Introduction
Originally discovered in the early to mid-1990's,
members of the CCN family are 30- to 40-kDa proteins that are
extremely cysteine-rich (10% by mass) (1). They are a group of secreted proteins
that specifically associate with the extracellular matrix. They are
comprised of cysteine-rich 61 (Cyr61/CCN1), connective tissue
growth factor (CTGF/CCN2) and nephroblastoma overexpressed
(NOV/CCN3). More recently, the Wnt-induced secreted proteins
(WISPs) have also been found to belong to the CCN family. Thus,
WISP-1 has been named CCN4, WISP-2 is CCN5 and WISP-3 is CCN6
(1–3). Initially it was believed that these
proteins were classical growth factors. However, following
extensive research we now appreciate that, although CCN proteins
indeed have some independent activity, they principally modify the
signalling of other molecules (1).
Their function is diverse – they stimulate mitosis, adhesion,
apoptosis, extracellular matrix (ECM) production, growth arrest and
migration, and regulate angiogenesis, tumour growth, placentation,
implantation, embryogenesis and endochondral ossification (1,2).
Target cells include fibroblasts, epithelial cells, endothelial
cells, smooth muscle cells and neuronal cells. CCN proteins have a
multi-modular mosaic structure, and the diversity in their function
is thought to be possibly due to the structural heterogeneity in
this modular configuration (1,2).
Despite the increased understanding of CCN proteins
in various biological functions, such as angiogenesis, bone
remodelling, cancer and endocrine pathways, to our knowledge their
effect on human wounds has not previously been investigated.
The aim of the present study was to assess the
expression of CCN family members Cyr61, CTGF and NOV in various
wounds, and to investigate whether correlations exist between the
CCN family and the nature of wound healing.
Materials and methods
Materials
Goat anti-human Cyr61, CTGF and NOV antibodies were
purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). RNA
extraction and RT kits were obtained from BioRad (Hemel Hemstead,
UK). PCR primers were designed using Beacon Designer (Biosoft
International, Palo Alto, CA, USA) and synthesized by Sigma Genesis
(Poole, Dorset, UK). Molecular biology-grade agarose and the DNA
ladder were obtained from Invitrogen. Mastermix for the routine and
quantitative PCR was from Sigma and BioRad, respectively.
Skin biopsies
Fresh frozen skin biopsies were retrieved from the
departmental tissue bank and processed as previously reported
(4,5). These samples were collected with the
approval of the South East Wales Local Research Ethics Committee
and stored in accordance to the Human Tissue Act (www.hta.gov.uk). Samples used included normal skin,
acute wound tissue and chronic wound tissue.
Normal skin
In order to obtain a comparison with wound tissue,
unwounded skin was analysed. Unwounded skin samples were obtained
from 10 healthy staff members in our department. Under local
anaesthesia, a 3-mm punch biopsy was extracted from the inner
region of the upper arm.
Acute wound tissue
Single punch biopsies were obtained from 10 patients
with acute wounds after undergoing excision of pilonidal disease.
These wounds were judged to be clinically non-infected by a medical
wound healing expert from a specialist wound healing unit. These
biopsies were obtained from the wound margin incorporating the
epidermis and dermis at the wound edge within 6 weeks from the
surgical excision.
Chronic wound tissue
Seventeen patients with chronic venous leg ulcers
(present for >6 months) showing no sign of healing for a
duration of 6 weeks were biopsied. Duplex ultrasonography was used
to confirm venous disease. These wounds were judged to be
clinically non-infected by a medical wound healing expert from a
specialist wound healing unit. Again biopsies were obtained from
the wound margin incorporating the epidermis and dermis at the
wound edge.
Immunohistochemical staining
Immunohistochemical staining was performed using a
standard technique employed in our laboratories (3,4). The
tissues were frozen-sectioned using a Leica cryostat, at an 8-μm
thickness. The sections were mounted on Super Frost Plus microscope
slides, air dried and then fixed in acetone (Fisher Scientific
Ltd., Loughborough, UK) for 15 min. Excess acetone was removed by
air drying the sections for 10 min before being washed three times
in Tris-buffered saline (TBS) for 5 min each time. Endogenous
peroxidase was blocked by further treating the slides in the
blocking solution (5 ml of 30% H2O2 in 300 ml
pure ethanol) for 15 min. Following rehydration, sections were
incubated at room temperature with normal blocking solution which
contained horse serum (Dako Ltd., High Wycombe, UK). Excess
blocking serum was removed, and the sections were probed with the
working dilution of primary antibody (produced in 1% TBS/BSA,
dilution 1:150).
