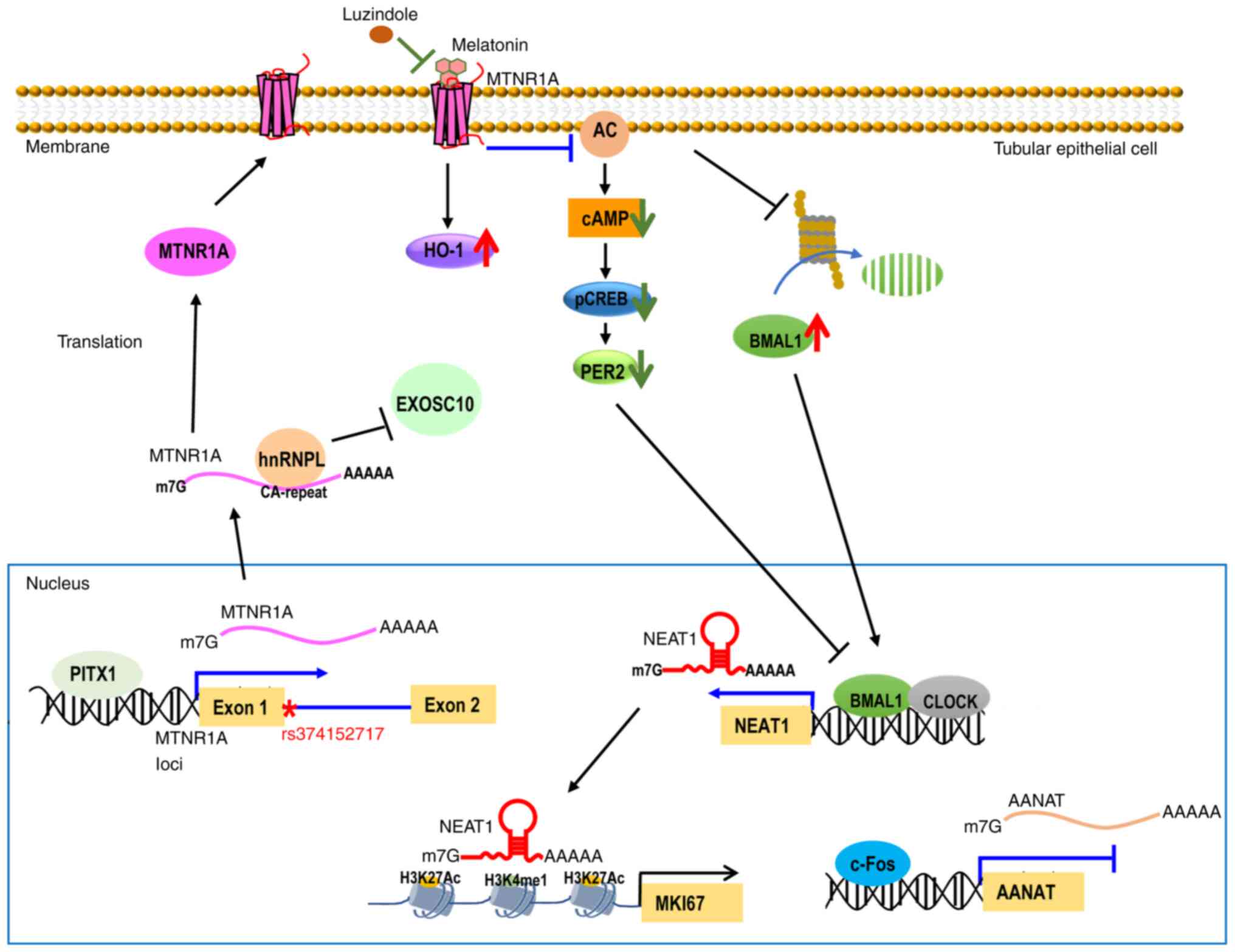 | Figure 3Schematic representation illustrates
the molecular mechanism regulating MTNR1A, NEAT1 and AANAT gene
expression in renal TECs. PITX1 transcriptionally upregulates
MTNR1A expression, while c-Fos transcriptionally downregulates
AANAT expression in the nucleus. AANAT is the rate-limiting factor
for melatonin synthesis. Albumin treatment reduced the viability of
TECs by decreasing PITX1 and increasing c-Fos. In the cytosol,
hnRNPL binds to MTNR1A transcripts via CA-repeat elements,
decreasing MTNR1A degradation by EXOSC10. Melatonin binding to
MTNR1A triggers upregulation of HO-1 levels and downregulation of
cAMP levels, phosphorylated CREB and PER2. Luzindole, an MTNR1A
antagonist, decreased the MTNR1A-mediated signaling pathway. The
long noncoding RNA NEAT1 is increased by melatonin and
exhibits circadian rhythm in TECs through whole gene
identification. Melatonin enhances clock-controlled NEAT1
expression in TECs by stabilizing the BMAL1 protein. Elevated
clock-controlled NEAT1 may regulate circadian genes, including
MKI67, by influencing H3K27Ac and H3K4me1 occupancy at enhancer
regions of target genes. Genomic location of MTNR1A single
nucleotide polymorphism rs374152717 (*), a donor splice site
variant in intron 1 near exon 1. MTNR1A, melatonin receptor 1A;
NEAT1, nuclear enriched abundant transcript 1; AANAT,
aryl-alkyl-amine N-acetyltransferase; TECs, tubular epithelial
cells; PITX1, pituitary homeobox-1; hnRNPL, heterogeneous nuclear
ribonucleoprotein L; HO-1, heme oxygenase-1; cAMP, cyclic adenosine
monophosphate; CREB, cAMP responsive element binding protein;
EXOSC10, exosome component 10. |
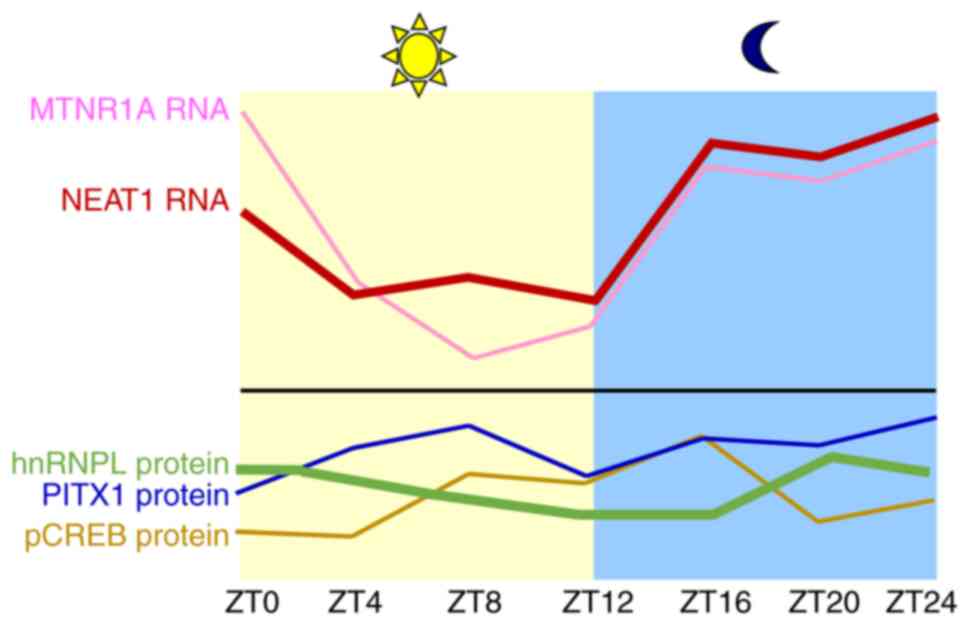
Although the daily rhythms of MTNR1A have been
extensively demonstrated in the SCN, adrenal gland, mammary gland
and liver, the diurnal change of MTNR1A in kidneys has been rarely
studied. The daily changes of MTNR1A, PITX1, hnRNPL, nuclear
