Introduction
There are 300,000 deaths and 400,000 new cases of
gastric cancer in China every year (1). Gastric cancer (GC) is the fourth most
common type of cancer worldwide. Although there has been a steady
decline in the risk of GC incidence and mortality over several
decades in most countries, it is the second most common cause of
cancer-related death (700,000 deaths annually) (2). The current staging system for GC is
inadequate for predicting the outcome of treatment.
The poor prognosis of patients with GC has been
attributed mostly to metastases and relapse (3,4).
Tumor cells utilize many cellular or biochemical mechanisms to
complete metastatic spread, and proteolytic enzymes play a key role
in the metastatic stage (5).
Mammalian proteolytic enzymes are divided into five classes
(aspartic, cysteine, metallo, serine and threonine), and the serine
protease (SP) family is the largest (6). It is widely accepted that SPs degrade
extracellular matrix and facilitate neoplastic progression. The
urokinase plasminogen activator, one of the SPs, promotes tumor
cell invasion (7) and the
kallikrein prostate-specific antigen predicts prognosis of prostate
cancer (8). KLK6 was found to be
significantly up-regulated in tissues and sera from patients with
colon cancer and was closely associated with a poor prognosis,
suggesting that KLK6 may be used as a potential biomarker and a
therapeutic target for colon cancer (9). SP inhibitors (serpins) are the
largest family of protease inhibitors identified to date. Most
serpins act as inhibitors of chymotrypsin-like SPs, others as
cross-class serine protease inhibitors of papain-like cysteine
proteases and caspases (10–13).
Certain serpins have been identified to be involved in the
progression of malignant tumor; these include SERPINB3 and SERPINB4
in squamous cell carcinoma (14,15),
SERPINB5 in human breast cancer (16), SERPINB13 in head and neck squamous
cell carcinoma (17), SERPINH1 in
head and neck carcinomas (18),
SERPINI2 in breast cancer (19),
SERPINB2 and SERPINE1 in breast cancer and other cancers (20), and SERPINB2, SERPINB3, SERPINB4,
SERPINB7, SERPINB11, SERPINB12 and SERPINB13 in oral squamous cell
carcinoma (21).
Yet, which SPs and their inhibitors are associated
with the progression of GC globally? Minimal research has been
carried out concerning this issue. In the present study, we focused
on SPs and serpins, the largest protease and protease inhibitor
families, since they carry out diverse functions, including
clotting, fibrinolytic cascades, complement activation,
fertilization, pro-hormone conversion, apoptosis, extracellular
matrix maintenance and remodeling, angiogenesis and tumor cell
invasion (22,23). We aimed to compare the difference
in gene expression profiles between cancerous and non-cancerous
gastric tissues by cDNA microarrays, and to identify the SPs and
serpins associated with the progression of GC. The results from the
microarrays were confirmed by real-time PCR and
immunohistochemistry (IHC). A survival prediction model was further
developed using the IHC data.
Materials and methods
Patients and clinical features
Human GC tissues and their paired normal gastric
tissues were obtained with informed consent from 140 patients who
underwent radical resection for GC in 2000 and 2003 at the
Department of Surgery, Ruijin Hospital, Shanghai, China. The
corresponding non-cancerous gastric tissue was obtained at least 6
cm from the tumor. The diagnoses were confirmed by histopathology.
Stage and grade were established using the tumor node metastases
(TNM) and World Health Organization classification systems. For
stage IIIB and stage IV by clinical staging, pre-operative
chemotherapy was offered. All of the patients, except stage IA of
surgical pathologic staging, received fluoropyrimidine-based
chemoradiotherapy. All patients were followed up systematically.
This follow-up included a complete history and physical examination
every 4 months for 3 years, then annually thereafter, complete
blood count, platelets, multichannel serum chemistry analysis and
other investigations (such as endoscopy and other radiologic
studies). Of these 140 patients, 100 were enrolled in 2000 and
included in the IHC analyses; 40 were enrolled in 2003 and
initially included in the microarray analyses. In total, 10 of the
40 patients in 2003 were later included in the real-time PCR
analyses. In addition to the 10 patients, half of the 40 patients
were involved in the IHC analyses.
The specimens were snap-frozen in liquid nitrogen
immediately after resection and then stored at −80°C until use or
fixed in 10% formalin and paraffin embedded by conventional
techniques for IHC staining. The Ethics Committee of Ruijin
Hospital approved the use of these tissues for research
purposes.
RNA preparation
Total RNAs were extracted from each tissue sample
using TRIzol reagent (Life Technologies, Grand Island, NY, USA) and
further purified with an RNeasy kit (Qiagen, Valencia, CA, USA)
according to the manufacturer's instructions. The quality of the
total RNA samples was determined by electrophoresis through
formaldehyde agarose gels, and the 18S and 28S RNA bands were
visualized under ultraviolet light.
Microarray assays and statistical
analysis
The cDNA microarray used in the present study
consisted of 13,824 genes/ESTs; this microarray was the same as
that used by Huang et al (24) and constructed by the National
Engineering Center for Biochips at Shanghai (Shanghai, China). The
microarray and the experimental procedure were confirmed to be
feasible by previous studies (24,25).
Thus, it was used instead of a new customized microarray containing
SP and serpin genes only. The microarray contained 50 common SPs
and serpin genes.
Total RNA (100 μg) was used to prepare
fluorescent dye-labeled first-strand cDNA, using SuperScript™ II
RNase H Reverse Transcriptase according to the relevant protocol
(Invitrogen, Carlsbad, CA, USA). Cy3-dCTP and Cy5-dCTP (Amersham,
Piscataway, NJ, USA) were used to label the GC specimens and the
corresponding non-cancerous gastric specimen, respectively. The
dye-labeled probes were purified by QIAquick Nucleotide Removal kit
(Qiagen) according to the specified protocol. A total of 30 pmol of
each probe was used in the two-color array hybridization. The
hybridization and scanning procedures were identical as those
described previously (24).
