Introduction
Hepatocellular carcinoma (HCC) is one of the most
common human malignancies in the world. Chronic infection with the
hepatitis B or C virus (HBV or HCV) is a major risk factor for HCC.
In Japan, approximately 85% of HCC cases are caused by HBV and HCV
infections (1). In particular,
HCV-positive HCC accounts for 72% of HCC cases. Although liver
resection has been established as a curative treatment for HCC,
recurrence is observed with a high frequency after resection. HCC
patients suffer relapses in 25, 50 and 80% of cases within 1, 2 and
5 years after liver resection, respectively (2,3).
Recurrence after liver resection for HCC occurs mostly in the
remnant liver. Vascular invasion, tumor size, multiple tumors,
cirrhosis, TNM classification, serum α-fetoprotein (AFP) level and
serum bilirubin level have been established as risk factors for HCC
recurrence, (3,4). However, the molecular pathogenesis of
HCC recurrence in the non-tumorous tissues of remnant livers is
controversial.
The recent introduction of DNA microarray technology
has opened a new road in medical science and has demonstrated the
critical molecular and biological characteristics of HCC (5). Comprehensive analyses of the
gene-expression profiles of HCC have been performed taking into
account various clinical characteristics, such as the differences
between tumor and non-tumorous tissues (6), causative hepatitis virus types
(6,7), the presence and absence of portal
invasion (6) and histological
differentiation grades (6), and
have revealed a group of significant genes. Moreover, genetic
predisposition towards recurrence is thought to be harbored in the
tumor environment, such as in a non-tumorous liver with chronic
hepatitis or liver cirrhosis. When liver cancer is surgically
resected, recurrence occurs in the remaining remnant liver or
extrahepatic lesion. In particular, recurrence after liver
resection for HCC occurs most frequently in the remnant liver. In
the present study, we focused on Japanese HCC patients, the
majority of whom presented with HCV-positive HCC. Gene expression
was assessed in the non-tumorous tissues of the remnant liver in
HCC patients who had an early recurrence compared to that in
patients who had late recurrence.
Materials and methods
Patients and tissue samples
From March 1995 to December 2006, a total of 309
patients with UICC TNM classification (8) stages I and II HCV-positive HCC
underwent liver resection at Nihon University Hospital, Japan.
Surgical samples were obtained from 39 of 222 patients who had only
a solitary tumor, and in whom no secondary cancerous mass was
clinically observed in the remnant liver and no extrahepatic
lesions were noted during the initial surgery. Non-tumorous tissues
were collected from the negative resected margin, which was
pathologically confirmed to be a non-cancerous region. Samples were
stored frozen at −80°C immediately after separation. Informed
consent was obtained from all of the patients in advance.
Recurrence was determined every 2–6 months during the follow-up
period.
RNA preparation and oligonucleotide
microarray
Total RNA was extracted from the frozen tissues
using TRIzol reagent (Invitrogen Corp., Carlsbad, CA, USA),
following the manufacturer’s protocol. The integrity of the RNA
obtained was assessed using an Agilent 2100 BioAnalyzer (Agilent
Technologies, Palo Alto, CA, USA). Two groups, non-tumorous cRNA
from early recurrence (<2 years) cases and that from late
recurrence (>4 years) cases, were analyzed. Total RNA (3.3 μg)
from 3 cases were pooled and used for the synthesis of biotinylated
cRNA from each group. Complementary DNA synthesis and in
vitro transcription from the cDNA to biotinylated cRNA were
performed following the Expression Analysis Technical Manual from
Affymetrix Inc. (Santa Clara, CA, USA). A GeneChip Human Genome
U133A oligonucleotide microarray (Affymetrix Inc.) included 22,283
human genes. In the present study, each group of subjects was
represented by a single microarray chip to screen candidate genes
differentially expressed between the groups.
Following the manufacturer’s instructions, the
microarray chips were subjected to pre-hybridization and
hybridization with the biotinylated cRNA. They were then washed and
stained. The fluorescence signal on the microarray chips was
detected using the Gene Array Scanner (Hewlett-Packard, Palo Alto,
CA, USA) and analyzed using GeneSpring version 5.0.3 software
(Silicon Genetics, Redwood City, CA, USA). The whole microarray
data set of each GeneChip was first normalized to the median of the
gene expression intensity values among the genes. The expression
signal intensity of each gene was then subjected to further
normalization to the median value among the GeneChips. The genes
were further screened with the criterion that at least one GeneChip
detection flag was present (‘P’). These pre-analysis
data-processing procedures identified 7,444 genes for expression
analysis, following the Data Analysis Fundamentals Manual from
Affymetrix Inc. The only criterion modified was the inclusion of
signal fold changes for genes that were up-regulated by at least
2.5 or down-regulated by at least −2.5 in the early recurrence
non-cancerous tissues compared to the late recurrence non-cancerous
tissues.