enriched abundant transcript 1 (NEAT1) and
phosphorylated-CREB (pCREB) were detected in renal tissue (26,66).
The data indicated that MTNR1A and NEAT1 expression
demonstrated a higher amplitude compared with hnRNPL, PITX1 and
pCREB proteins (Fig. 4). These
findings suggest that MTNR1A and NEAT1 RNAs may
function as clock-controlled genes within the kidney. Furthermore,
the similar pattern of period and amplitude observed for
MTNR1A and NEAT1 transcript levels suggests a
correlation between NEAT1 and MTNR1A. Consistent with expectations,
melatonin enhances NEAT1 expression in a manner dependent on the
MTNR1A receptor in TECs (66).
Our data further demonstrated that MTNR1A
expression decreases between ZT0 and ZT8, followed by a rapid
increase between ZT12 and ZT16 (Fig.
4). Among the tested proteins, only pCREB displayed a clear
diurnal rhythm, peaking at ZT16 (Fig.
4). Notably, pCREB levels negatively correlated with MTNR1A
transcript levels between ZT4 and ZT12, suggesting that reduced
MTNR1A expression may lead to diminished CREB activation.
The detailed mechanisms by which PITX1 and hnRNPL regulate MTNR1A
expression in TECs, depending on timing and genomic context, remain
unclear.
The pathological lesions in MN include TECs' damage
(67). The reason is abnormal
glomerular permeability to proteins, which causes renal tubular
cell dysfunction (68). Therefore,
MTNR1A expression in TECs was investigated using an albumin-injury
cell model and an experimental model of MN. The results indicated
that albumin triggered downregulation of MTNR1A and PITX1 levels in
the TEC cell line HK-2 and in MN kidneys. Mechanistic findings
revealed low PITX1 expression in this albumin-induced TEC injury
model, leading to reduced recruitment of PITX1 to the MTNR1A
promoter (49). Notably, PITX1
levels decreased and associated with MTNR1A expression in clinical
MN kidney (49). These results
suggest a potential molecular mechanism of low MTNR1A levels in TEC
underlying proteinuria damage.
