Introduction
Thyroid cancer is the most common endocrine
malignancy, accounting for approximately 1% of all cancers. It is
the most rapidly increasing cancer among women and the second most
rapidly increasing cancer among men. Histologically, thyroid cancer
is classified as papillary thyroid cancer (PTC), follicular thyroid
cancer, medullary thyroid cancer and undifferentiated or anaplastic
thyroid cancer. Among them, PTC is the most common type and
accounts for 85–90% of all thyroid malignancies (1,2).
Psammoma bodies (PBs), very fine calcifications, are
a well-known characteristic of PTC and have diagnostic value
(3,4). Although the molecular mechanisms of
calcification in these non-osseous tissues are poorly
characterized, collagen production by the tumor cells plays an
important role in the formation of PBs in PTC, ovarian cancer and
meningioma (5–7). Indeed, Tsuchida et al
(7) reported that round bodies
with concentric laminations that were frequently detected in
meningiomas were composed mainly of collagen fibers that emerged
from the surrounding tumor cells, suggesting that they could be
precursors of PBs.
Collagen type XI α1 (COL11A1) is a minor fibril
component of cartilage (8). COL11A
is expressed mainly in articular cartilage and controls collagen
type II (9,10). It functions in skeletal
morphogenesis, fibrillogenesis, chondrocyte maturation and bone
mineralization (11,12). In addition, COL11A1 was found to be
expressed at a low level in a wide variety of normal adult human
tissues, including lung tissue, with the highest expression
detected in the parotid gland (13). Little is known about the function
of COL11A1 aside from its demonstrated importance in regulating the
assembly, organization and development of cartilage. However,
importantly, previous studies have found overexpression of the
COL11A1 gene in various types of cancers, such as non-small cell
lung (NSCLC), ovarian and oral cavity and colorectal cancers
(13–16). In particular, overexpression of the
COL11A1 gene was found to be correlated with invasion and
metastasis of these cancers (13–16).
In addition, the association of COL11A1 gene polymorphisms with
esophageal squamous cell carcinoma through genome-wide analysis of
chromosomal alterations has been reported (17). These studies implicated the COL11A1
gene as a candidate marker of these cancers.
For PTC, several risk factors, such as ionizing
radiation and nodular disease of the thyroid, have been
established. Genetic influence is also an established risk factor
for PTC (18). The familial risk
for PTC is 3 and 6 when a parent and a sibling are diagnosed with
thyroid cancers, respectively (19). In addition, recent studies have
implicated variants on 1p12, 8q24, 9q22.33 and in the pre-miR146a
at 5q33 in the disease (20–23).
However, information concerning the possible variants that affect
the risk of PTC is limited. Given the important role of collagen in
the formation of PBs and the involvement of COL11A1 in various
types of cancers, we speculated that COL11A1 may play a role as a
candidate gene having an association with PTC. In this study, we
investigated whether COL11A1 gene polymorphisms are associated with
susceptibility to PTC in a Korean population.
Materials and methods
Subjects
Ninety-eight PTC patients [mean age ± standard
deviation (SD) 52.8±12.2 years; male/female 29/69] and 366 control
subjects (59.9±10.6 years; male/female 158/208) were enrolled in
this study. PTC diagnoses and the presence of cervical regional
lymph node metastasis were confirmed by pathological examination.
The specimens that were diagnosed as follicular variants, diffuse
sclerosing and tall cell variants were excluded. None of the
controls were diagnosed with cancer or thyroid disease at the time
of enrollment. All PTC patients and control subjects were recruited
at the Kyung Hee Medical Center, Seoul, Korea, and were of Korean
background. Written informed consent was obtained from each
subject. The study was approved by the Ethics Review Committee of
the Medical Research Institute, Kyung Hee University Medical
Center, Seoul, Korea.