Real-time RT-PCR
To assess the risk of recurrence using the
non-cancerous tissue, RNA was isolated with TRIzol reagent
according to the manufacturer’s instructions, and the RNA was
treated with RNase-free DNase before cDNA synthesis. First-strand
cDNA was synthesized using a SuperScript™ First-Strand
kit (Invitrogen Corp.). Real-time RT-PCR (Thermal Cycler Dice® Real
Time System TP800; Takara Bio Inc., Shiga, Japan) was carried out
using a SYBR® Premix Ex Taq™ kit or SYBR® Premix Ex
Taq™ II (Takara Bio Inc.). To perform an
optimal-condition real-time PCR for each gene, the optimum amount
of cDNA was determined and used for each gene. The 25-μl two-step
RT-PCR mixture consisted of 12.5 μl of SYBR Premix Ex Taq, 0.5 μl
each of a forward and reverse primer, 10.5 μl of RNase-free water
and 1 μl of template cDNA for the PSMB8, RALGDS, APOL3, GBP1,
RPS14, CXCL9, DKFZp564F212, CYP1B1, TNFSF10, NROB2, AKR1B10, MAFB,
BF530535, MRPL24, TSC22D3, QPRT, VNN1, FMO5, DCN and
GAPDH genes, respectively. The 25-μl two-step RT-PCR mixture
consisted of 12.5 μl of SYBR Premix Ex Taq II, 1.0 μl each of a
forward and reverse primer, 9.5 μl of RNase-free water and 1 μl of
template cDNA for the ALB and IRS2 genes. The
real-time cycler conditions consisted of 95°C for 10 sec followed
by 40 cycles at 95°C for 5 sec and 60°C for 30 sec. The relative
quantity for these gene expression levels was normalized to
GAPDH expression.
Each cDNA sample was subjected to triplicate RT-PCR
reactions, and the results were processed by absolute quantitative
analysis based on a standard curve determined by serial 5-fold
dilutions of an appropriate cDNA. Multiple RQ software, ver. 1.00
(Takara Bio Inc.) was used for data analysis. The primer sequences
of the internal standard and target genes are shown in Table I.
 | Table I.Recurrence-related candidate genes
tested by real-time RT-PCR. |
Table I.
Recurrence-related candidate genes
tested by real-time RT-PCR.
| Genes | Primer sequence
|
|---|
| Forward (5′-3′) | Reverse (5′-3′) |
|---|
| Early
recurrence-related | | |
| ALB |
CAAAGCATGGGCAGTAGCTC |
CAAGCAGATCTCCATGGCAG |
| NROB2 |
TCTTCAACCCCGATGTGCCA |
AGGCTGGTCGGAATGGACTT |
| AKR1B10 |
CTTGGAAGTCTCCTCTTGGC |
ATGAACAGGTCCTCCCGCTT |
| MAFB |
TCTGGGCCTGCGCTAATTG |
TTGGTTCAGTGCAGTGTCTGCTTAC |
|
BF530535 |
AGTGGGATGGATCAGCTGTGAA |
TGGTGAGGCGACCATCAATTAG |
|
MRPL24 |
AAGGAAGGTTTCGAGCGTTT |
GAGATGGGTTCCACAACCAC |
|
TSC22D3 |
AACAGGCCATGGATCTGGTG |
AGGACTGGAACTTCTCCAGC |
| QPRT |
CTGACTTCGCTCTGAAGGTGGA |
CACAGCCACACTCGGGAACT |
| VNN1 |
GCTGGAACTTCAACAGGGAC |
CTGAGGATCACTGGTATCGC |
| IRS2 |
GCATTCCAGCCCCTATGTTA |
AGTGTCGAGGGAGCAGAAAA |
|
FMO51 |
ACACAGAGCTCTGAGTCAGC |
TCCAGGTTAGGAGGGAAGAC |
|
FMO52 |
GAAAGGACTGATGACATCG |
TGAAATACTCCAGGACCTGG |
| DCN |
CCTCAAGGTCTTCCTCCTTC |
CACCAGGTACTCTGGTAAGC |
| Late
recurrence-related | | |