6. Single nucleotide polymorphisms (SNPs) in
MTNR1A
Previously, eight SNPs, namely rs216666, rs4862705,
rs684769, rs117287777, rs6553010, rs1946977, rs7687823 and
rs13140012, in the MTNR1A gene region were genotyped in 489
patients with type 1 diabetes (69). The authors evaluated the
associations of these eight SNPs with the decline in renal function
over a median follow-up period of 8 years (70). The results indicated that only the A
allele of rs4862705 was observed at a higher frequency in patients
with renal function decline compared with non-decliners (69). rs4862705 is located in 3' of MTNR1A
gene and located in LOC105377596 gene. The two genes overlap,
suggesting that rs4862705 affects LOC105377596 expression, then
affecting MTNR1A transcription. This mechanism is similar with the
X chromosome inactivation (XCI) process, two non-coding RNA X
inactive specific transcript (Xist) and Xist antisense RNA
transcribed in a mutually exclusive manner to mediated XCI in
female cells (71,72). Further studies are necessary to
confirm this molecular mechanism.
A recent study has identified the MTNR1A variant
rs374152717 as a genetic determinant of idiopathic osteoporosis, a
rare form of early-onset osteoporosis characterized by unexplained
bone loss (73). Specifically,
MTNR1A variant rs374152717, which disrupts the 5'consensus donor
splice site in one allele, results in alternative splicing variants
and subsequent partial translational deficiency (Fig. 3), leading to dysregulation of
melatonin signaling (73). The
mouse model of the rs374152717 variant and Mtnr1a+/-
reproduced the low bone mass and hypercalciuria phenotype of
young-adult patients with idiopathic osteoporosis (70). Collectively, the rs374152717 variant
leads to reduced MTNR1A protein levels and decreased bone mass.
This research establishes MTNR1A as a critical genetic factor in
osteoporosis, offering new avenues for diagnosis, genetic
counseling and therapeutic development.
A previous study indicated that MN mice treated with
the MTNR1A antagonist luzindole exhibited a marked increase in
proteinuria and hypercholesterolemia, along with a marked decrease
in serum albumin (49). These
results suggest that blocking MTNR1A receptor-mediated signaling
worsened renal function in this experimental MN model. Consistent
with this, decreased MTNR1A levels were observed in clinical MN
specimens. Given that the MTNR1A variant rs374152717 reduces its
expression (73), it warrants
investigation whether this variant is present in clinical MN
kidneys.
7. Exogenous melatonin treatment in MN
Elevated melatonin levels can attenuate
environmental stress-induced damage through the regulation of
complex interactions between immune cells, cytokines and signaling
pathways (74). Exogenous melatonin
exhibits pleiotropic therapeutic effects in various kidney
diseases, including those related to hypertension, diabetes
mellitus, acute kidney injury, CKD and MN (30,56).
Taking MN as an example, the renoprotective mechanisms of exogenous
melatonin include antioxidant, anti-apoptotic and anti-inflammatory
effects (56). In MN mice treated
with exogenous melatonin (MN-melatonin), elevated levels of
CD19+ B cells were observed, while T cell levels
remained unchanged. Additionally, heme oxygenase-1 (HO-1)
expression was higher in both glomerular and TECs in the
MN-melatonin mice compared with MN mice (Fig. 3). Of note, the renoprotective effect
of melatonin was reduced by treatment with a HO-1 inhibitor zinc
protoporphyrin (ZnPP) in an experimental MN model (56). Although the role of elevated HO-1 in
glomerular and TECs is unknown in experimental MN kidney, its
mediated protective mechanisms in podocytes and TECs have been
demonstrated in various mouse models (75). Induction of HO-1 against apoptosis
activity in podocytes treated with high glucose has been observed
and in diabetic kidneys (76).
Moreover, HO-1 inhibits inflammation by suppressing pyroptosis in
lipopolysaccharide (LPS)-exposed TECs (77). These results suggest that HO-1
expression protects podocytes and TECs against diabetic and
LPS-induced conditions (77). Given
that ZnPP inhibit both HO-1 and HO-2(78), these results cannot definitively
determine whether the protective effect in podocytes and TECs was
due to the blockade of HO-1, HO-2, or both activities. Further
studies should include the specific HO-1 target inhibitor.
The physiological conditions of the kidney can be
modulated by endogenous or exogenous melatonin through the
regulation of clock-controlled coding genes (79). However, there have been no studies
indicating which long non-coding RNAs (lncRNAs) are
clock-controlled and regulated by melatonin in the kidney.
Recently, unpublished results by the authors demonstrated that
exogenous melatonin upregulated clock-controlled lncRNAs, including
nuclear enriched abundant transcript 1 (NEAT1), which is
important for maintaining paraspeckles (66,80).
The molecular mechanism discovered that melatonin enhances
NEAT1 transactivation by increasing the stabilization of
BMAL1, which targets the NEAT1 promoter via the MTNR1A receptor
(Fig. 3). Furthermore, melatonin
enhances cell viability by increasing the occupancy of H3K27ac and
H3K4me1 at the upstream regions of proliferation gene
Ki-67(81), counteracting
albumin-induced injury to TECs. Collectively, these findings
suggest that melatonin treatment ameliorates experimental MN
through multiple pathways, including the elevation of HO-1 and
NEAT1 levels in TECs and the augmentation of
CD19+ B cells.