Antibody localisation was subsequently identified
using a standard streptavidin-biotin peroxidase technique using the
Vector Elite ABC kit (Vector Laboratories). This involved
incubation with a biotinylated secondary antibody for 30 min,
followed by incubation for an additional 30 min with the
avidin-biotin complex reagent provided in the kit. Diaminobenzidine
(DAB) (0.005%) was then added to the sections, which were incubated
in the dark for 5 min. The sections were then rinsed in TBS,
followed by tap water and then counterstained with Ehrlrich's
haematoxylin solution (BDH-Merck, Poole, UK) for 30 sec and then
washed again in tap water for 5 min. The sections were then
dehydrated through a graded series of alcohol solutions (BDH-Merck)
and mounted in DPX medium (BDH-Merck) under a coverslip.
Positive staining was noted as a brown deposit, and
non-stained cells as blue counterstained nucleated cells with no
associated brown DAB stain. Images were obtained using a digital
camera. Slides were evaluated by two independent researchers.
RNA extraction and complimentary DNA
(cDNA) synthesis
Individual biopsies were rapidly thawed and
homogenised using the Ultra-Turrax T8 (IKA Labortechnik, Staufen,
Germany) in RNA extraction buffer (AbGene). Total cellular RNA was
quantified using a spectrophotometer (WPA UV 1101; Biotech
Photometer, Cambridge, UK). Reverse transcriptase was performed
using 0.5 μg of the RNA sample using oligo-dT primer according to
the manufacturer's instructions and a reverse transcription kit
(Sigma, Poole, Dorset, UK). cDNA was prepared by heating the
samples at 47°C for 60 min, followed by incubation at 75°C for 10
min to inactivate any reverse transcriptase.
Quantitative analysis of CCN family
members
The transcript levels of the CCN family members from
the prepared cDNA were determined using a real-time quantitative
PCR, based on the Ampliflour™ technology to determine the
expression levels of Cyr61, CTGF, NOV and GAPDH transcripts in
acute and chronic wound tissue and in normal skin. The primers used
are listed in Table I.
 | Table I.PCR primers used. |
Table I.
PCR primers used.
| Cyr61 |
5′-GGGCTGGAATGCAACTTC-3′ |
|
5′-AACTGAACCTGACCGTACACGTTTTGGTAGATTCTGGAG-3′ |
| (spanning the third
intron; GenBank accession no. AF307860) |
| CTGF |
5′-GAGTGGGTGTGTGACGAG-3′ |
|
5′-ACTGAACCTGACCGTACAGGCAGTTGGCTCTAATCATA-3′ |
| (spanning the fourth
intron; NM_001901) |
| NOV |
5′-CTGTGAACAAGAGCCAGAG-3′ |
|
5′-ACTGAACCTGACCGTACACTTGAACTGCAGGTGGAT-3′ |
| (spanning positions
848-849; NM-002514) |
| GAPDH |
5′-CTGAGTACGTCGTGGAGTC-3′ |
|
5′-ACTGAACCTGACCGTACACAGAGATGACCCTTTTG-3′ |
The reaction was carried out using conditions as
previously reported (6,7): Hot-Start Q-Mastermix (Abgene), 10
pmol of specific forward primer, 10 pmol of reverse primer which
contains the Z sequence, 100 pmol of 6 carboxyfluorescein
(FAM)-tagged probe (Integren), and cDNA from ∼50 ng RNA (calculated
from the initial RNA in the reverse transcriptase reaction). The
reaction was carried out using IcyclerIQ™ (BioRad) which was
equipped with an optic unit that allowed real-time detection of 96
reactions using the following conditions: 94°C for 12 min, 50
cycles at 94°C for 15 sec, 55°C for 40 sec and 72°C for 20 sec. The
levels of the transcripts were generated from an internal standard
(8) that was simultaneously
amplified with the samples.
Results
Protein expression of CCN family members
in acute and chronic wound tissues and normal skin tissue
The staining pattern of CCN family members was
evaluated according to the spatial distribution. Subdivisions of
skin adjacent to the wound edge (WE) and 2 mm from the wound edge
moving towards normal skin were analysed in acute and chronic
wounds to allow comparison of the dermis at intervals from the
wound bed.
Cyr61
In normal skin, Cyr61 staining was found to be most
intense in the dermal layer of the skin. In acute wounds this
staining of the dermal layer of the skin was unaltered. However, in
chronic wounds the staining in the dermal layer was increased,
i.e., was more intensely stained possibly suggesting increased
levels of Cyr61 (Fig. 1A).