Patient subgroups
To determine the nature of the relationship between
COL11A1 gene polymorphisms and the clinicopathological
characteristics of PTC, the patients were divided into subgroups
according to the size of the cancers (<1 and ≥1 cm), the number
of cancers (unifocality and multifocality) and the location of the
cancers (one lobe and both lobes). In addition, the PTC patients
were also subgrouped into extrathyroidal invasion (+) and (−)
groups based on pathological findings. Finally, the PTC patients
were further subgrouped into lymph node metastasis (+) and (−)
groups to evaluate the contribution of COL11A1 gene polymorphisms
to cancer metastasis. Demographic characteristics of the PTC
patients are summarized in Table
I; small differences in subgroup numbers were caused by loss of
certain clinical data.
 | Table IClinical characteristics of the study
population. |
Table I
Clinical characteristics of the study
population.
| PTC (n=98) | Control (n=366) |
|---|
| Age (years) | | |
| Mean ± SD | 52.8±12.2 | 59.9±10.6 |
| Gender (n) | | |
| Male/female | 29/69 | 158/208 |
| Cancer size (n) | | |
| <1 cm | 49 | |
| ≥1 cm | 47 | |
| No. of cancer
(n) | | |
| Unifocality | 64 | |
| Multifocality | 31 | |
| Location of cancer
(n) | | |
| One lobe | 65 | |
| Both lobes | 30 | |
| Extrathyroidal
invasion (n) | | |
| Absent | 46 | |
| Present | 49 | |
| Cervical lymph node
metastasis (n) | | |
| Absent | 64 | |
| Present | 27 | |
| Angiolymphatic
invasion (n) | | |
| Absent | 88 | |
| Present | 5 | |
SNP selection and genotyping
Coding region single nucleotide polymorphisms
(cSNPs) for the COL11A1 gene were selected from the SNP database
(dbSNP; www.ncbi.nlm.nih.gov/SNP, dbSNP BUILD
129) of the National Center of Biotechnology Information. The cSNPs
without genotype frequency data and with a heterozygosity ≤0.05 or
a minor allele frequency ≤0.05 in Han Chinese and Japanese
populations were excluded. The cSNPs with various genotypes >3
were also excluded. Finally, four cSNPs were selected from COL11A1:
rs12731843 (Lys276Asn), rs3753841 (Pro1335Leu), rs1763347
(Gly1516Gly) and rs2229783 (Ile1602Ile). DNA was isolated from a
peripheral blood sample using the DNA isolation kit for blood
(Roche, Indianapolis, IN, USA). SNP genotyping was conducted with
direct sequencing using the following primers for each SNP:
rs12731843 (sense, 5′-TTTCACTTTTGCTTAGCCTTCC-3′; antisense,
5′-AAGGTGATCCCAAATGTATGGA-3′), rs3753841 (sense,
5′-CAGCAGGTTTTGTCATGACTTT-3′; antisense,
5′-GGATTTTCCTGCATTTGCAATT-3′), rs1763347 (sense,
5′-CACCATGGAAAAATGTTTAAGC-3′; antisense,
5′-CCTTGAGGACCCTACAAAATGC-3′) and rs2229783 (sense,
5′-CCTTTACCAATCTTGTCCTCCA-3′; antisense,
5′-TAGAATGAATGAGCTGCCAATG-3′). The PCR products were sequenced
using an ABI PRISM 3730XL analyzer (PE Applied Biosystems, Foster
City, CA, USA). Sequence data were analyzed using SeqManII software
(DNAStar, Madison, WI, USA).
Statistical analysis
Continuous variables are presented as the means ± SD
and were analyzed by the independent t-test and Chi-square test
using SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). For the
analysis of the genetic data, SNPAnalyzer Pro (Istech, Goyang,
Korea) and SNPStats (http://bioinfo.icon-cologia.net/index.php) were used.