To avoid systematic error for each microarray
sample, a space and intensity-dependent normalization based on a
LOWESS program was employed (26)
to normalize the logarithm transformed background corrected signal
intensities for the dual-channel data. Thereafter, normalized data
were withdrawn for each SP and serpin gene for each specimen. The
difference in C (GC specimen) and N (corresponding non-cancerous
gastric specimen) was estimated by the ln C-ln N and T test
performed using SAS to compare the difference between the GC
specimen and its corresponding non-cancerous gastric specimen
transcript. All ratios were filtered using a P-value of ≤0.05.
Real-time PCR
Real-time PCR reactions were performed according to
a previously reported protocol (27). The primer sequences were as
follows: sense 5′-GTTCCAGACATTCT CGCTTC-3′ and anti-sense
5′-ATAGTAGCCTGAGCAT GTGC-3′ for SERPINB5 (107 bp); sense
5′-CCTGCTCCA GCATCACTATC-3′ and anti-sense 5′-GGTCCAGTCCAGC
ACATATC-3′ for KLK10 (93 bp); sense 5′-AGCTCTCCAGC
CTCATCATC-3′ and anti-sense 5′-CAACAGCCTTCTTCTG CATC-3′ for
SERPINH1 (120 bp); sense 5′-CAGAAGTGTGA GAACGCCTAC-3′ and
anti-sense 5′-CCTTGAAGAGA CTGGTTACAG-3′ for KLK11 (131 bp);
sense 5′-GTCATCTC CGTGTGTGATTG-3′ and anti-sense 5′-TCATAGCGAA
GGCTGACTTG-3′ for HPN (149 bp); sense 5′-AACGCCAG
ACTTCTATCCTC-3′ and anti-sense 5′-CAACAATAAGGC CAGTCAGG-3′ for
SPINK1 (102 bp); sense 5′-GATGAAGAA GAGAGTCGAGG-3′ and
anti-sense 5′-GAAGAAGATGTT CTGGCTGG-3′ for SERPINA5 (124
bp); sense 5′-CAAGGAAG CCTATGAGGTCAAG-3′ and anti-sense
5′-TGAGTTGGA GGAGTGCAAT-3′ for PRSS8 (146 bp); sense
5′-AAGCAC TGTGCATCACCTTG-3′ and anti-sense 5′-CAGAGTTGG
AGCACTTGCTG-3′ for TMPRSS2 (102 bp); sense 5′-GGA
CCTGACCTGCCGTCTAG-3′ and anti-sense 5′-GTAGCC CAGGATGCCCTTGA-3′ for
GAPDH (100 bp).
The results of the real-time PCR are presented as Ct
values. The relative changes in gene expression were calculated by
the ΔΔCt method (28).
Immunohistochemistry
IHC was performed according to our previously
reported protocol (29). The
primary antibodies used in the present study were Hepsin polyclonal
antibody (Abcam, Cambridge, MA, USA), anti-human Kallikrein 11
antibody (R&D Systems, Minneapolis, MN, USA), goat polyclonal
antibody against KLK10 (Santa Cruz, Santa Cruz, CA USA) and mouse
monoclonal antibody to SERPINB5 (Novocastra, Newcastle-upon-Tyne,
UK). Negative control slides were treated without the primary
antibody under equivalent conditions. For the secondary developing
reagents, EnVision™ System labeled Polymer-HRP/M/R (DakoCytomation,
Glostrup, Denmark) and UltraSensitive™ S-P (Goat) kit (Maixin Bio,
Fuzhou, China) were used. Slides were developed with
diaminobenzidine (DAB; DakoCytomation) and counterstained with
hematoxylin.
Pathologists without knowledge of the patient
outcomes scored the immunostained slides independently as
previously described (30). In
brief, a proportion (proportion of positive-staining tumor cells)
score (0, none; 1, <1/100; 2, 1/100-1/10; 3, 1/10-1/3; 4,
1/3-2/3; and 5, >2/3), and an intensity score (0, none; 1, weak;
2, intermediate; and 3, strong) were assigned. These two scores
were then added to obtain a total score for each slide.
Statistical analysis
Results of the real-time PCR were evaluated by the
Wilcoxon signed rank test. Associations between gene expression
profiles as assessed by real-time PCR and IHC were analyzed using
non-parametric Spearman rank correlation coefficients. Survival
curves were computed with the Kaplan-Meier method and were compared
using the log-rank test. A two-sided Fisher's exact test was used
in univariate analysis of potential prognostic factors regarding
overall survival. Stepwise regression analysis and the best subset
regression were used to develop a prediction model of survival. A
P-value <0.05 was taken as the level of significance.
Statistical analyses were performed using the SAS 6.12 software
(SAS Inc., Cary, NC, USA) or GraphPad Prism version 5.00 for
Windows (GraphPad Software, San Diego, CA, USA).