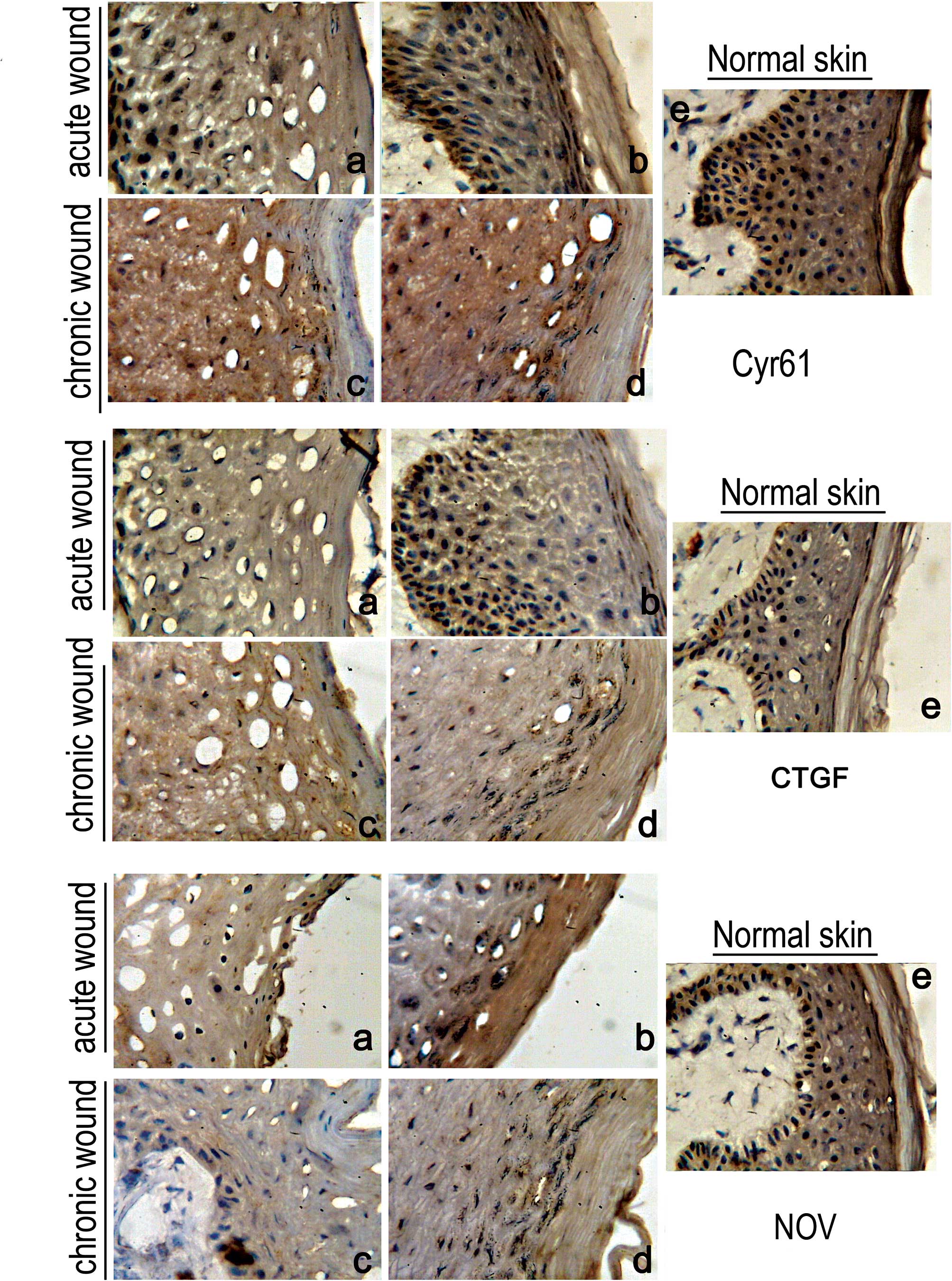 | Figure 1.Immunohistochemical staining of
Cyr61/CCN1, CTGF/CCN2 and NOV/CCN3. (A) Cyr61 staining: a, acute
wound - wound edge; b, acute wound - 2 mm from wound edge in
direction towards normal skin; c, chronic wound - wound edge; d,
chronic wound - 2 mm from wound edge in direction towards normal
skin; e, normal skin. (B) CTGF/CCN2 staining: a, acute wound -
wound edge; b, acute wound - 2 mm from wound edge in direction
towards normal skin; c, chronic wound - wound edge; d, chronic
wound - 2 mm from wound edge in direction towards normal skin; e,
normal skin. (C) NOV/CCN3 staining: a, acute wound - wound edge; b,
acute wound - 2 mm from wound edge in direction towards normal
skin; c, chronic wound - wound edge; d, chronic wound - 2 mm from
wound edge in direction towards normal skin; e, normal skin. |
CTGF
In normal skin, CTGF staining was found to be most
intense in the dermal layer of the skin. However, in comparison to
Cyr61, the level of staining was weaker. In acute wounds, the
staining of the dermal layer was again weaker and even weaker in
chronic wounds possibly suggesting the loss of CTGF (Fig. 1B).