8. Exploring the clinical utility of
melatonin in kidney disease management
The promising use of exogenous melatonin has been
demonstrated in preclinical studies across various experimental
models, including adriamycin-induced nephropathy, 5/6 nephrectomy,
unilateral ureteral obstruction, spontaneously hypertensive rats
and MN (49,82). Moreover, increasing evidence
suggests a correlation between melatonin levels and renal function
(83). Abnormalities in serum
melatonin amplitude and rhythm are associated with the severity of
CKD in patients (83). Nighttime
urinary 6-sulphatoxymelatonin (6-SMT) levels, a major metabolite of
melatonin, were found to be lower in patients with stage 5 CKD
compared with those in other CKD stages (84). Decreased urinary 6-SMT levels were
positively associated with renal function parameters (84). These findings support the hypothesis
that exogenous melatonin could potentially slow CKD progression and
benefit other kidney diseases in clinical practice. A small
clinical trial (NCT04336566) evaluated the impact of a 5 mg daily
dose of melatonin on renal function in CKD, but the results are
pending. While melatonin itself is not directly metabolized by the
kidneys, its metabolites are renally excreted. Therefore, CKD
patients with renal insufficiency or those on dialysis should use
melatonin at a low recommended daily dose of 0.5-3 mg.
The absence of published results on ClinicalTrials.gov concerning exogenous melatonin use
in non-malignant kidney disease treatment can be attributed to
several factors. Primarily, melatonin's natural and largely
unpatentable nature makes it difficult to attract industry
sponsorship for extensive, high-quality trials in kidney disease.
Furthermore, the typically slow progression of CKD contributes to
the time-consuming and costly nature of such studies. Consequently,
the development of novel drugs aimed at enhancing melatonin
expression or melatonin receptor levels represents a promising new
direction for melatonin signaling therapy.
Previous clinical studies have indicated that after
a 12-week administration period, exogenous melatonin has beneficial
effects on glycemic control, high-density lipoprotein cholesterol
levels, and peroxisome proliferator-activated receptor gamma
expression in patients with diabetic nephropathy (DN) (85). However, it did not affect other
metabolic parameters related to renal functions. (85). It was hypothesized that the lack of
improvement in renal function with exogenous melatonin treatment
may be attributed to the downregulation of melatonin receptors in
DN. Therefore, combined melatonin and elevated MTNR1A drug
treatment could be beneficial for these patients. Hence, drugs that
enhance MTNR1A activity or expression may also enhance melatonin's
renoprotective functions.
9. Conclusion
The melatonin signaling pathway is a functionally
conserved and broad-spectrum physiological modulator found in
vertebrates. In the present study, the expression levels of
melatonin synthesis components and melatonin receptors in renal TEC
were summarized. The highest expression of DDC and MTNR1A compared
with other genes involved in the melatonin pathway in the kidney
was demonstrated. Using experimental models of MN and albumin
injury, reduced MTNR1A expression in renal TEC was further
identified, leading to upregulation of PER2 gene expression within
the MTNR1A/CREB axis. The molecular mechanism demonstrates that
MTNR1A is upregulated by PITX1 in a transcriptional manner and by
hnRNPL in a post-transcriptional manner in TECs. MTNR1A is reduced
in TEC and correlated with PITX1 in clinical MN samples. Exogenous
melatonin treatment alleviated the severity of experimental MN by
upregulating CD19+ B cells and HO-1 expression in both
glomerular and TECs. Moreover, melatonin enhances cell viability by
increasing NEAT1 transactivation through MTNR1A receptor and its
mediated Ki-67 expression in response to albumin-induced injury in
TECs. These results suggest that low MTNR1A in TECs worsens renal
function in experimental MN models. However, the clinical relevance
of decreased MTNR1A in MN progression should be investigated in the
future.
10 Future directions
Primary MN is an autoimmune glomerular disease
typically caused by circulating autoantibodies targeting podocytes,
predominantly anti-phospholipase A2 receptor autoantibodies
(70-75%) (86). After immune
complexes form and deposit, complement activation ensues, leading
to podocyte damage and alteration of the glomerular basement
membrane. The toxic effects of filtered proteinuria on renal TECs
exacerbate injury and promote TEC death, contributing to tubular
atrophy and interstitial fibrosis, which drive progression of CKD
toward ESRD (87). Consistent with
this notion, ~1/3 of patients with MN will progress to ESRD within
10 years (88,89). However, the current therapeutic
approach for MN patients involves the use of renin-angiotensin
system blockers and immunosuppressive agents targeting activated B
cells, effector T cells, complement activation and cytokine
production, without a specific focus on treating TECs (90). New drugs that specifically target
TECs to counteract proteinuria-induced fibrosis in MN should be
explored. Evidence suggests that MNTR1A levels in TECs are
associated with MN severity, indicating that MTNR1A could be a
potential therapeutic target for treating CKD progression.
Therefore, exploring drugs that enhance MTNR1A expression in TECs
could leverage endogenous melatonin signaling to improve renal
function in patients with MN and CKD.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported in part by the
Tri-Service General Hospital (grant nos. TSGH-PH-E-111001,
TSGH-PH-E-112016, TSGH-C02-112030, TSGH-C03-113038 and
TSGH-PH-E-113009) and the National Science and Technology Council
(grant no. MOST111-2314-B-016-038-MY3).
Availability of data and materials
Not applicable.
Authors' contributions
YSH and CCW designed and wrote the review. SMK, KCL
and AC revised and provided comments during all stages of writing
the manuscript. All authors read and approved the final version of
the manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tan DX, Zheng X, Kong J, Manchester LC,
Hardeland R, Kim SJ, Xu X and Reiter RJ: Fundamental issues related
to the origin of melatonin and melatonin isomers during evolution:
Relation to their biological functions. Int J Mol Sci.