Results
Selection and confirmation of the serine
protease-related genes for the prediction model
Nine serpins or SP genes were determined to be
differentially expressed by the microarray experiments. The gene
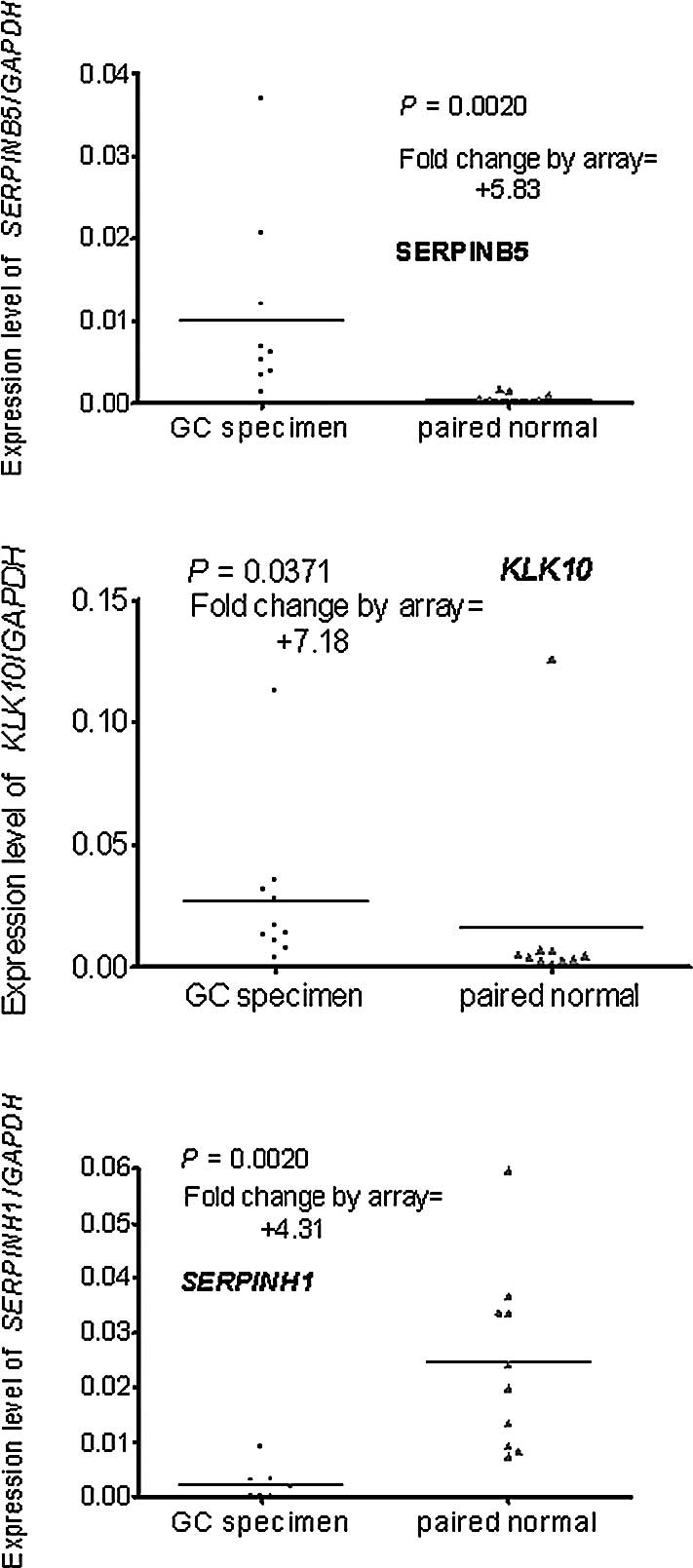
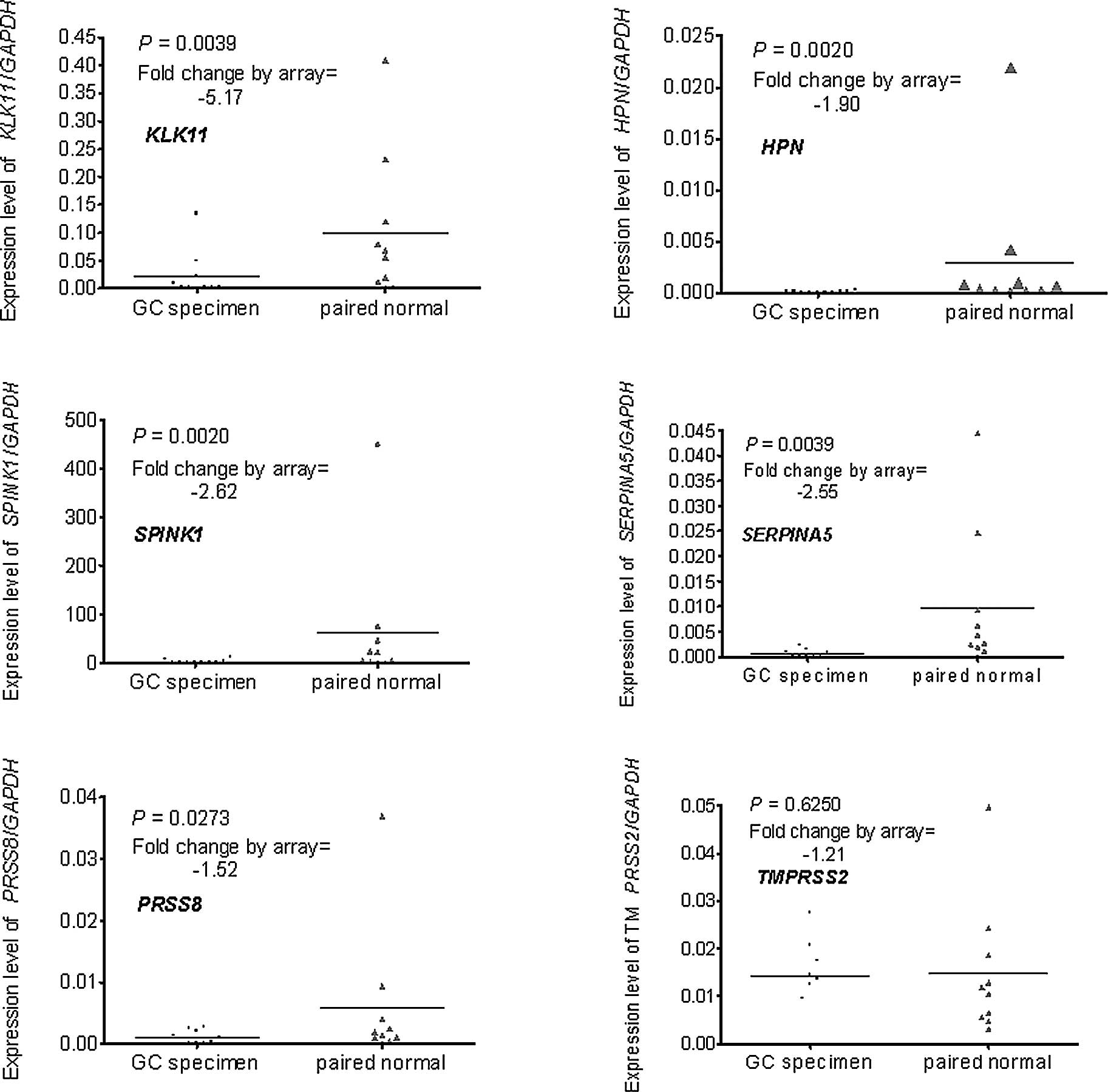
list and fold changes are shown in Table I; it includes three up-regulated
and six down-regulated genes. The up-regulated genes included two
serpins (SERPINB5 and SERPINH1) and one SP
(KLK10); the six down-regulated genes included two Serpins
(SPINK1 and SERPINA5) and four SPs (KLK11,
HPN, PRSS8 and TMPRSS2).
 | Table I.Differentially expressed genes as
determined by the microarray. |
Table I.