| PSMB8 |
AGACTGTCAGTACTGGGAGC |
GTCCAGGACCCTTCTTATCC |
|
RALGDS |
TGCCGCTCTACAACCAGCAG |
GAATCTGCAGCAGCTCATAGTCCTC |
| APOL3 |
AATTGCCCAGGGATGAGGCA |
TGGACTCCTGGATCTTCCTC |
| GBP1 |
AGAGGACCCTCGCTCTTAAACTTC |
TTATGGTACATGCCTTTCGTCGTCT |
| RPS14 |
GACGTGCAGAAATGGCACCT |
CAGTCACACGGCAGATGGTT |
| CXCL9 |
CCTGCATCAGCACCAACCAA |
TGGCTGACCTGTTTCTCCCA |
|
DKFZp564F212 |
CCTGGGCAAGTGAGGTCTTC |
TCTCTGGCAGGTTGTTCCTGA |
|
CYP1B1 |
CCTCTTCACCAGGTATCCTG |
CCACAGTGTCCTTGGGAATG |
|
TNFSF10 |
GCTGAAGCAGATGCAGGACA |
CTAACGAGCTGACGGAGTTG |
| Internal control
gene | | |
| GAPDH |
GCACCGTCAAGGCTGAGAAC |
TGGTGAAGACGCCAGTGGA |
Immunohistochemistry
To confirm the localization of early
recurrence-related gene products in the non-tumorous liver, 4-μm
sections were routinely prepared from archived paraffin-embedded
blocks of two selected cases that clearly showed low or high mRNA
expression by RT-PCR. Deparaffinized sections were autoclaved in
0.01 M sodium citrate buffer (pH 6.0) at 121°C for 15 min. After
routine pre-treatments, sections were incubated at 4°C overnight
with an anti-GBP1 monoclonal antibody (clone 4D10; Abnova, Taipei,
Taiwan) at a 1:300 dilution and anti-TSC22D3 (TSC22 domain family,
member 3) antibody (clone 3A5; Abnova) at a 1:300 dilution.
Histofine simple stain Max PO (Multi) (Nichirei Co., Tokyo, Japan)
was used as the secondary antibody, and was applied for 30 min at
room temperature. After visualization with Vector® SG substrate
(Vector Laboratories, Burlingame, CA, USA) the sections were
counterstained with nuclear fast red. A negative control slide
without primary antibody was included in each staining. Nuclei
positive for TSC22D3 staining were counted in ten high-power
fields.
Classification of early and late
recurrence groups
The disease-free survival rate was calculated using
the Kaplan-Meier method. To distinguish the early from the late
recurrence group, segmented linear regression analysis with two
segments was used to determine a crosspoint of threshold values
above and below which it was possible to divide the two groups
according to the distribution of the interval of recurrence for
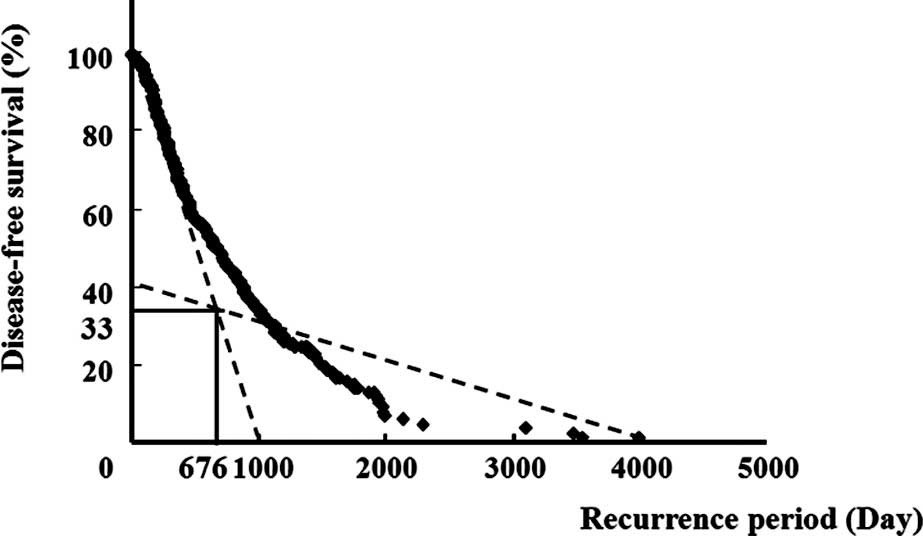
either the early or the late recurrence group (Fig. 1). The y1 = −0.097x +
98.84 line for an interval of x ≤676 days (crosspoint), and the
y2 = −0.012x + 41.16 line for an interval of x >676
days (crosspoint) are shown. The goodness of fit was estimated by
R2. R2 values for the intervals of x ≤676
days and >676 days were 0.98 and 0.81, respectively, suggesting
that each group fit a distinct linear regression separated at a
crosspoint of 676 days. Thus, the early recurrence group was
defined as having an interval of recurrence less than the
cross-point, and the late recurrence group, an interval of
recurrence greater than the crosspoint (676 days) (Fig. 1).