The associations between genotypes of cSNPs and PTC, as well as any
associations between cSNPs and the PTC subgroups, were estimated by
computing the odds ratios (ORs) and their 95% confidence intervals
(CIs) with logistic regression analyses, controlling for age and
gender as covariables. In logistic regression analysis for each
cSNP, models assuming either codominant inheritance (that is, the
relative hazard differed between subjects with one minor allele and
those with two minor alleles), dominant inheritance (that is,
subjects with one or two minor alleles had the same relative hazard
for the disease), or recessive inheritance (that is, only subjects
with two minor alleles were at increased risk of the disease) were
used. Linkage disequilibrium (LD) was tested using Haploview
version 4.2 (Broad Institute, Cambridge, MA, USA). The LD block was
constructed using Gabriel's method (24). The association of SNPs and
haplotypes was analyzed using HapAnalyzer version 1.0 (http://hap.ngri.go.kr/). To avoid chance findings due
to multiple testing, the Bonferroni correction was applied by
lowering the significance levels to p=0.013 (p=0.05/4) for the four
SNPs.
Results
Four cSNPs of the COL11A1 gene were polymorphic, and
the genotype distributions of the cSNPs were in Hardy-Weinberg
equilibrium in the control group (p>0.05; data not shown). The
power of the sample size was calculated to verify the data using a
genetic power calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/cc2.html). In
the present study, the sample powers of the SNPs were 0.872
(rs12731843, number of effective sample for 80% power = 81), 0.863
(rs3753841, number of effective sample for 80% power = 83), 0.868
(rs1763347, number of effective sample for 80% power = 82) and
0.862 (rs4753426, number of effective sample for 80% power = 83),
respectively (α=0.05, genotype relative risk 2-fold). We therefore
have statistical confidence in our results.
As shown in Table
II, rs1763347 and rs2229783 of the COL11A1 gene showed a
significant association with PTC. Association of rs1763347 with PTC
was detected in the dominant model (CT/TT vs. CC; p=0.0042,
OR=0.50, 95% CI 0.31–0.81). Frequencies of the genotype containing
the T allele (CT and TT genotypes) were lower in the PTC patients
(37.8%) compared to the control subjects (52.2%). Allele frequency
analysis also revealed an association between the T allele and a
reduced risk of PTC (p=0.010, OR=0.61, 95% CI 0.42–0.89).
Frequencies of the C and T alleles were 69.7 and 30.3% in the
control subjects, and 79.1 and 20.9% in the PTC patients,
respectively. This result indicated that the T allele was
associated with a reduced risk of PTC. As well, rs2229783 was
associated with PTC in the codominant (TT vs. CC; p=0.017, OR=0.30,
95% CI 0.13–0.73), dominant (p=0.014, OR=0.55, 95% CI 0.35–0.89)
and recessive models (p=0.013, OR=0.38, 95% CI 0.16–0.87). However,
the significance of the association did not remain after Bonferroni
correction. In the allele analysis, a significant association was
demonstrated (p=0.007, OR=0.62, 95% CI 0.44–0.88). The frequency of
the T allele was lower in the PTC patients (28.1%) than in the
control subjects (38.5%). This result indicated that the T allele
contributed to a decreased risk of PTC. No association of
rs12731843 and rs3753841 with PTC was detected.
 | Table IIFrequencies of the genotypes and
alleles of COL11A1 polymorphisms in control subjects and patients
with PTC. |
Table II
Frequencies of the genotypes and
alleles of COL11A1 polymorphisms in control subjects and patients
with PTC.