Differentially expressed genes as
determined by the microarray.
| Gene name | GeneBank no. | Fold changea |
|---|
| Up-regulated | | |
| KLK10 | NM_002776 | 7.181442890 |
|
SERPINB5 | NM_002639 | 5.827452523 |
|
SERPINH1 | NM_001235 | 4.313578371 |
| Down-regulated | | |
| TMPRSS2 | NM_005656 | 0.823943603 |
| PRSS8 | NM_002773 | 0.659704511 |
| HPN | NM_002151 | 0.527439252 |
|
SERPINA5 | NM_000624 | 0.392424461 |
| SPINK1 | NM_003122 | 0.381989951 |
| KLK11 | NM_006853 | 0.193303929 |
Real-time PCR was used to verify the differential
expression of the genes selected by the microarray. A total of 10
pairs of RNA stock used in the real-time PCR were the same as those
in the microarray, the remaining 30 had no remaining RNA or the RNA
was of inadequate quality. The results of the real-time PCR were
compared to those of the microarray. One of the three up-regulated
genes (SERPINH1) was excluded as it was down-regulated in
the real-time PCR assay (Fig.
1A–C). One of the six down-regulated genes (TMPRSS2) was
excluded as the difference did not reach statistical significance
(P=0.6250) (Fig. 2A–F).
Therefore, seven genes were further analyzed. Four
genes (SERPINB5, KLK10, KLK11 and HPN),
which exhibited the highest differential fold change and for which
a primary antibody for IHC was commercially available, were
included in the IHC assay.
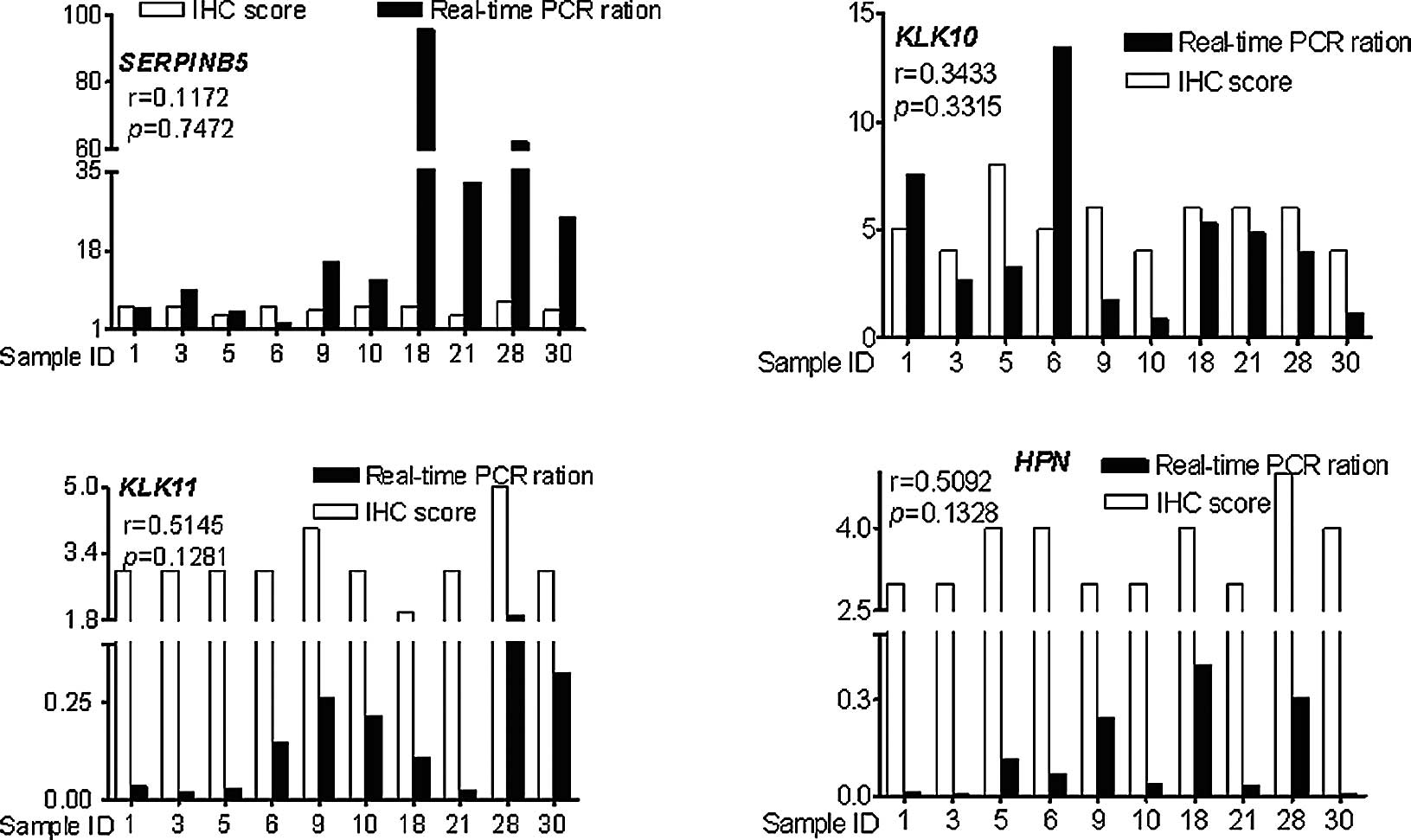
Correlation of the RNA and protein
expression profiles of SERPINB5, KLK10, KLK11 and HPN
The expression profiles of the aforementioned four
genes were assayed by real-time PCR and IHC. The RNA expression
profiles were positively correlated with the protein expression
profiles (r=0.1172 for SERPINB5, r= 0.3433 for KLK10, r= 0.5145 for
KLK11, r=0.5092 for HPN), but the correlations did not reach
statistical significance (P=0.7472 for SERPINB5, P=0.3315 for
KLK10, P=0.1281 for KLK11, P=0.1328 for HPN) (Fig. 3).