Statistical analysis
The disease-free survival rate was calculated from
the period begining with the date of surgery until the date of
recurrence. Differences in gene expression between the two groups
were evaluated using the Mann-Whitney U-test. The Chi-square and
Fisher’s exact probability tests were used for discrete variables,
and the Student’s t-test for continuous variables. A step-wise
discriminant analysis was applied to the expression levels of 21
genes isolated from microarray analysis, and a gene combination
suitable for distinguishing between the early and late recurrence
groups was extracted. The best combination of genes associated with
recurrence risk within 2 years after resection was also determined
by a backward step-wise multivariate logistic regression analysis
using gene expression levels from the non-tumorous livers. Akaike’s
information criterion (AIC) was used to evaluate the
goodness-of-fit of a logistic regression model in each step.
Statistical analysis was performed using SPSS, version 15.0 (SPSS
Inc., Chicago, IL, USA). A P-value of <0.05 was considered to
indicate statistical significance.
Results
Comparison of the clinicopathological
features between the early and late recurrence groups
Using the Kaplan-Meier method, 676 days was selected
as the crosspoint for distinguishing between the early and late
recurrence groups. As shown in Fig.
1, the early recurrence group was defined as patients having
disease recurrence before 676 days. Among the 39 cases, early
recurrence occurred in 22 cases and late recurrence in 17 cases.
Table II shows a comparison of
background clinicopathological features between the two groups. No
significant differences were found among variables, including
gender, age, tumor size, histological grade, background liver,
indocyanin green retention rate at 15 min, total bilirubin,
albumin, aspartate aminotransferase, alanine aminotransferase,
platelets, AFP and vascular invasion.
 | Table II.Comparison of clinicopathological
features between the early and late recurrence groups. |
Table II.
Comparison of clinicopathological
features between the early and late recurrence groups.
| Variable | Early group
(n=22) | Late group
(n=17) | P-value |
|---|
| Gender | | | 0.70 |
| Male | 17 | 14 | |
| Female | 5 | 3 | |
| Age (years) | | | 0.81 |
| Median | 65 | 66 | |
| Range | 50–78 | 48–73 | |
| Tumor size
(cm) | | | 0.50 |
| <5 | 20 | 17 | |
| ≥5 | 2 | 0 | |
| Histological
grade | | | 0.52 |
| W/D | 9 | 5 | |
| M/D, P/D | 13 | 12 | |
| Background | | | 0.21 |
| Non-LC | 7 | 9 | |
| LC | 15 | 8 | |
| ICG15R (%) | | | 0.52 |
| <10 | 9 | 5 | |
| ≥10 | 13 | 12 | |
| T.Bil (mg/dl) | | | 0.50 |
| <1.2 | 20 | 17 | |
| ≥1.2 | 2 | 0 | |
| Alb (g/dl) | | | 1.00 |
| <3.5 | 6 | 4 | |
| ≥3.5 | 16 | 13 | |
| AST (U/l) | | | 1.00 |
| <40 | 6 | 5 | |
| ≥40 | 16 | 12 | |
| ALT (U/l) | | | 0.21 |
| <40 | 7 | 9 | |
| ≥40 | 15 | 8 | |
| PLT
(109/l) | | | 0.73 |
| <15 | 16 | 11 | |
| ≥15 | 6 | 6 | |
| AFP (ng/ml) | | | 0.18 |
| <20 | 6 | 9 | |
| ≥20 | 16 | 8 | |
| Vascular
invasion | | | 1.00 |
| (+) | 8 | 7 | |
| (−) | 14 | 10 | |
Recurrence-related genes in the
non-tumorous liver
Two pooled samples of cRNA each from three
representative cases from the early or late non-tumorous recurrence
groups were applied to the oligonucleotide microarray expression
analysis of non-tumorous liver mRNA. The data analysis referred to
a signal of the control gene BioB for an intrinsically fixed
quantity after normalization as a detection limit. Probes that had
differences in expression of ≥2.5-fold between the two groups were
extracted; finally, only probes having P flags on all of the
microarrays with increased expression were selected. In this way,
21 of the 22,283 genes were identified as being differentially
expressed between the non-tumorous livers of the early and late
recurrence groups. Among these genes, 9 were up-regulated in the
late recurrence group and 12 were up-regulated in the early
recurrence group (Table I).
Prediction of early recurrence genes
To examine whether the 21 candidate genes were
differentially expressed between the early and late HCC recurrence
groups, mRNAs from the 21 genes in the non-tumorous tissues of all
39 HCC cases were quantified. Between the early and late recurrence
group, 2 genes were recognized to exhibit a difference in
expression. Significantly low expression of the GBP1 gene
and significantly high expression of the TSC22D3 gene were
noted in the early recurrence group (Table III). No significant difference in
expression was noted for the other genes.
 | Table III.Genes exhibiting differential
expression between the early and late recurrence groups by the
Mann-Whitney U-test. |
Table III.
