Introduction
In the last decade, the zebrafish (Danio
rerio) was identified as a new genetic system in which to
analyze hematopoietic development. Hematopoiesis in zebrafish is
similar to that in mammals and other higher vertebrates whose
representative blood cell types include the erythroid,
thrombocytic, myeloid and lymphoid lineages. In mammals, primitive
hematopoiesis occurs outside the embryo, in the blood islands of
the yolk sac. Later in development, it moves to the
aorta-gonadmesonephros (AGM) region, fetal liver and ultimately the
bone marrow. By contrast, primitive hematopoietic stem cells (HSCs)
in the zebrafish embryo are born intra-embryonically in ventral
mesoderm-derived tissue called the intermediate cell mass (ICM).
During this wave, the anterior part of the embryo generates myeloid
cells, while the posterior part generates mostly erythrocytes and a
number of myeloid cells. From 24 h post-fertilization (hpf), these
primitive blood cells start to circulate throughout the embryo.
Subsequently, the definitive HSCs emerge from the ventral wall of
the dorsal aorta and migrate to the posterior region in the tail
called the caudal hematopoietic tissue (CHT). From 3 days
post-fertilization (dpf), lymphopoiesis initiates in the thymi. By
4 dpf, HSCs seed the kidney marrow, which is equivalent to bone
marrow in mammals (1).
Myelopoiesis is the process of producing all types
of myeloid cells including granulocytes and monocytes/macrophages.
A number of zebrafish myelopoietic genes are expressed in patterns
consistent with their mammalian orthologs, including
myeloperoxidase (mpo), an enzyme that is a major component
of human neutrophil and eosinophil granules. It is also a marker
for zebrafish granulocytes (2).
Myeloid cells have a wide spectrum of activities, including immune
surveillance and tissue remodeling. Irregularities in myeloid cell
development and their function are associated with the onset and
the progression of a variety of human disorders, including leukemia
(3). An in-depth study of
mpo expression and function in zebrafish is likely to
improve our ability to identify, isolate and culture hematopoietic
cells to enhance our ability to use this simple organism to address
disease biology.
Whole-mount in situ hybridization (WISH) is
the method of choice to characterize the spatial distribution of
gene transcripts during embryonic development. Initial protocols
used non-radioactive digoxigenin (DIG)-labeled probes that permit
for the first time visualization of global gene expression patterns
in Drosophila embryos (4).
We present a modified protocol, using a DIG-labeled probe to detect
the spatio-temporal spectrum of mpo expression in zebrafish,
which reduces the number of steps and obtains signal
enhancement.
Materials and methods
Animals
The AB zebrafish strain was maintained at 28.5°C as
described by Westerfield (5).
Embryos were staged as described by Kimmel et al (6). Developmental stages refer to hpf or
dpf. This study was approved by the Institutional Animal Care and
Use Committee (IACUC) of Shanghai Research Center for Model
Organisms (Shanghai, China) with approval ID 2012-0008.
Experimental materials
Various restriction enzymes and the TOP10
Escherichia coli strain were purchased from Takara Bio Inc.
(Japan). T4-DNA ligase was purchased from Promega Corporation
(Madison, WI, USA) and DNA high fidelity polymerase KOD-Plus was
purchased from Toyobo Co., Ltd. (Japan). The
DIG-deoxyribonucleotide triphosphate (dNTP) labeling kit, blocking
reagents, anti-DIG-AP and BM purple were purchased from Roche
Diagnostics (Indianapolis, IN, USA). RNase inhibitor was purchased
from Ambion (USA), levamisole, heparin and yeast RNA were purchased
from Sigma (St. Louis, MO, USA). TRIzol reagent was purchased from
Invitrogen Life Technologies (Carlsbad, CA, USA) and the plasmid
Maxiprep kit was purchased from Qiagen (Hilden, Germany). The
first-strand cDNA Quantscript RT kit was purchased from Tiangen
Biotech Co., Ltd. (Beijing, China). A fluorescence microscope
(SMZ-1500; Nikon, Japan), ultra-violet spectrophotometer (ASP-3700;
ACT Gene, Piscataway, NJ, USA), Biometra T personal polymerase
chain reaction (PCR) amplification instrument (Goettingen,
Germany), gel imaging analysis system (Tanon 3500, Shanghai, China)
and SPX biochemical incubator (GNP-9160; Shanghai Jinghong
Laboratory Instrument Co., Ltd., China) were also used.
Cloning and mpo/pBK-CML plasmid
construction
Total RNA was extracted from 40 embryos at 48 hpf,
using TRIzol reagent according to the manufacturer’s instructions.