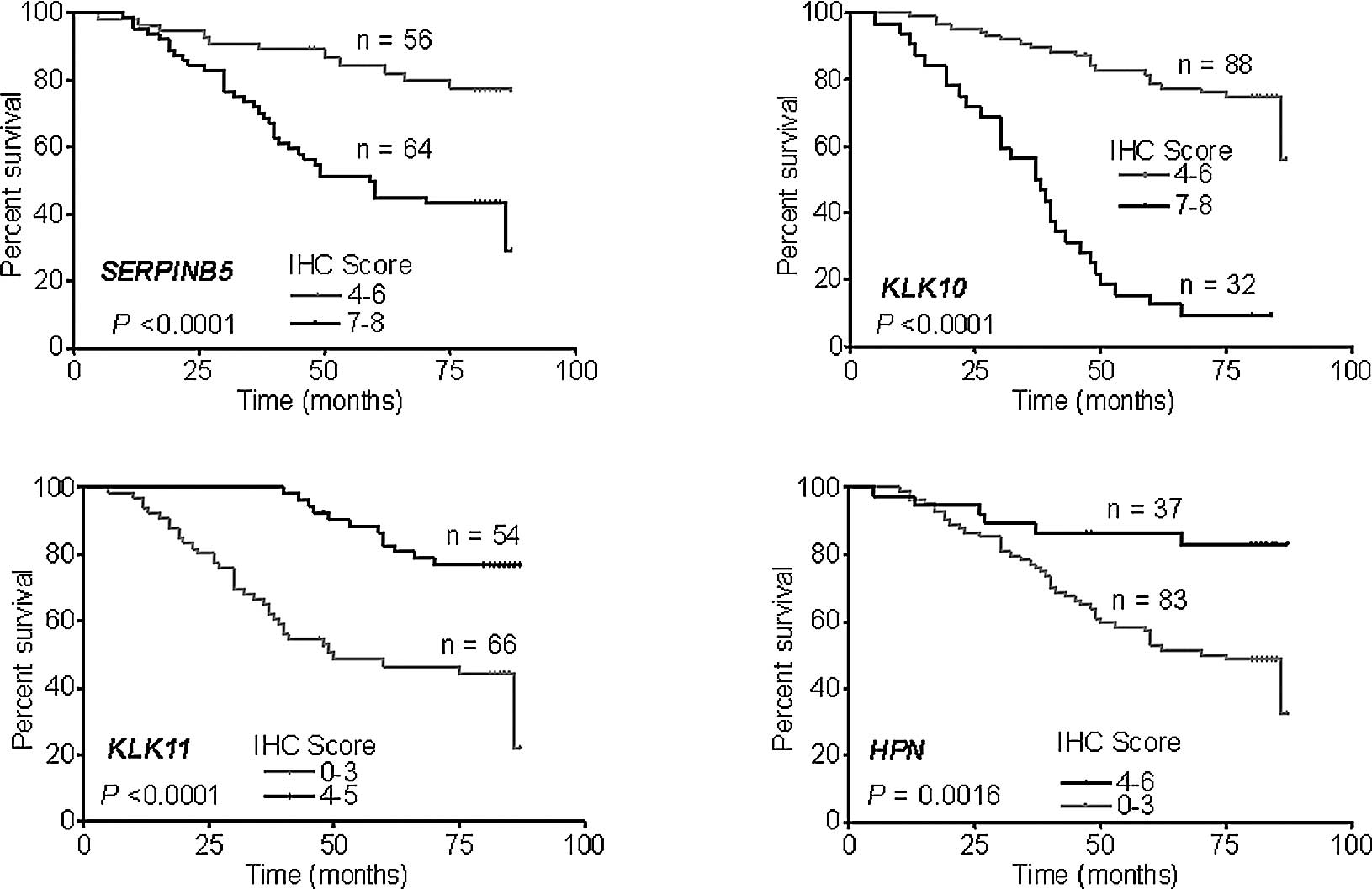
Prediction of survival using the IHC
score and clinical and pathological characteristics
IHC was performed in 120 patients, including 100
patients who underwent GC surgery in 2000 and 20 patients in 2003
who were enrolled in the microarray. An IHC score <3 was
recognized as negative, 4–6 as weakly positive and 7–8 as positive.
SERPINB5 and KLK10 were positively expressed in all of the patients
(IHC score >4), while the patients with weak positive expression
of SERPINB5 and KLK10 had a more favorable prognosis (Fig. 4A and B). KLK11 and HPN were
negatively expressed in most of the patients. The number of
patients with negative expression of KLK11 and HPN was 66 and 83,
respectively, and negative expression of these genes indicated a
worse survival (Fig. 4C and
D).
To elucidate the factors contributing to improved
survival, an analysis was carried out to identify the prognostic
factors for overall survival using univariate analyses. A total of
10 factors were considered in the analyses, including patient age
and gender, tumor size, WHO classification and differentiation of
the tumor, TNM score (e.g., score of T3N3M0 = 3+3+0 = 6), and IHC
scores for SERPINB5, KLK10, KLK11 and HPN. Tumor size, TNM score,
IHC scores for SERPINB5, KLK10, KLK11 and HPN were shown to be
indicative of patient survival (Table
II); these six factors were used in further analyses.
 | Table II.Univariate analysis of significant
prognostic factors for overall survival in the GC patients. |
Table II.