15:15858–15890. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Emet M, Ozcan H, Ozel L, Yayla M, Halici Z
and Hacimuftuoglu A: A review of melatonin, its receptors and
drugs. Eurasian J Med. 48:135–141. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Slominski AT, Zmijewski MA, Semak I, Kim
TK, Janjetovic Z, Slominski RM and Zmijewski JW: Melatonin,
mitochondria, and the skin. Cell Mol Life Sci. 74:3913–3925.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Liu J, Clough SJ, Hutchinson AJ,
Adamah-Biassi EB, Popovska-Gorevski M and Dubocovich ML: MT1 and
MT2 melatonin receptors: A therapeutic perspective. Annu Rev
Pharmacol Toxicol. 56:361–383. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Reiter RJ, Mayo JC, Tan DX, Sainz RM,
Alatorre-Jimenez M and Qin L: Melatonin as an antioxidant: Under
promises but over delivers. J Pineal Res. 61:253–278.
2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Armstrong SM: Melatonin and circadian
control in mammals. Experientia. 45:932–938. 1989.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hardeland R: Melatonin and
inflammation-Story of a double-edged blade. J Pineal Res.
65(e12525)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tordjman S, Chokron S, Delorme R, Charrier
A, Bellissant E, Jaafari N and Fougerou C: Melatonin: Pharmacology,
functions and therapeutic benefits. Curr Neuropharmacol.
15:434–443. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xia Y, Chen S, Zeng S, Zhao Y, Zhu C, Deng
B, Zhu G, Yin Y, Wang W, Hardeland R and Ren W: Melatonin in
macrophage biology: Current understanding and future perspectives.
J Pineal Res. 66(e12547)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hardeland R: Aging, melatonin, and the
Pro- and anti-inflammatory networks. Int J Mol Sci.
20(1223)2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kopustinskiene DM and Bernatoniene J:
Molecular mechanisms of Melatonin-mediated cell protection and
signaling in health and disease. Pharmaceutics.
13(129)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Nikolaev G, Robeva R and Konakchieva R:
Membrane melatonin receptors activated cell signaling in physiology
and disease. Int J Mol Sci. 23(471)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tan DX, Reiter RJ, Manchester LC, Yan MT,
El-Sawi M, Sainz RM, Mayo JC, Kohen R, Allegra M and Hardeland R:
Chemical and physical properties and potential mechanisms:
Melatonin as a broad spectrum antioxidant and free radical
scavenger. Curr Top Med Chem. 2:181–197. 2002.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Okamoto HH, Cecon E, Nureki O, Rivara S
and Jockers R: Melatonin receptor structure and signaling. J Pineal
Res. 76(e12952)2024.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Klosen P: Thirty-seven years of MT1 and
MT2 melatonin receptor localization in the brain: Past and future
challenges. J Pineal Res. 76(e12955)2024.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Afsar B, Elsurer Afsar R, Sag AA, Kanbay
A, Korkmaz H, Cipolla-Neto J, Covic A, Ortiz A and Kanbay M: Sweet
dreams: Therapeutic insights, targeting imaging and physiologic
evidence linking sleep, melatonin and diabetic nephropathy. Clin
Kidney J. 13:522–530. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ekmekcioglu C: Melatonin receptors in
humans: Biological role and clinical relevance. Biomed
Pharmacother. 60:97–108. 2006.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Cagampang FR and Bruce KD: The role of the
circadian clock system in nutrition and metabolism. Br J Nutr.
108:381–392. 2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sanchez-Hidalgo M, de la Lastra CA,
Carrascosa-Salmoral MP, Naranjo MC, Gomez-Corvera A, Caballero B
and Guerrero JM: Age-related changes in melatonin synthesis in rat
extrapineal tissues. Exp Gerontol. 44:328–334. 2009.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Thul PJ and Lindskog C: The human protein
atlas: A spatial map of the human proteome. Protein Sci.
27:233–244. 2018.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Karlsson M, Zhang C, Mear L, Zhong W,
Digre A, Katona B, Sjöstedt E, Butler L, Odeberg J, Dusart P, et
al: A single-cell type transcriptomics map of human tissues. Sci
Adv. 7(eabh2169)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Acuna-Castroviejo D, Rahim I,
Acuna-Fernandez C, Fernández-Ortiz M, Solera-Marín J, Sayed RKA,
Díaz-Casado ME, Rusanova I, López LC and Escames G: Melatonin,
clock genes and mitochondria in sepsis. Cell Mol Life Sci.
74:3965–3987. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Konturek SJ, Konturek PC, Brzozowska I,
Pawlik M, Sliwowski Z, Cześnikiewicz-Guzik M, Kwiecień S,
Brzozowski T, Bubenik GA and Pawlik WW: Localization and biological
activities of melatonin in intact and diseased gastrointestinal
tract (GIT). J Physiol Pharmacol. 58:381–405. 2007.PubMed/NCBI
|
|
24
|
Fagerberg L, Hallstrom BM, Oksvold P,
Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S,
Danielsson A, Edlund K, et al: Analysis of the human
tissue-specific expression by genome-wide integration of
transcriptomics and Antibody-based proteomics. Mol Cell Proteomics.