| SNP | Model/allele | Type | Control n (%) | PTC n (%) | OR (95% CI) | p-value |
|---|
| rs12731843 | Codominant | A/A | 311 (85.0) | 87 (88.8) | Ref | |
| Lys276Asn | | A/C | 52 (14.2) | 11 (11.2) | 0.78 (0.38–1.61) | 0.430 |
| C/C | 3 (0.8) | 0 (0.0) | 0.00 (0.00-NA) | |
| Dominant | AA | 311 (85.0) | 87 (88.8) | Ref | |
| AC/CC | 55 (15.0) | 11 (11.2) | 0.71 (0.35–1.46) | 0.340 |
| Recessive | AA/AC | 363 (99.0) | 98 (100.0) | Ref | |
| CC | 3 (0.8) | 0 (0.0) | 0.00 (0.00-NA) | 0.130 |
| Allele | A | 674 (92.1) | 185 (94.4) | Ref | |
| C | 58 (7.9) | 11 (5.6) | 0.69 (0.36–1.34) | 0.280 |
| rs3753841 | Codominant | T/T | 158 (43.2) | 53 (54.1) | Ref | |
| Pro1335Leu | | T/C | 165 (45.1) | 37 (37.7) | 0.69 (0.42–1.13) | 0.100 |
| C/C | 43 (11.7) | 8 (8.2) | 0.45 (0.19–1.04) | 0.160 |
| Dominant | TT | 158 (43.2) | 53 (54.1) | Ref | |
| TC/CC | 208 (56.8) | 45 (45.9) | 0.63
(0.39–1.00) | 0.050 |
| Recessive | TT/TC | 323 (88.2) | 90 (91.8) | Ref | |
| CC | 43 (11.8) | 8 (8.2) | 0.53
(0.23–1.20) | 0.110 |
| Allele | T | 481 (65.7) | 143 (73.0) | Ref | |
| C | 251 (34.3) | 53 (27.0) | 0.71
(0.50–1.01) | 0.060 |
| rs1763347 | Codominant | C/C | 175 (47.8) | 61 (62.2) | Ref | |
| Gly1516Gly | | C/T | 160 (43.7) | 33 (33.7) | 0.52
(0.32–0.86) | 0.030 |
| T/T | 31 (8.5) | 4 (4.1) | 0.38
(0.12–1.14) | 0.070 |
| Dominant | CC | 175 (47.8) | 61 (62.2) | Ref | |
| CT/TT | 191 (52.2) | 37 (37.8) | 0.50
(0.31–0.81) | 0.0042 |
| Recessive | CC/CT | 335 (91.5) | 94 (95.9) | Ref | |
| TT | 31 (8.5) | 4 (4.1) | 0.49
(0.17–1.47) | 0.170 |
| Allele | C | 510 (69.7) | 155 (79.1) | Ref | |
| T | 222 (30.3) | 41 (20.9) | 0.61
(0.42–0.89) | 0.010 |
| rs2229783 | Codominant | CC | 139 (38.0) | 50 (51.0) | Ref | |
| Ile1602Ile | | CT | 172 (47.0) | 41 (41.8) | 0.65
(0.39–1.06) | 0.090 |
| TT | 55 (15.0) | 7 (7.2) | 0.30
(0.13–0.73) | 0.017 |
| Dominant | CC | 139 (38.0) | 50 (51.0) | Ref | |
| CT/TT | 227 (62.0) | 48 (49.0) | 0.55
(0.35–0.89) | 0.014 |
| Recessive | CC/CT | 311 (85.0) | 91 (92.9) | Ref | |
| TT | 55 (15.0) | 7 (7.1) | 0.38
(0.16–0.87) | 0.013 |
| Allele | C | 450 (61.5) | 141 (71.9) | Ref | |
| T | 282 (38.5) | 55 (28.1) | 0.62
(0.44–0.88) | 0.007 |
We performed a further analysis according to the
clinical symptoms of PTC, including the size of the cancers (<1
and ≥1 cm), the number of cancers (unifocality and multifocality),
location of the cancers (one lobe and both lobes), invasion (+/−)
and lymph node metastasis (+/−). In the analysis according to
cancer size of PTC, a weak association of rs1763347 was revealed
with PTC patients with cancer size of ≥1 cm in the codominant (CT
vs. CC; p=0.044, OR=0.39, 95% CI 0.16–0.97) and dominant models
(CT/TT vs. CC; p=0.046, OR=0.42, 95% CI 0.18–1.00). In the
codominant model, the frequencies of the CC, CT and TT genotypes
were 53.1, 42.9 and 4.1%, respectively, in the PTC patients with
cancer size of <1 cm, and 72.3, 23.4 and 4.3%, respectively, in
the PTC patients with cancer size of ≥1 cm (data not shown).
However, this significance did not remain after the Bonferroni
correction.
In the analysis according to other symptoms, we were
not able to find any association with polymorphisms of COL11A1
(data not shown).