Univariate analysis of significant
prognostic factors for overall survival in the GC patients.
| Variable | Overall survival
|
|---|
| n | Events | P-value |
|---|
| Age at diagnosis
(years) | | | |
| ≤65 | 87 | 31 | 0.311 |
| >65 | 33 | 17 | |
| Gender | | | |
| Male | 77 | 31 | 0.959 |
| Female | 43 | 17 | |
| Tumor size
(cm) | | | |
| ≤5 | 86 | 26 | 0.03 |
| >5 | 34 | 22 | |
| WHO
classification | | | |
| 1 | 104 | 38 | 0.469 |
| 2 | 7 | 4 | |
| 3 | 9 | 6 | |
| TNM score | | | |
| 1 | 14 | 2 | 0.006 |
| 2 | 31 | 4 | |
| 3 | 22 | 5 | |
| 4 | 32 | 19 | |
| 5 | 16 | 13 | |
| 6 | 5 | 5 | |
|
Differentiation | | | |
| 1 | 13 | 3 | 0.612 |
| 2 | 63 | 25 | |
| 3 | 44 | 20 | |
| SERPINB5 | | | |
| Weakly
positive | 56 | 11 | 0.005 |
| Positive | 64 | 37 | |
| KLK10 | | | |
| Weakly
positive | 88 | 19 | 0.001 |
| Positive | 32 | 29 | |
| KLK11 | | | |
| Negative | 66 | 36 | 0.016 |
| Positive | 56 | 12 | |
| HPN | | | |
| Negative | 84 | 42 | 0.014 |
| Positive | 36 | 6 | |
Translation of expression of SERPINB5,
KLK10, KLK11 and HPN together with tumor size and TNM score in the
prediction model of survival
Complete data from 42 patients who underwent GC
surgery in 2000 were used to develop a prediction model of survival
by stepwise regression analysis and the best subset regression. It
identified 63 models totally using the data of expression of
SERPINB5, KLK10, KLK11 and HPN together with tumor size and TNM
score. The following was identified as the most accurate model:
Survival time (months) = 88.8607 + 2.6395 SERPINB5 − 12.0772 KLK10
+ 13.7562 KLK11 − 7.0318 TNM, where SERPINB5, KLK10 and KLK11 are
the IHC scores of these genes and TNM indicates the TNM score. This
model had the highest adjusted R2 value (0.9556) and the
least CP value (4.4748).
Survival prediction for the different
groups of GC patients
The prediction model consisting of SERPINB5, KLK10,
KLK11 and TNM was applied to the different test groups of GC
patients. This was considered to be accurately predictive, for
complete data, when the prediction value was in the range of actual
survival time ± 5 months; for censored data, the prediction value
was higher than the actual survival time. Survival of 47 patients
out of 58 patients of 2000 was correctly predicted, and yielded a
sensitivity of 0.8103. In particular, this prediction model
correctly predicted 5 death events and 14 survivals for 20 patients
of 2003 (Table III). A highly
predictive power was achieved for this prediction model for the GC
patients in the independent test group.
 | Table III.Frequency distribution of accuracy in
all of the patients of 2003 (n=20). |
Table III.
Frequency distribution of accuracy in
all of the patients of 2003 (n=20).
| Predicted
status | Clinical status
| Total |
|---|
| Death | Survival |
|---|
| Death | 5 | 0 | 5 |
| Survival | 1 | 14 | 15 |
| Total | 6 | 14 | 20 |
Discussion
Microarray technology has rapidly evolved during the
past decade. The main objectives of microarray studies are i) to
identify homogeneous subtypes of a disease on the basis of gene
expression, ii) to find genes that are differentially expressed in
tumors with different characteristics, or iii) to develop a rule on
the basis of gene expression allowing the prediction of patient
prognosis or of the response to a particular treatment (31). Using cDNA microarray, a classifier
containing 153 genes with weights was generated (32). Microarrays can also be used to
identify leukemia subtypes (33).
However, these significant results have seldom been put into
clinical application as they rely on massive data output gathered
from a relatively large number of genes, sophisticated algorithms
for analyzing the data and costly experiment platforms, let alone
the technical variance in the hybridization-based systems. Studies
have demonstrated that the combination of microarray and RT-PCR
technologies is a highly efficient and reliable approach for the
identification of clinically important diagnostic and prognostic
biomarkers, as well as for the identification of novel therapeutic
target candidates in pancreatic cancer (34). However, protein is mainly the
effective molecule in cells. We found that the correlations between
the RNA and protein expression profiles did not reach statistical
significance. In our study, the cDNA microarray was used to
identify differentially expressed genes, and a survival prediction
model of clinical applicability was developed using the IHC data of
these genes.
Nine SPs and serpin genes were found to be
differentially expressed between the GC and non-cancerous gastric
tissues after the microarray assay, but two genes were excluded
after confirmation by real-time PCR. Therefore, seven genes were
further analyzed: SERPINB5, KLK10, KLK11,
HPN, SPINK1, SERPINA5 and PRSS8. The
first four genes were included in the IHC assay. Finally, three,
SERPINB5, KLK10 and KLK11, were identified to
be involved in the most effective prediction model.
SERPINB5 (maspin, mammary serine protease
inhibitor) was identified in 1994 by subtractive hybridization
analysis of normal mammary tissue and breast cancer cell lines
(16). SERPINB5 regulates the
invasive activity of tumor cells (35), inhibits angiogenesis (36), primary tumor growth as well as
invasion and metastasis (37).
However, high expression of maspin is associated with early tumor
relapse in breast cancer (38),
and SERPINB5 may contribute to gastric carcinogenesis and have a
potential role in tumor metastasis in GC (39,40).
Our microarray data, real-time PCR and IHC assay demonstrated that
the expression of SERPINB5 was up-regulated at the RNA and protein
levels in the GC tissues. These may indicate that SerpineB5 is a
‘bad’ gene for survival. However, the final model indicated
expression of SerpineB5 to be a good predictor for survival. The
predictive effectiveness of SerpineB5 may be poor; further studies
are required to explain this discrepancy.
KLK10 and KLK11 belong to the kallikreins which are
enzymes which historically release vasoactive peptides from high
molecular weight precursors. In humans, there are two categories of
kallikreins: plasma and tissue kallikreins. Human tissue
kallikreins have attracted increased attention due to their role as
biomarkers for the screening, diagnosis, prognosis and monitoring
of various types of cancers, including those of the prostate,
ovarian, breast, testicular and lung (41). KLK10 is significantly up-regulated
in pancreatic, colon, ovarian and gastric cancer (42,43);
this is consistent with our findings concerning KLK10. KLK11 is
down-regulated in renal cell carcinoma and prostate cancer
(44,45), similar to our findings in GC. It
appears that these genes have only limited predictive power in
isolation, but we get a more efficient survival prediction model
combining the expression profiles of these genes with the TNM
status of the tumor.