Genes exhibiting differential
expression between the early and late recurrence groups by the
Mann-Whitney U-test.
| Gene | Early group median
(range) | Late group median
(range) | P-value |
|---|
| GBP1 | 0.45
(0.24–1.48) | 1.23
(0.49–4.23) | 0.04 |
| TSC22D3 | 1.07
(0.12–2.38) | 0.81
(0.17–2.30) | 0.04 |
The best combination of genes associated with
recurrence risk within 2 years after resection was also determined
by a backward step-wise multivariate logistic regression analysis
using gene expression levels from the non-tumorous livers. The
expression of GBP1 and TSC22D3 was associated with
the rate of recurrence. The GBP1 gene was identified as
being associated with a reduced risk of early recurrence [odds
ratio (OR)=0.20], while the TSC22D3 gene was identified as
being associated with an increased risk of early recurrence
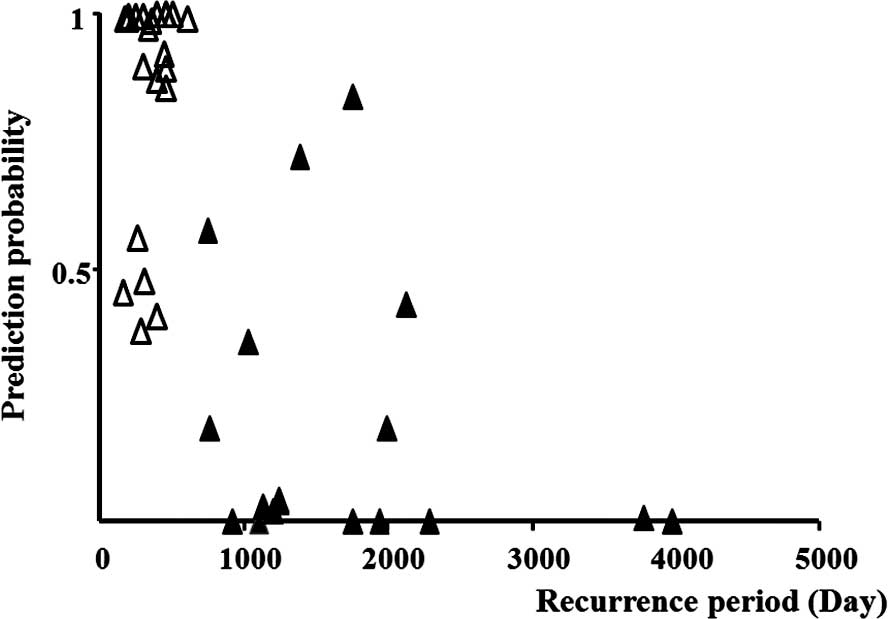
(OR=19.6). A 95% confidence interval (CI) is indicated in Table IV. These two genetic combinations
were identified as useful parameters for the prediction of
recurrence by multivariate logistic regression analysis (Fig. 2).
 | Table IV.Multivariate logistic regression
analysis of early recurrence risk. |
Table IV.
Multivariate logistic regression
analysis of early recurrence risk.