Four SNPs of the COL11A1 gene were analyzed for LD
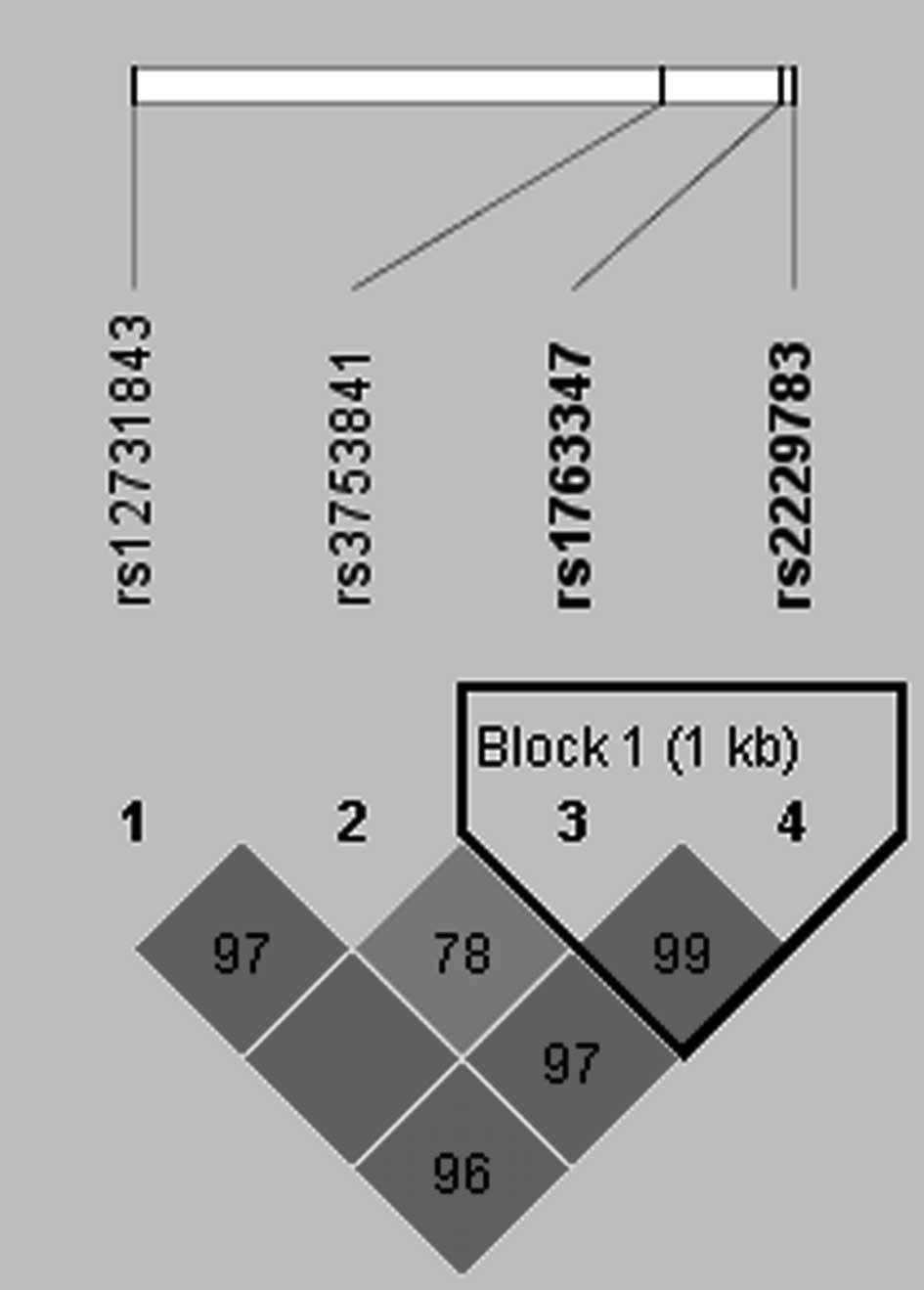
and haplotypes using Haploview 4.2. As shown in Fig. 1, LD block was constructed between
rs1763347 and rs2229783 (D'≥0.95, r2≥0.8). Thus, we
analyzed the association of haplotypes consisting of rs1763347 and
rs2229783 with PTC. Haplotype analysis revealed that the CC
haplotype was associated with PTC in the codominant (p=0.011,
OR=1.56, 95% CI 1.11–2.21) and recessive models (p=0.020, OR=1.70,
95% CI 1.09–2.66) (Table III). The
TT haplotype was also associated with PTC in the codominant model
(p=0.006, OR=0.58, 95% CI 0.39–0.85). The frequency of the CC
haplotype was higher in the PTC patients (0.71) compared to the
control subjects (0.61), whereas that of the TT haplotype was lower
(0.20 in PTC patients and 0.30 in control subjects). This result
indicated that the CC and TT haplotypes may be related to the risk
of PTC.