The prognostic model presented here may predict the
survival of GC patients after radical resection; this may influence
the clinical treatment of GC patients by aiding in the selection of
patients for adjuvant therapy and developing meaningful clinical
trials for new regimens.
Although the genes in our model were selected by
microarray, the model is independent of the microarray platform. It
requires only IHC, which is a common assay and can be performed
easily in any laboratory or pathological department of any
hospital. Certainly, our model is developed from the data of a
relatively small population of patients and from one digestive
surgery center, therefore it may require various modifications in
future practice.
Acknowledgements
The authors would like to thank Mr Qi
Li and Mr Xi-Jian Chen from the Geminix Informatics Co., Ltd.,
Shanghai, China, for the assistance in the microarray statistical
analysis; Ms. Qu Cai from the Shanghai Institute of Digestive
Surgery for the assistance in the real-time PCR experiments; Ms.
Yue-Mei Sun from the Shanghai Institute of Digestive Surgery for
the assistance in the patient follow-up; Mr Jun Ji from the
Shanghai Institute of Digestive Surgery for the assistance in the
IHC experiments; Mr Kang Chen from the Department of Pathology,
Ruijin Hospital, for the assistance in the preparation of the
sections for the IHC experiments. This study was supported in part
by the China National ‘863’ R&D High-Tech Key Project
(2006AA02A301 and 2007AA02Z179), and the National Natural Science
Foundation of China (30772107).
References
|
1.
|
L YangIncidence and mortality of gastric
cancer in ChinaWorld J Gastroenterol1217202006
|
|
2.
|
DM ParkinF BrayJ FerlayP PisaniGlobal
cancer statistics, 2002CA Cancer J
Clin5574108200510.3322/canjclin.55.2.74
|
|
3.
|
I ChauAR NormanD CunninghamJS WatersJ
OatesPJ RossMultivariate prognostic factor analysis in locally
advanced and metastatic esophago-gastric cancer – pooled analysis
from three multicenter, randomized, controlled trials using
individual patient dataJ Clin Oncol2223952403200415197201
|
|
4.
|
P HohenbergerS GretschelGastric
cancerLancet362305315200310.1016/S0140-6736(03)13975-X
|
|
5.
|
S CurranGI MurrayMatrix metalloproteinases
in tumour invasion and metastasisJ
Pathol189300308199910.1002/(SICI)1096-9896(199911)189:3%3C300::AID-PATH456%3E3.0.CO;2-C10547590
|
|
6.
|
ND RawlingsFR MortonAJ BarrettMEROPS: the
peptidase databaseNucleic Acids
Res34D270D272200610.1093/nar/gkj08916381862
|
|
7.
|
C QuemenerEE GabisonB NaimiExtracellular
matrix metalloproteinase inducer up-regulates the urokinase-type
plasminogen activator system promoting tumor cell invasionCancer
Res67915200710.1158/0008-5472.CAN-06-244817210677
|
|
8.
|
CV ObiezuEP DiamandisHuman tissue
kallikrein gene family: applications in cancerCancer
Lett224122200510.1016/j.canlet.2004.09.02415911097
|
|
9.
|
JT KimEY SongKS ChungUp-regulation and
clinical significance of serine protease kallikrein 6 in colon
cancerCancer117112201121656738
|
|
10.
|
JC WhisstockSP BottomleyPI BirdRN PikeP
CoughlinSerpins 2005 – fun between the beta-sheets. Meeting report
based upon presentations made at the 4th International Symposium on
Serpin Structure, Function and Biology (Cairns, Australia)FEBS
J272486848732005
|
|
11.
|
JA IrvingRN PikeAM LeskJC
WhisstockPhylogeny of the serpin superfamily: implications of
patterns of amino acid conservation for structure and
functionGenome Res1018451864200010.1101/gr.GR-1478R11116082
|
|
12.
|
JA IrvingSS ShushanovRN PikeInhibitory
activity of a heterochromatin-associated serpin (MENT) against
papain-like cysteine proteinases affects chromatin structure and
blocks cell proliferationJ Biol
Chem2771319213201200210.1074/jbc.M108460200
|
|
13.
|
C SchickD BrommeAJ BartuskiY UemuraNM
SchechterGA SilvermanThe reactive site loop of the serpin SCCA1 is
essential for cysteine proteinase inhibitionProc Natl Acad Sci
USA951346513470199810.1073/pnas.95.23.134659811823
|
|
14.
|
Y SuminamiS NagashimaA MurakamiSuppression
of a squamous cell carcinoma (SCC)-related serpin, SCC antigen,
inhibits tumor growth with increased intratumor infiltration of
natural killer cellsCancer Res6117761780200111280721
|
|
15.
|
Y SuminamiS NagashimaNL VujanovicK
HirabayashiH KatoTL WhitesideInhibition of apoptosis in human
tumour cells by the tumour-associated serpin, SCC antigen-1Br J
Cancer82981989200010.1054/bjoc.1999.102810732775
|
|
16.
|
Z ZouA AnisowiczMJ HendrixMaspin, a serpin
with tumor-suppressing activity in human mammary epithelial
cellsScience263526529199410.1126/science.82909628290962
|
|
17.
|
TD ShellenbergerA MazumdarY
HendersonHeadpin: a serpin with endogenous and exogenous
suppression of angiogenesisCancer
Res651150111509200510.1158/0008-5472.CAN-05-226216357159
|
|
18.
|
GJ OllinsN NikitakisK NorrisC HerbertH
SiavashJJ SaukThe production of the endostatin precursor collagen
XVIII in head and neck carcinomas is modulated by
CBP2/Hsp47Anticancer Res2219771982200212174873
|
|
19.