13:397–406. 2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Huang YS, Lo CH, Tsai PH, Hou YC, Chang
YT, Guo CY, Hsieh HY, Lu KC, Shih HM and Wu CC: Downregulation of
AANAT by c-Fos in tubular epithelial cells with membranous
nephropathy. Biochem Biophys Res Commun. 584:32–38. 2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Huang YS, Lu KC, Chao HW, Chen A, Chao TK,
Guo CY, Hsieh HY, Shih HM, Sytwu HK and Wu CC: The MTNR1A mRNA is
stabilized by the cytoplasmic hnRNPL in renal tubular cells. J Cell
Physiol. 236:2023–2035. 2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Gazon H, Barbeau B, Mesnard JM and
Peloponese JM Jr: Hijacking of the AP-1 Signaling pathway during
development of ATL. Front Microbiol. 8(2686)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hardeland R, Tan DX and Reiter RJ:
Kynuramines, metabolites of melatonin and other indoles: The
resurrection of an almost forgotten class of biogenic amines. J
Pineal Res. 47:109–126. 2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ma X, Idle JR, Krausz KW and Gonzalez FJ:
Metabolism of melatonin by human cytochromes p450. Drug Metab
Dispos. 33:489–494. 2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Tang KS, Ho CY, Hsu CN and Tain YL:
Melatonin and Kidney Health: From Fetal Stage to Later Life. Int J
Mol Sci. 24(8105)2023.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Cecon E, Oishi A and Jockers R: Melatonin
receptors: Molecular pharmacology and signalling in the context of
system bias. Br J Pharmacol. 175:3263–3280. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Dubocovich ML, Delagrange P, Krause DN,
Sugden D, Cardinali DP and Olcese J: International union of basic
and clinical pharmacology. LXXV. Nomenclature, classification, and
pharmacology of G protein-coupled melatonin receptors. Pharmacol
Rev. 62:343–380. 2010.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu L, Labani N, Cecon E and Jockers R:
Melatonin target proteins: Too many or not enough? Front Endocrinol
(Lausanne). 10(791)2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Smirnov AN: Nuclear melatonin receptors.
Biochemistry (Mosc). 66:19–26. 2001.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Slominski RM, Reiter RJ,
Schlabritz-Loutsevitch N, Ostrom RS and Slominski AT: Melatonin
membrane receptors in peripheral tissues: Distribution and
functions. Mol Cell Endocrinol. 351:152–166. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Takahashi JS: Transcriptional architecture
of the mammalian circadian clock. Nat Rev Genet. 18:164–179.
2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Jockers R, Delagrange P, Dubocovich ML,
Markus RP, Renault N, Tosini G, Cecon E and Zlotos DP: Update on
melatonin receptors: IUPHAR Review 20. Br J Pharmacol.
173:2702–2725. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hardeland R, Cardinali DP, Srinivasan V,
Spence DW, Brown GM and Pandi-Perumal SR: Melatonin-a pleiotropic,
orchestrating regulator molecule. Prog Neurobiol. 93:350–384.
2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Reppert SM, Weaver DR and Godson C:
Melatonin receptors step into the light: Cloning and classification
of subtypes. Trends Pharmacol Sci. 17:100–102. 1996.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Tao L and Zhu Y: Melatonin regulates
CRE-dependent gene transcription underlying osteoblast
proliferation by activating Src and PKA in parallel. Am J Transl
Res. 10:86–100. 2018.PubMed/NCBI
|
|
41
|
Alexander SP, Davenport AP, Kelly E,
Marrion N, Peters JA, Benson HE, Faccenda E, Pawson AJ, Sharman JL,
Southan C, et al: The Concise Guide to PHARMACOLOGY 2015/16: G
protein-coupled receptors. Br J Pharmacol. 172:5744–5869.
2015.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Brydon L, Roka F, Petit L, de Coppet P,
Tissot M, Barrett P, Morgan PJ, Nanoff C, Strosberg AD and Jockers
Rl: Dual signaling of human Mel1a melatonin receptors via G(i2),
G(i3), and G(q/11) proteins. Mol Endocrinol. 13:2025–2038.
1999.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Tosini G, Owino S, Guillaume JL and
Jockers R: Understanding melatonin receptor pharmacology: Latest
insights from mouse models, and their relevance to human disease.
Bioessays. 36:778–787. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Tocharus C, Puriboriboon Y, Junmanee T,
Tocharus J, Ekthuwapranee K and Govitrapong P: Melatonin enhances
adult rat hippocampal progenitor cell proliferation via ERK
signaling pathway through melatonin receptor. Neuroscience.
275:314–321. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Xia AY, Zhu H, Zhao ZJ, Liu HY, Wang PH,
Ji LD and Xu J: Molecular mechanisms of the melatonin receptor
pathway linking circadian rhythm to type 2 diabetes mellitus.
Nutrients. 15(1406)2023.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Li DY, Smith DG, Hardeland R, Yang MY, Xu
HL, Zhang L, Yin HD and Zhu Q: Melatonin receptor genes in
vertebrates. Int J Mol Sci. 14:11208–11223. 2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Reppert SM: Melatonin receptors: Molecular
biology of a new family of G protein-coupled receptors. J Biol
Rhythms. 12:528–531. 1997.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Inoue T and Koike H: High-resolution
low-temperature scanning electron microscopy for observing
intracellular structures of quick frozen biological specimens. J
Microsc. 156:137–147. 1989.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Huang YS, Lu KC, Chao TK, Chen JS, Chen A,
Guo CY, Hsieh HY, Shih HM, Sytwu HK and Wu CC: Role of melatonin
receptor 1A and pituitary homeobox-1 coexpression in protecting
tubular epithelial cells in membranous nephropathy. J Pineal Res.