 | Table IIIHaplotype analysis of COL11A1 gene
polymorphisms in PTC patients and control subjects. |
Table III
Haplotype analysis of COL11A1 gene
polymorphisms in PTC patients and control subjects.
| Haplotype | Type | Control
| PTC
| Model | OR (95% CI) | p-value |
|---|
| Freq | n | % | Freq | n | % |
|---|
| CC | H/H | 0.61 | 139 | 38.0 | 0.71 | 50 | 51.0 | Codominant | 1.56
(1.11–2.21) | 0.011 |
| H/− | | 172 | 47.0 | | 40 | 40.8 | Dominant | 1.99
(0.91–4.33) | 0.080 |
| −/− | | 55 | 15.0 | | 8 | 8.2 | Recessive | 1.70
(1.09–2.66) | 0.020 |
| TT | H/H | 0.30 | 31 | 8.5 | 0.20 | 3 | 3.1 | Codominant | 0.58
(0.39–0.85) | 0.006 |
| H/− | | 160 | 43.7 | | 34 | 34.7 | Dominant | 0.56
(0.35–0.88) | 0.080 |
| −/− | | 175 | 47.8 | | 61 | 62.2 | Recessive | 0.34
(0.10–1.14) | 0.600 |
| CT | H/H | 0.08 | 3 | 0.8 | 0.08 | 0 | 0.0 | Codominant | 0.93
(0.51–1.67) | 0.800 |
| H/− | | 54 | 17.8 | | 15 | 15.3 | Dominant | 0.98
(0.53–1.82) | 0.950 |
| −/− | | 309 | 84.4 | | 83 | 84.7 | Recessive | 0.00 (0.00-NA) | 1.000 |
Discussion
The results of the present study revealed an
association between the COL11A1 gene and PTC in a Korean
population. The rs1763347 and rs2229783 polymorphisms of the
COL11A1 gene were associated with PTC, but not rs12731843 and
rs3753841. In particular, the T allele of rs1763347 and rs2229783
was associated with a decreased risk of PTC. Moreover, the CC and
TT haplotypes consisting of rs1763347 and rs2229783 were related to
PTC.
Notably, in previous studies, overexpression of the
COL11A1 gene was demonstrated in various types of cancers, such as
NSCLC, ovarian, oral cavity and colorectal cancers. COL11A1 was
proposed as a marker for NSCLC (13,14).
Additionally, Chong et al (14) found that overexpression of COL11A1
was correlated with pathological stage, lymph node metastasis and
poor prognosis of NSCLC. Moreover, overexpression of the COL11A1
gene was correlated with invasion and metastasis of ovarian, oral
cavity and colorectal cancers (15,16).
Considering its overexpression in various types of cancers and,
moreover, the contribution of collagen in the formation of PBs in
PTC (5–7), we postulate that the COL11A1 level
may be increased in PTC similar to other cancers.
In a previous study, the T allele of rs1676486 (C/T)
of the COL11A1 gene exhibited a significant association with an
increased risk of lumbar disc herniation (LDH) in a Japanese
population (25). In addition, the
mRNA expression level of COL11A1, which was decreased according to
the severity of degeneration in patients with LDH, was lower in the
intervertebral discs of patients with minor allele T allele than
with major allele C allele (25).