|
G XiaoYE LiuR GentzSuppression of breast
cancer growth and metastasis by a serpin myoepithelium-derived
serine proteinase inhibitor expressed in the mammary myoepithelial
cellsProc Natl Acad Sci USA9637003705199910.1073/pnas.96.7.3700
|
|
20.
|
MJ DuffyC DugganThe urokinase plasminogen
activator system: a rich source of tumour markers for the
individualised management of patients with cancerClin
Biochem37541548200415234235
|
|
21.
|
M ShiibaH NomuraK ShinozukaDown-regulated
expression of SERPIN genes located on chromosome 18q21 in oral
squamous cell carcinomasOncol
Rep24241249201010.3892/or_0000085220514468
|
|
22.
|
GA SilvermanPI BirdRW CarrellThe serpins
are an expanding superfamily of structurally similar but
functionally diverse proteins. Evolution, mechanism of inhibition,
novel functions, and a revised nomenclatureJ Biol
Chem2763329333296200110.1074/jbc.R100016200
|
|
23.
|
GM YousefEP DiamandisTissue kallikreins:
new players in normal and abnormal cell growth?Thromb
Haemost90716200312876620
|
|
24.
|
J HuangHH ShengT ShenCorrelation between
genomic DNA copy number alterations and transcriptional expression
in hepatitis B virus-associated hepatocellular carcinomaFEBS
Lett58035713581200610.1016/j.febslet.2006.05.032
|
|
25.
|
XR XuJ HuangZG XuInsight into
hepatocellular carcinogenesis at transcriptome level by comparing
gene expression profiles of hepatocellular carcinoma with those of
corresponding noncancerous liverProc Natl Acad Sci
USA981508915094200110.1073/pnas.241522398
|
|
26.
|
YH YangS DudoitP LuuNormalization for cDNA
microarray data: a robust composite method addressing single and
multiple slide systematic variationNucleic Acids
Res30e15200210.1093/nar/30.4.e1511842121
|
|
27.
|
KF LeiYF WangXQ ZhuIdentification of MSRA
gene on chromosome 8p as a candidate metastasis suppressor for
human hepatitis B virus-positive hepatocellular carcinomaBMC
Cancer7172200710.1186/1471-2407-7-17217784942
|
|
28.
|
KJ LivakTD SchmittgenAnalysis of relative
gene expression data using real-time quantitative PCR and the
2(−Delta Delta C(T)) MethodMethods254024082001
|
|
29.
|
Y QuJF LiQ CaiOver-expression of FRZB in
gastric cancer cells suppresses proliferation and induces
differentiationJ Cancer Res Clin
Oncol134353364200810.1007/s00432-007-0291-017680269
|
|
30.
|
JM HarveyGM ClarkCK OsborneDC
AllredEstrogen receptor status by immunohistochemistry is superior
to the ligand-binding assay for predicting response to adjuvant
endocrine therapy in breast cancerJ Clin Oncol17147414811999
|
|
31.
|
S MichielsS KoscielnyC HillInterpretation
of microarray data in cancerBr J
Cancer9611551158200710.1038/sj.bjc.660367317342085
|
|
32.
|
QH YeLX QinM ForguesPredicting hepatitis B
virus-positive metastatic hepatocellular carcinomas using gene
expression profiling and supervised machine learningNat
Med9416423200310.1038/nm843
|
|
33.
|
EJ YeohME RossSA ShurtleffClassification,
subtype discovery, and prediction of outcome in pediatric acute
lymphoblastic leukemia by gene expression profilingCancer
Cell1133143200210.1016/S1535-6108(02)00032-612086872
|
|
34.
|
S StreitCW MichalskiM ErkanH FriessJ
KleeffConfirmation of DNA microarray-derived differentially
expressed genes in pancreatic cancer using quantitative
RT-PCRPancreatology9577582200910.1159/00021208419657213
|
|
35.
|
A DokrasLM GardnerDA KirschmannEA SeftorMJ
HendrixThe tumour suppressor gene maspin is differentially
regulated in cytotrophoblasts during human placental
developmentPlacenta23274280200210.1053/plac.2001.078411969337
|
|
36.
|
M ZhangO VolpertYH ShiN BouckMaspin is an
angiogenesis inhibitorNat Med6196199200010.1038/72303
|
|
37.
|
HY ShiW ZhangR LiangModeling human breast
cancer metastasis in mice: maspin as a paradigmHistol
Histopathol18201206200312507299
|
|
38.
|
KM JoensuuMH LeideniusLC AnderssonPS
HeikkilaHigh expression of maspin is associated with early tumor
relapse in breast cancerHum
Pathol4011431151200910.1016/j.humpath.2009.02.00619427667
|
|
39.
|
HJ SonTS SohnSY SongJH LeeJC RheeMaspin
expression in human gastric adenocarcinomaPathol
Int52508513200210.1046/j.1440-1827.2002.01381.x12366809
|
|
40.
|
M TerashimaC MaesawaK OyamaGene expression
profiles in human gastric cancer: expression of maspin correlates
with lymph node metastasisBr J
Cancer9211301136200510.1038/sj.bjc.660242915770218
|
|
41.
|
M PaliourasC BorgonoEP DiamandisHuman
tissue kallikreins: the cancer biomarker familyCancer
Lett2496179200710.1016/j.canlet.2006.12.01817275179
|
|
42.
|
GM YousefCA BorgonoC PopalisIn-silico
analysis of kallikrein gene expression in pancreatic and colon
cancersAnticancer Res244351200415015574
|
|
43.
|
GM YousefNM WhiteIP MichaelIdentification
of new splice variants and differential expression of the human
kallikrein 10 gene, a candidate cancer biomarkerTumour
Biol26227235200510.1159/00008737716103744
|
|
44.
|
CD PetrakiAK GregorakisMM
VaslamatzisPrognostic implications of the immunohistochemical
expression of human kallikreins 5, 6, 10 and 11 in renal cell
carcinomaTumour Biol2717200610.1159/00009015016340244
|
|
45.
|
X BiH HeY YeAssociation of TMPRSS2 and
KLK11 gene expression levels with clinical progression of human
prostate cancerMed
Oncol27145151201010.1007/s12032-009-9185-019242826
|