| Gene | Odds ratio | 95% CI | P-value |
|---|
| GBP1 | 0.2 | 0.06–0.73 | 0.02 |
| TSC22D3 | 19.6 | 1.14–337.2 | 0.04 |
Localization of recurrence-related gene
products
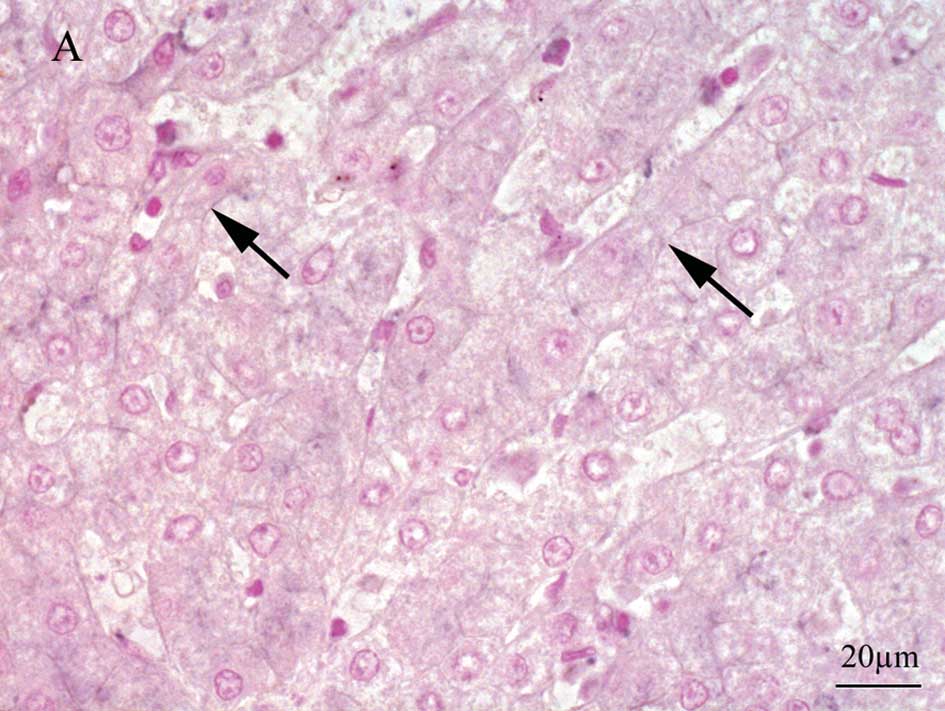
GBP1 immunostaining revealed strong membranous
expression in hepatocytes demonstrating a reticular pattern in the
late recurrence group, while relatively weak membranous staining
was noted in the early recurrence group (Fig. 3A and B). Upon TSC22D3 staining,
endothelial cells, some lymphocytes and regenerating bile ductuli
exhibited nuclear positivity throughout the sections. The mean
values of the positive nuclei were 9.1 and 4.5 in the early and
late recurrence groups, respectively. A few nuclei of hepatocytes
were positively stained for TSC22D3 in the early recurrence group
(Fig. 3C), while there were no
positive cells noted in the late recurrence group (Fig. 3D).
Discussion
Hoshida et al demonstrated the genome-wide
expression profiling of formalin-fixed, paraffin-embedded tissues
and showed that a reproducible gene expression signature
correlating with survival is present in liver tissues adjacent to
the tumor in HCC patients (9).
Tsuchiya et al reported that various genes in non-tumoral
tissues of HCV-associated HCC cases were associated with late
recurrence (10). Here, we
identified the early and late recurrence-related genes in
non-tumorous tissues from HCV-positive HCC cases. We examined
whether the early and late recurrence groups had different patient
and tumor characteristics. Although clinically there was no
difference when comparing the two groups, we found that the gene
expression of two genes could be used to distinguish between
them.
Previously, researchers have reported various risk
factors for recurrence after liver resection, such as the degree of
liver fibrosis (11), hepatitis
virus infection type (12), number
of tumor nodules (13,14), presence of AFP mRNA in the
circulation (15), serum albumin
level, ICG-15 level, tumor location (16), presence of venous invasion
(4,17) and tumor size (14).
In light of such processes, we aimed to ascertain
whether there is a ‘bud of recurrence’ in the remnant liver, namely
in the non-tumorous tissues, and whether this can be defined in
postoperative early or late recurrence groups through a gene
expression study. Here, non-tumorous tissues were used to
investigate whether variations in the expression of HCC
recurrence-related genes affect the recurrence interval. To date,
there have been various reports on the gene expression of HCC, but
most of the comparisons were carried out between the gene
expression of tumor vs. non-tumorous tissues (5–7). Our
data clearly showed that the TSC22D3 and GBP1 genes
were differentially expressed in the remnant liver tissues of the
early and late recurrence groups. Of these, GBP1 exhibited
high expression in the late recurrence group, while TSC22D3
demonstrated high expression in the early recurrence group.
GBP1, human guanylate binding protein-1, is a member of the
large GTPase protein family. Expression of GBP1 is induced
by inflammatory cytokines (ICs) in endothelial cells (ECs).
GBP1 was found to mediate the angiostatic effects of ICs on
EC proliferation in vitro (18). Moreover, in vivo GBP1
expression was shown to decrease the angiogenic activity of ECs
(18,19). Genes associated with HCV-induced
HCC, such as GPC3, HSP70 and TSAP6, have been
investigated, while the TSC22D3 and GBP1 genes have
not been studied using tumor materials (20,21).
Knockdown of GBP1, IFI-6-16 and
IFI-27 by short hairpin RNA was found to result in an
increase in HCV replication (22).
Based on our analysis of GBP1 localization, GBP1 appears to be
produced by hepatocytes in the non-tumorous tissue of the remnant
liver. GBP1 protein expression appeared to be further increased in
the late recurrence group than in the early recurrence group.