These authors also showed that the decrease in the expression of
the T allele resulted from transcription degradation induced in the
T allele (25). The rs1676486 with
rs1763347 was in strong LD (r2≥0.8) in the HapMap
database for Chinese and Japanese population (http://www.hapmap.org/; genome build 36). Thus,
rs1763347 as well as rs1676486 may also influence the expression of
the COL11A1 gene, and minor allele T alleles of rs1763347 may
decrease the level of COL11A1. Moreover, rs1763347 and rs2229783
were in strong LD block in our study (r2≥0.8). Thus, T
alleles of both rs1763347 and rs2229783 may cause a decrease in
COL11A1 expression.
In our data, T alleles of rs1763347 and rs2229783
were associated with a reduced risk of PTC. The CC and TT
haplotypes comprised of these SNPs also showed a significant
association with PTC with decreased frequency of the TT haplotype
in PTC patients. Additionally, rs1763347 was associated with cancer
size, and T allele frequencies of rs1763347 were lower in PTC
patients with a cancer size of ≥1 cm than in those with a cancer
size of <1 cm, although the significance did not remain after
Bonferroni correction. While to our knowledge there has not been
any report concerning the expression of COL11A1 in PTC, we
speculate that the T alleles of both rs1763347 and rs2229783 may
induce a relative decrease in COL11A1 expression in PTC compared to
the C allele, and this decrease may be associated with a reduced
risk of PTC. Further study is required to assess whether the
expression of COL11A1 is up-regulated in PTC, and whether the
expression level is related to alleles of rs1763347 and rs2229783
of the COL11A1 gene. In addition, due to the relatively small
number of subjects, our findings are considered preliminary and
need to be validated in further studies using larger sample sizes
of these subpopulations.
In conclusion, rs1763347 and rs2229783 polymorphisms
of the COL11A1 gene are significantly associated with PTC. In
particular, the T alleles of rs2229783 and rs1763347 may be
implicated as a protective factor against PTC. These findings
provide evidence that the COL11A1 gene may play a role in the
pathophysiology of PTC.
Acknowledgements
This study was supported by a grant
from the Kyung Hee University in 2011 (KHU-20110061).
References
|
1
|
Hundahl SA, Fleming ID, Fremgen AM and
Menck HR: A National Cancer Data Base report on 53,856 cases of
thyroid carcinoma treated in the U.S., 1985–1995. Cancer.
83:2638–2648. 1998.PubMed/NCBI
|
|
2
|
Dohán O, Baloch Z, Bánrévi Z, Livolsi V
and Carrasco N: Rapid communication: predominant intracellular
overexpression of the Na+/I− symporter (NIS)
in a large sampling of thyroid cancer cases. J Clin Endocrinol
Metab. 86:2697–2700. 2001.PubMed/NCBI
|
|
3
|
Carcangiu MI, Zampi G, Pupi A, Castognoli
A and Rosai J: Papillary carcinoma of the thyroid: a
clinicopathologic study of 241 cases treated at the University of
Florence, Italy. Cancer. 55:805–828. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johannessen JV and Sobrinho-Simões M: The
origin and significance of thyroid psammoma bodies. Lab Invest.
43:287–296. 1980.PubMed/NCBI
|
|
5
|
Endo T, Ohta K and Kobayashi T: Expression
and function of Cbfa-1/Runx2 in thyroid papillary carcinoma cells.
J Clin Endocrinol Metab. 93:2409–2412. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kiyozuka Y, Nakagawa H, Senzaki H, et al:
Bone morphogenetic protein-2 and type IV collagen expression in
psammoma body-forming ovarian cancer. Anticancer Res. 21:1723–1730.
2001.PubMed/NCBI
|
|
7
|
Tsuchida T, Matsumoto M, Schirayama Y,
Kasai H and Kawamoto K: Observation of psammoma bodies in cultural
meningiomas: analysis of three-dimensional structure using scanning
and transmission electron microscopy. Ultrastruct Pathol.
20:241–247. 1996. View Article : Google Scholar
|
|
8
|
Morris NP and Bächinger HP: Type XI
collagen is a heterotrimer with the composition (1α, 2α, 3α)
retaining non-triple-helical domains. J Biol Chem. 262:11345–11350.