65(e12482)2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Couser WG: Primary membranous nephropathy.
Clin J Am Soc Nephrol. 12:983–997. 2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
AlYousef A, AlSahow A, AlHelal B, Alqallaf
A, Abdallah E, Abdellatif M, Nawar H and Elmahalawy R:
Glomerulonephritis histopathological pattern change. BMC Nephrol.
21(186)2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Hill SM, Cheng C, Yuan L, Mao L, Jockers
R, Dauchy B, Frasch T and Blask DE: Declining melatonin levels and
MT1 receptor expression in aging rats is associated with enhanced
mammary tumor growth and decreased sensitivity to melatonin. Breast
Cancer Res Treat. 127:91–98. 2011.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Adi N, Mash DC, Ali Y, Singer C, Shehadeh
L and Papapetropoulos S: Melatonin MT1 and MT2 receptor expression
in Parkinson's disease. Med Sci Monit. 16:BR61–ER67.
2010.PubMed/NCBI
|
|
54
|
Huang YS, Hsieh HY, Shih HM, Sytwu HK and
Wu CC: Urinary Xist is a potential biomarker for membranous
nephropathy. Biochem Biophys Res Commun. 452:415–421.
2014.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Wu CC, Lu KC, Lin YF, Chen JS, Huang CF,
Chen CC, Lin SH, Chu P and Sytwu HK: Pathogenic role of effector
cells and immunoglobulins in cationic bovine serum albumin-induced
membranous nephropathy. J Clin Immunol. 32:138–149. 2012.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Wu CC, Lu KC, Lin GJ, Hsieh HY, Chu P, Lin
SH and Sytwu HK: Melatonin enhances endogenous heme oxygenase-1 and
represses immune responses to ameliorate experimental murine
membranous nephropathy. J Pineal Res. 52:460–469. 2012.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Wu CC, Chen JS, Huang CF, Chen CC, Lu KC,
Chu P, Sytwu HK and Lin YF: Approaching biomarkers of membranous
nephropathy from a murine model to human disease. J Biomed
Biotechnol. 2011(581928)2011.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Wu CC, Chen JS, Lin SH, Chen A, Sytwu HK
and Lin YF: Experimental model of membranous nephropathy in mice:
Sequence of histological and biochemical events. Lab Anim.
42:350–359. 2008.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Wu CC, Chen JS, Chen SJ, Lin SH, Chen A,
Chang LC, Sytwu HK and Lin YF: Kinetics of adaptive immunity to
cationic bovine serum albumin-induced membranous nephropathy.
Kidney Int. 72:831–840. 2007.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Koyanagi S, Hamdan AM, Horiguchi M,
Kusunose N, Okamoto A, Matsunaga N and Ohdo S: cAMP-response
element (CRE)-mediated transcription by activating transcription
factor-4 (ATF4) is essential for circadian expression of the
Period2 gene. J Biol Chem. 286:32416–32423. 2011.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Stuparevic I, Novacic A, Rahmouni AR,
Fernandez A, Lamb N and Primig M: Regulation of the conserved
3'-5'exoribonuclease EXOSC10/Rrp6 during cell division, development
and cancer. Biol Rev Camb Philos Soc. 96:1092–1113. 2021.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Schmid M and Jensen TH: The exosome: A
multipurpose RNA-decay machine. Trends Biochem Sci. 33:501–510.
2008.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Johnston JD, Klosen P, Barrett P and
Hazlerigg DG: Regulation of MT melatonin receptor expression in the
foetal rat pituitary. J Neuroendocrinol. 18:50–56. 2006.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Johnston JD, Schuster C, Barrett P and
Hazlerigg DG: Regulation of the ovine MT1 melatonin receptor
promoter: Interaction between multiple pituitary transcription
factors at different phases of development. Mol Cell Endocrinol.
268:59–66. 2007.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Kim M, de la Pena JB, Cheong JH and Kim
HJ: Neurobiological functions of the period circadian clock 2 gene,
Per2. Biomol Ther (Seoul). 26:358–367. 2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Huang YS, Lu KC, Chang YT, Ka SM, Guo CY,
Hsieh HY, Shih HM, Sytwu HK and Wu CC: Melatonin alleviates
Albumin-induced tubular cell injury by activating Clock-controlled
nuclear enriched abundant transcript 1-mediated proliferation. ACS
Pharmacol Transl Sci. 7:3607–3617. 2024.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Alexopoulos E, Papagianni A and
Papadimitriou M: Is membranous nephropathy only a glomerular
disease? Ren Fail. 20:1–6. 1998.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Remuzzi G, Ruggenenti P and Benigni A:
Understanding the nature of renal disease progression. Kidney Int.
51:2–15. 1997.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Albert FW and Kruglyak L: The role of
regulatory variation in complex traits and disease. Nat Rev Genet.