Therefore, increased expression of GBP1 in hepatocytes may suppress
HCV replication. GBP1 protein translation may be suppressed in
hepatocytes by a degradation pathway, and this pathway may be
deregulated in the HCC carcinogenesis process, specifically in
early recurrence. The 5-year survival rate was found to be
significantly increased in GBP1-positive colorectal cancer
patients. GBP1 may be a novel biomarker and an active
component of the T-helper 1 angiostatic immune reaction in
colorectal cancer (23). In this
way, GBP1 may also control angiostatic effects in the
liver.
The expression of TSC22D3 (TSC22 domain
family, member 3) is stimulated by glucocorticoids and
interleukin-10, and appears to play a key role in the
anti-inflammatory and immunosuppressive effects of this steroid and
chemokine. TSC22D3, also known as glucocorticoid-induced
leucine zipper protein (GILZ), is expressed in a variety of
mammalian cells. According to a recent report, this gene
up-regulated cyclin D1 and phosphorylated retinoblastoma,
down-regulated cyclin-dependent kinase inhibitor p21, and promoted
entry into the S phase of the cell cycle in epithelial ovarian
cancer HCCs are hypervascular tumors and angiogenesis plays a
crucial role in the process of neoplasia (25). In culture, VEGF was found to
enhance cyclin D1 expression and HCC cell growth. The restraint of
cyclin D1 control was found to decrease angiogenesis (26). In the present study, a few
TSC22D3-positive hepatocytes were scattered in the non-tumorous
tissue of the early recurrence group, while no positive cells were
scattered in the late recurrence group. The physiological role of
TSC22D3 has not been clarified, and no previous report on
hepatocytes producing TSC22D3 has been published. This seems to
suggest that cyclin D1 up-regulated by GILZ and angiogenesis were
promoted in the non-tumorous tissues of the early recurrence
group.
Here, we investigated recurrence-related genes in
the non-tumorous tissues of HCV-positive HCC cases, and
demonstrated that GBP1 is associated with a decreased risk
of recurrence while TSC22D3 is associated with an increased
risk of recurrence. Although there are no reports that these two
genes are differentially expressed during liver carcinogenesis,
this gene pair appears to be useful as a parameter in the
assessment of risk of HCC recurrence.
In conclusion, a combination of high TSC22D3
expression and low GBP1 expression may be considered a risk
factor for the early recurrence of HCC after liver resection.
Acknowledgements
This study was supported by the Nihon
University Multidisciplinary Research Grant for 2007; the Academic
Frontier Project for 2006 Project for Private Universities; and a
matching fund subsidy from MEXT to H.N. The authors thank their
colleagues in the Department of Digestive Surgery, and Dr Masako
Mitsumata and Dr Norimichi Nemoto of the Department of Pathology,
Nihon University School of Medicine. They also thank Ms. Sawako
Matsumoto and Dr Hideyo Yasuda, Central Research Institute, Nippon
Flour Mills Co., Ltd., for their assistance.
References
|
1.
|
Ikai I, Arii S, Okazaki M, et al: Report
of the 17th Nationwide follow-up survey of primary liver cancer in
Japan. Hepatol Res. 37:676–691. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Makuuchi M, Takayama T, Kubota K, et al:
Hepatic resection for hepatocellular carcinoma – Japanese
experience. Hepatogastroenterology. 45(Suppl 3): 1267–1274.
1998.
|
|
3.
|
Imamura H, Matsuyama Y, Tanaka E, et al:
Risk factors contributing to early and late phase intrahepatic
recurrence of hepatocellular carcinoma after hepatectomy. J
Hepatol. 38:200–207. 2003. View Article : Google Scholar
|
|
4.
|
Poon RT, Fan ST, Ng IO, Lo CM, Liu CL and
Wong J: Different risk factors and prognosis for early and late
intrahepatic recurrence after resection of hepatocellular
carcinoma. Cancer. 89:500–507. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Honda M, Kaneko S, Kawai H, Sirota Y and
Kobayashi K: Differential gene expression between chronic hepatitis
B and C hepatic lesion. Gastroenterology. 120:955–966. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Okabe H, Satoh S, Kato T, et al:
Genome-wide analysis of gene expression in human hepatocellular
carcinomas using cDNA microarray: identification of genes involved
in viral carcinogenesis and tumor progression. Cancer Res.
61:2129–2137. 2001.
|
|
7.
|
Iizuka N, Oka M, Yamada-Okabe H, et al:
Differential gene expression in distinct virologic types of
hepatocellular carcinoma: association with liver cirrhosis.
Oncogene. 22:3007–3014. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Sobin LH and Wittekind C: TNM
Classification of Malignant Tumours (UICC). 6th edition.