NOV
In normal skin, NOV staining was found to be most
intense in the dermal layer of the skin. However, similar to CTGF,
the staining in comparison to Cyr61 was weaker. In acute and
chronic wounds, staining of the dermal layer of the skin was
unaltered, possibly suggesting no change in NOV expression in the
wounded states (Fig. 1C).
Analysis of CCN mRNA in normal skin,
acute and chronic wound tissues
Both conventional RT-PCR and quantitative real-time
RT-PCR were used to analyze the presence and quantity of messages
of Cyr61/CCN1, CTGF/CCN2 and NOV/CCN3. Comparisons were made
between acute and chronic wound tissues.
Over 70% of the chronic wound tissues showed a high
level of Cyr61 compared to 40% of the acute wounds. The difference
was nonetheless not statistically significant (p=0.13) (Table II). The most significant finding
was the levels of CTGF/CCN2 in chronic wounds. All (14/14) of the
chronic tissues were found to exhibit low levels of the CTGF/CCN2
transcript, compared to only 60% of acute wounds (p=0.0016;
Table III). A trend similar to
CTGF/CCN2 was observed for NOV/CCN3; a higher percentage of chronic
wound tissues had low levels of the NOV transcript, although this
did not reach statistical significance (p=0.075; Table IV).
 | Table II.Expression of Cyr61/CCN1 transcript in
wound tissues. |
Table II.
Expression of Cyr61/CCN1 transcript in
wound tissues.
| Normal skin
n=10 | Acute wound
tissue
n=10 | Chronic wound
tissue
n=14 |
|---|
| High | 4 | 4 | 10 |
| Low | 6 | 6 | 4 |
 | Table III.Expression of CTGF/CCN2 transcript in
wound tissues. |
Table III.
Expression of CTGF/CCN2 transcript in
wound tissues.
| Normal skin
n=10 | Acute wound
tissue
n=10 | Chronic wound
tissue
n=14 |
|---|
| High | 7 | 6 | 0 |
| Low | 3 | 4 | 14 |
 | Table IV.Expression of NOV/CCN3 transcript in
wound tissues. |
Table IV.
Expression of NOV/CCN3 transcript in
wound tissues.
| Normal skin
n=10 | Acute wound
tissue
n=10 | Chronic wound
tissue
n=14 |
|---|
| High | 1 | 5 | 2 |
| Low | 9 | 5 | 12 |
Discussion
Non-healing wounds cause great distress to both
patients and health professionals. It is estimated that chronic
wounds cost the NHS £2.3–3.1 billion/year in the UK (9). Therefore, identifying factors which
cause wounds to become chronic may result in the development of
possible future therapeutic targets. The present study demonstrated
a differential expression of Cyr61/CCN1, CTGF/CCN2 and NOV/CCN3 in
acute wound tissue, chronic wound tissue and normal skin
particularly for CTGF/CCN2.
The most important finding in the present study was
the significant change in CTGF/CCN2 expression in chronic wounds.
Virtually all of the tissues showed low levels of CTGF/CCN2
expression. CTGF/CCN2, also known as insulin-like growth factor
binding protein-8 (IGFBP8), was initially identified from
endothelial cells and is thought to play a role in connective
tissue biology (10). Similar to
other IGFBPs, CTGF/CCN2 is known to be involved in the action of
IGF (10). Cyr61/CCN1, CTGF/CCN2
and NOV/CCN3 were also found to be induced during tissue repair in
adults; however, no comparisons of their expression levels have
been evaluated in acute and chronic wound tissues. Elevated levels
of CTGF/CCN2 were also found to be a hallmark of fibrosis (2,10).
It has been suggested that an inappropriate overexpression of
CTGF/CCN2 creates an environment allowing other stimuli to induce
potent fibrotic responses (9). It
has also been postulated that CTGF/CCN2 is required for maximal
adhesive signalling in fibroblasts undergoing active tissue
remodelling, such as in embryogenesis, fibrotic cells or tumour
cells (10). Therefore, it would
follow that underexpression of CTGF/CCN2 creates an environment
that impedes wound healing and lack of adhesive signalling in
fibroblasts leading to chronicity of wounds as suggested in the
present study. This important finding further suggests a
therapeutic role of CTGF in chronic wound healing.