1987.
|
|
9
|
So CL, Kaluarachchi K, Tam PP and Cheah
KS: Impact of mutations of cartilage matrix genes on matrix
structure, gene activity and chondrogenesis. Osteoarthritis
Cartilage. 9:S160–S173. 2001.PubMed/NCBI
|
|
10
|
Yoshioka H, Greenwel P, Inoguchi K, et al:
Structural and functional analysis of the promoter of the human α1
(XI) collagen gene. J Biol Chem. 270:418–424. 1995.
|
|
11
|
Li Y, Lacerda DA, Warman ML, et al: A
fibrillar collagen gene, Col11a1, is essential for skeletal
morphogenesis. Cell. 80:423–430. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Seegmiller R, Fraser FC and Sheldon H: A
new chondrodystrophic mutant in mice. Electron microscopy of normal
and abnormal chondrogenesis. J Cell Biol. 48:580–593. 1971.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang KK, Liu N, Radulovich N, et al: Novel
candidate tumor marker genes for lung adenocarcinoma. Oncogene.
21:7598–7604. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chong IW, Chang MY, Chang HC, et al: Great
potential of a panel of multiple hMTH1, SPD, ITGA11 and COL11A1
markers for diagnosis of patients with non-small cell lung cancer.
Oncol Rep. 16:981–988. 2006.PubMed/NCBI
|
|
15
|
Kim H, Watkinson J, Varadan V and
Anastassiou D: Multi-cancer computational analysis reveals
invasion-associated variant of desmoplastic reaction involving
INHBA, THBS2 and COL11A1. BMC Med Genomics. 3:512010. View Article : Google Scholar
|
|
16
|
Schmalbach CE, Chepeha DB, Giordano TJ, et
al: Molecular profiling and the identification of genes associated
with meta-static oral cavity/pharynx squamous cell carcinoma. Arch
Otolaryngol Head Neck Surg. 130:295–302. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chattopadhyay I, Singh A, Phukan R, et al:
Genome-wide analysis of chromosomal alterations in patients with
esophageal squamous cell carcinoma exposed to tobacco and betel
quid from high-risk area in India. Mutat Res. 696:130–138. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kondo T, Ezzat S and Asa SL: Pathogenetic
mechanisms in thyroid follicular-cell neoplasia. Nat Rev Cancer.
6:292–306. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zatelli MC, Trasforini G, Leoni S, et al:
BRAF V600E mutation analysis increases diagnostic accuracy for
papillary thyroid carcinoma in fine-needle aspiration biopsies. Eur
J Endocrinol. 161:467–473. 2009. View Article : Google Scholar
|
|
20
|
Gudmundsson J, Sulem P, Gudbjartsson DF,
et al: Common variants on 9q22.33 and 14q13.3 predispose to thyroid
cancer in European populations. Nat Genet. 41:460–464. 2009.
View Article : Google Scholar
|
|
21
|
Baida A, Akdi M, González-Flores E,
Galofré P, Marcos R and Velázquez A: Strong association of
chromosome 1p12 loci with thyroid cancer susceptibility. Cancer
Epidemiol Biomarkers Prev. 17:1499–1504. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jazdzewski K, Murray EL, Franssila K,
Jarzab B, Schoenberg DR and de la Chapelle A: Common SNP in
pre-miR-146a decreases mature miR expression and predisposes to
papillary thyroid carcinoma. Proc Natl Acad Sci USA. 105:7269–7274.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He H, Nagy R, Liyanarachchi S, et al: A
susceptibility locus for papillary thyroid carcinoma on chromosome
8q24. Cancer Res. 69:625–631. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gabriel SB, Schaffner SF, Nguyen H, et al:
The structure of haplotype blocks in the human genome. Science.
296:2225–2229. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mio F, Chiba K, Hirose Y, et al: A
functional polymorphism in COL11A1, which encodes the α1 chain of
type XI collagen, is associated with susceptibility to lumbar disc
herniation. Am J Hum Genet. 81:1271–1277. 2007.PubMed/NCBI
|