Wiley-Liss; New York: pp. 81–83. 2002
|
|
9.
|
Hosida Y, Villanueva A, Kobayashi M, et
al: Gene expression in fixed tissues and outcome in hepatocellular
carcinoma. N Engl J Med. 359:2045–2047. 2008.PubMed/NCBI
|
|
10.
|
Tsuchiya M, Parker JS, Kono H, Matsuda M,
Fujii H and Rusyn I: Gene expression in nontumoral liver tissue and
recurrence-free survival in hepatitis C virus-positive
hepatocellular carcinoma. Mol Cancer. 9:742010. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Ko S, Kanehiro H, Hisanaga M, Nagao M,
Ikeda N and Nakajima Y: Liver fibrosis increases the risk of
intrahepatic recurrence after hepatectomy for hepatocellular
carcinoma. Br J Surg. 89:57–62. 2002. View Article : Google Scholar
|
|
12.
|
Wakai T, Shirai Y, Yokoyama N, Nagakura S
and Hatakeyama K: Hepatitis virus status affects the pattern of
intrahepatic recurrence after resection for hepatocellular
carcinoma. Eur J Surg Oncol. 29:266–271. 2003. View Article : Google Scholar
|
|
13.
|
Shah SA, Cleary SP, Wei AC, et al:
Recurrence after liver resection for hepatocellular carcinoma: risk
factors, treatment, and outcomes. Surgery. 141:330–339. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Regimbeau JM, Abdalla EK, Vauthey JN, et
al: Risk factors for early death due to recurrence after liver
resection for hepatocellular carcinoma: results of a multicenter
study. J Surg Oncol. 85:36–41. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Ijichi M, Takayama T, Matsumura M,
Shiratori Y, Omata M and Makuuchi M: α-fetoprotein mRNA in the
circulation as a predictor of postsurgical recurrence of
hepatocellular carcinoma: a prospective study. Hepatology.
35:853–860. 2002.
|
|
16.
|
Chen JY, Chau GY, Lui WY, Tsay SH, King KL
and Wu CW: Clinicopathologic features and factors related to
survival of patients with small hepatocellular carcinoma after
hepatic resection. World J Surg. 27:294–298. 2003. View Article : Google Scholar
|
|
17.
|
Shirabe K, Kanematsu T, Matsumata T,
Adachi E, Akazawa K and Sugimachi K: Factors linked to early
recurrence of small hepatocellular carcinoma after hepatectomy:
univariate and multivariate analyses. Hepatology. 14:802–805. 1991.
View Article : Google Scholar
|
|
18.
|
Guenzi E, Töpolt K, Lubeseder-Martellato
C, et al: The guanylate binding protein-1 GTPase controls the
invasive and angiogenic capability of endothelial cells through
inhibition of MMP-1 expression. EMBO J. 15:3772–3782. 2003.
View Article : Google Scholar
|
|
19.
|
Guenzi E, Töpolt K, Cornali E, et al: The
helical domain of GBP-1 mediates the inhibition of endothelial cell
proliferation by inflammatory cytokines. EMBO J. 20:5568–5577.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Llovet JM, Chen Y, Wurmbach E, et al: A
molecular signature to discriminate dysplastic nodules from early
hepatocellular carcinoma in HCV cirrhosis. Gastroenterology.
131:1758–1767. 2006. View Article : Google Scholar
|
|
21.
|
Smith MW, Yue ZN, Geiss GK, et al:
Identification of novel tumor markers in hepatitis C
virus-associated hepatocellular carcinoma. Cancer Res. 63:859–864.
2003.PubMed/NCBI
|
|
22.
|
Itsui Y, Sakamoto N, Kurosaki M, et al:
Expressional screening of interferon-stimulated genes for antiviral
activity against hepatitis C virus replication. J Viral Hepat.
13:690–700. 2006. View Article : Google Scholar
|
|
23.
|
Naschberger E, Croner RS, Merkel S, et al:
Angiostatic immune reaction in colorectal carcinoma: impact on
survival and perspectives for antiangiogenic therapy. Int J Cancer.
123:2120–2129. 2008. View Article : Google Scholar
|
|
24.
|
Redjimi N, Gaudin F, Touboul C, et al:
Identification of glucocorticoid-induced leucine zipper as a key
regulator of tumor cell proliferation in epithelial ovarian cancer.
Mol Cancer. 8:832009. View Article : Google Scholar
|
|
25.
|
Wu XZ, Xie GR and Chen D: Hypoxia and
hepatocellular carcinoma: the therapeutic target for hepatocellular
carcinoma. J Gastroen Hepatol. 22:1178–1182. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Yasui M, Yamamoto H, Ngan CY, et al:
Antisense to cyclin D1 inhibits vascular endothelial growth factor
– stimulated growth of vascular endothelial cells: implication of
tumor vascularization. Clin Cancer Res. 12:4720–4729. 2006.
|