16:197–212. 2015.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Daher G, Santos-Bezerra DP, Cavaleiro AM,
Pelaes TS, Admoni SN, Perez RV, Machado CG, do Amaral FG,
Cipolla-Neto J and Correa-Giannella ML: Rs4862705 in the melatonin
receptor 1A gene is associated with renal function decline in type
1 diabetes individuals. Front Endocrinol (Lausanne).
15(1331012)2024.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Loos F, Maduro C, Loda A, Lehmann J,
Kremers GJ, Ten Berge D, Grootegoed JA and Gribnau J: Xist and tsix
transcription dynamics is regulated by the X-to-autosome ratio and
semistable transcriptional states. Mol Cell Biol. 36:2656–2667.
2016.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Lee JT, Davidow LS and Warshawsky D: Tsix,
a gene antisense to Xist at the X-inactivation centre. Nat Genet.
21:400–404. 1999.PubMed/NCBI View
Article : Google Scholar
|
|
73
|
Bisikirska B, Labella R, Cuesta-Dominguez
A, Luo N, De Angelis J, Mosialou I, Lin CS, Beck D, Lata S, Shyu
PT, et al: Melatonin receptor 1A variants as genetic cause of
idiopathic osteoporosis. Sci Transl Med.
16(eadj0085)2024.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Watt SM: The long and winding road:
Homeostatic and disordered haematopoietic microenvironmental
niches: A narrative review. Biomater Transl. 3:31–54.
2022.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Zhai H, Ni L and Wu X: The roles of heme
oxygenase-1 in renal disease. Front Nephrol.
3(1156346)2023.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Lee SC, Han SH, Li JJ, Lee SH, Jung DS,
Kwak SJ, Kim SH, Kim DK, Yoo TH, Kim JH, et al: Induction of heme
oxygenase-1 protects against podocyte apoptosis under diabetic
conditions. Kidney Int. 76:838–848. 2009.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Li HB, Mo YS, Zhang XZ, Zhou Q, Liang XD,
Song JN, Hou LN, Wu JN, Guo Y, Feng DD, et al: Heme oxygenase-1
inhibits renal tubular epithelial cell pyroptosis by regulating
mitochondrial function through PINK1. Exp Ther Med.
25(213)2023.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Rublevskaya I and Maines MD: Interaction
of Fe-protoporphyrin IX and heme analogues with purified
recombinant heme oxygenase-2, the constitutive isozyme of the brain
and testes. J Biol Chem. 269:26390–26395. 1994.PubMed/NCBI
|
|
79
|
Stow LR and Gumz ML: The circadian clock
in the kidney. J Am Soc Nephrol. 22:598–604. 2011.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Sasaki YT, Ideue T, Sano M, Mituyama T and
Hirose T: MENepsilon/beta noncoding RNAs are essential for
structural integrity of nuclear paraspeckles. Proc Natl Acad Sci
USA. 106:2525–2530. 2009.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Sun X and Kaufman PD: Ki-67: More than a
proliferation marker. Chromosoma. 127:175–186. 2018.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Rahman A, Hasan AU and Kobori H: Melatonin
in chronic kidney disease: A promising chronotherapy targeting the
intrarenal renin-angiotensin system. Hypertens Res. 42:920–923.
2019.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Koch BC, van der Putten K, Van Someren EJ,
Wielders JP, Ter Wee PM, Nagtegaal JE and Gaillard CA: Impairment
of endogenous melatonin rhythm is related to the degree of chronic
kidney disease (CREAM study). Nephrol Dial Transplant. 25:513–519.
2010.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Ishigaki S, Ohashi N, Isobe S, Tsuji N,
Iwakura T, Ono M, Sakao Y, Tsuji T, Kato A, Miyajima H and Yasuda
H: Impaired endogenous nighttime melatonin secretion relates to
intrarenal renin-angiotensin system activation and renal damage in
patients with chronic kidney disease. Clin Exp Nephrol. 20:878–884.
2016.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Satari M, Bahmani F, Reiner Z, Soleimani
A, Aghadavod E, Kheiripour N and Asemi Z: Metabolic and
Anti-inflammatory response to melatonin administration in patients
with diabetic nephropathy. Iran J Kidney Dis. 1:22–30.
2021.PubMed/NCBI
|
|
86
|
Hoxha E, Reinhard L and Stahl RAK:
Membranous nephropathy: New pathogenic mechanisms and their
clinical implications. Nat Rev Nephrol. 18:466–478. 2022.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Makhammajanov Z, Gaipov A, Myngbay A,
Bukasov R, Aljofan M and Kanbay M: Tubular toxicity of proteinuria
and the progression of chronic kidney disease. Nephrol Dial
Transplant. 39:589–599. 2024.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Hanset N, Esteve E, Plaisier E, Johanet C,
Michel PA, Boffa JJ, Fievet P, Mesnard L, Morelle J, Ronco P and
Dahan K: Rituximab in patients with phospholipase A2
Receptor-associated membranous nephropathy and severe CKD. Kidney
Int Rep. 5:331–338. 2020.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Trujillo H, Alonso M and Praga M: New ways
of understanding membranous nephropathy. Nephron. 144:261–271.
2020.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Rojas-Rivera JE, Ortiz A and Fervenza FC:
Novel treatments paradigms: Membranous nephropathy. Kidney Int Rep.
8:419–431. 2023.PubMed/NCBI View Article : Google Scholar
|