Notably, NOV/CCN3 is also present at low levels in
chronic wound tissues. Initially discovered from
myeloblastosis-associated virus-induced nephroblastoma, the
molecule also shares some homology with the IGFBP family (11). Although the functions of NOV/CCN3
are not clear, it has been suggested that the molecule is a keen
regulator of haematopoietic stem cells and is associated with
tumourigenicity in animal models (12). The result from the present study,
although they did not reach statistical significance (due largely
to a small sample size), points to a potential role of NOV in
abnormal wound healing. This finding warrants further
investigation. Although Cyr61/CCN1 has been shown to be strongly
linked to cell migration, matrix adhesion and angiogenesis
(1,2), one reason to examine this family
further is that the present study failed to show such a connection.
In contrast to our initial hypothesis that this factor may be
reduced in chronic wounds, and in clear contrast to CTGF/CCN2 and
NOV/CCN3, Cyr61/CCN1 appeared to be present at marginally higher
levels in chronic wounds. Although this must be further confirmed
in a larger study, it does indicate that Cyr61/CCN1 may have a
differential role in the healing process.
In conclusion, the present study demonstrated that
CTGF/CCN2 and NOV/CCN3, to a smaller extent, are aberrantly
expressed in wound tissues and are linked to the outcome of
clinical wound healing. The findings from the present study
indicate that these molecules are potential therapeutic targets in
chronic wound healing.
Acknowledgements
The authors wish to thank Dr Kevin
Conway for his assistance in the processing of the tissues.
References
|
1.
|
Brigstock DR: The CCN family: a new
stimulus package. J Endocrinol. 178:169–175. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Schutze N, Noth U, Schneidereit J,
Hendrich C and Jakob F: Differential expression of CCN-family
members in primary human bone marrow-derived mesenchymal stem cells
during osteogenic, chondrogenic and adipogenic differentiation.
Cell Commun Signal. 3:52005. View Article : Google Scholar
|
|
3.
|
Jiang WG, Watkins G, Fodstad O,
Douglas-Jones A, Mokbel K and Mansel RE: Differential expression of
the CCN family memebers Cyr61, CTGF and Nov in human breast cancer.
Endoc Relat Cancer. 11:781–791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Conway K, Ruge F, Price P, Harding KG and
Jiang WG: Hepatocyte growth factor regulation: an integral part of
why wounds become chronic. Wound Repair Reg. 15:683–692. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Jiang WG and Harding KG: Enhancement of
wound tissue expansion and angiogenesis by matrix-embedded
fibroblast (dermagraft), a role of hepatocyte growth factor/scatter
factor. Int J Mol Med. 2:203–210. 1998.PubMed/NCBI
|
|
6.
|
Parr C and Jiang WG: Quantitative analysis
of lymphangiogenic markers in human colorectal cancer. Int J Oncol.
23:533–539. 2003.PubMed/NCBI
|
|
7.
|
Jiang WG, Ye L, Patel G and Harding KG:
Expression of WAVEs, the WASP (Wiskott-Aldrich syndrome protein)
family of verprolin homologous proteins in human wound tissues and
the biological influence on human keratinocytes. Wound Repair Reg.
8:594–604. 2011.
|
|
8.
|
Jiang WG, Douglas-Jones A and Mansel RE:
Levels of expression of lipoxygenases and cyclooxygenase-2 in human
breast cancer. Prostaglandin, Leukot Essent Fatty Acids.
69:275–281. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Posnett J and Franks PJ: The costs of skin
breakdown and ulceration in the UK. Skin Breakdown: The Silent
Epidemic. Smith & Nephew Foundation; Hull: pp. 6–12. 2007
|
|
10.
|
Kim HS, Nagalla SR, Oh Y, Wilson E,
Roberts CT and Rosenfeld RG: Identification of a family of
low-affinity insulin-like growth factor binding proteins (IGFBPs):
characterization of connective tissue growth factor as a member of
the IGFBP superfamily. Proc Nat Acad Sci USA. 94:12981–12986. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Doghman M, Arhatte M, Thibout H, et al:
Nephroblastoma overexpressed/cysteine-rich protein 61/connective
tissue growth factor/nephroblastoma overexpressed gene-3
(NOV/CCN3), a selective adrenocortical cell preapoptotic factor, is
down-regulated in childhood adrenocortical tumors. J Clin Endocr
Metab. 92:3253–3260. 2007. View Article : Google Scholar
|
|
12.
|
Bork P: The modular architecture of a new
family of growth regulators related to connective tissue growth
factor. FEBS Lett. 327:125–130. 1993. View Article : Google Scholar : PubMed/NCBI
|