The cDNA was synthesized from 1 μg total RNA using the
first-strand cDNA Quantscript RT kit. The specific primers were
designed according to the zebrafish mpo (zmpo)
genomic sequence on the National Center for Biotechnology
Information (NCBI) web site (gene ID, 337514); forward primer:
5’-TTCAAGTCCAGAACCAGTGAGCCT-3’ and reverse primer:
5’-TTTAGCAGTGGCAGGAAGGATGGA-3’. The length of the amplified PCR
product was 2442 bp, including two restriction enzyme cutting sites
(Xho I at 216 bp and EcoRI at 2384 bp) and the probe
sequence (at 737 bp). The probe sequence was designed to span exon
borders of the gene. PCR was performed as follows: 95°C for 10 min;
then 35 cycles at 95°C for 30 sec, 58°C for 30 sec and 72°C for 60
sec. The integrity of the PCR product was examined by 1% agarose
gel electrophoresis. The purity was analyzed based on the
absorbance ratio at 260 and 280 nm (A260/280). Then the zmpo
fragment and the pBK-CML vector were digested with XhoI and
EcoRI enzymes and connected at the same sticky end with the
T4 ligase, which resulted in the construct of an mpo/pBK-CML
plasmid.
Plasmid linearization and probe
incubation
The mpo/pBK-CML plasmid was linearized with
the SalI restriction enzyme. DIG-antisense RNA probes were
generated by T7 in vitro transcription (1 μg
linearization template DNA, 1 μl DIG-dNTP mix, 2 μl
100 mM dithiothreitol (DTT), 4 μl 5X transcription buffer, 1
μl RNase inhibitor, 1 μl T7 RNA polymerase and
diethylpyrocarbonate (DEPC) H2O to make 20 μl,
incubated for 2 h at 37°C). Then, 1 μl DNaseI was added and
incubated at 37°C for 20 min to purify the product. The final
precipitation was stored at −20°C.
Embryo preparation
The required developmental stage of the embryos was
selected according to the somite. Embryos younger than 48 hpf were
dechorionated. The embryos were digested in 1 mg/ml pronase for
∼2–10 min. The digestion was stopped when >10% embryos were free
from their chorions. The chorions were broken with air from a pipe.
If the chorions were difficult to remove, they were manually broken
with a pair of tweezers. Embryos older than 24 hpf were decolored
in 5% hydrogen peroxide and 5% potassium peroxide fish water for 15
min. Embryos (up to 40 embryos per 1.5 ml eppendorf tube) were
fixed in fresh 4% paraformaldehyde (PFA) in phosphate-buffered
saline (PBS) and agitated overnight at 4°C. Then, the embryos were
washed twice in PBS with Tween-20 (PBST) for 5 min each and
dehydrated with gradient (25, 50, 75 and 100%) methanal/PBST for 10
min each at room temperature (RT), then stored at −20°C.
First day of hybridization
The embryos were removed from the −20°C storage and
rehydrated with gradient (75, 50, 25 and 0%) methanal/PBST. The
embryos were fixed with 4% PFA for 20 min and rinsed twice with
PBST for 5 min at RT. The embryos were digested with an appropriate
concentration of proteinase K (10 μg/ml in PBST) at 37°C for
3 min and then fixed and rinsed as above. They were then incubated
at 68°C in negative hybrid liquid (HYB−, 50% formamide
in 5X SSC buffer) for 15 min and prehybridized at 68°C in 300
μl positive hybrid liquid (HYB+, 0.5 mg/ml yeast
RNA and 50 μg/ml heparin in HYB−) for 4 h.
Following this, the HYB+ was replaced with fresh
HYB+ containing the DIG-labeled probe (concentration, ∼1
ng/μl) and incubated overnight at 68°C.
Second day of hybridization
The probe was removed and the embryos were washed
twice for 30 min each with 50% formamide in 2X SSC buffer, then for
15 min in 2X SSC buffer and twice for 30 min each in 0.2X SSC
buffer at 68°C. Then, the embryos were rinsed three times for 5 min
each at RT in MABT (100 mM maleic acid, 150 mM NaCl and 0.1%
Tween-20; pH 7.5) and blocked for 1 h at RT with blocking solution
(10% blocking reagent and 50% lamb serum in MAB). Anti-DIG-AP
(1:4000 dilution in blocking solution) was added and agitated
overnight at 4°C.
Third day of hybridization
The embryos were washed with blocking solution for
30 min and MABT for 1 h at RT. Then, the embryos were soaked three
times for 5 min each in staining buffer (100 mM Tris, 50 mM
MgCl2, 100 mM NaCl, 0.1% Tween-20 and 1 mM levamisole;
pH 9.5). The embryos were transferred to a 12-well plate, incubated
in 1 ml/well BM purple stain for 30 min, monitored with a
dissecting microscope every 30 min. The reaction was terminated by
being washed twice in PBST and fixed with 4% PFA at 4°C to store.
Images were taken in 3% methyl cellulose.
Results
mpo/pBK-CML plasmid construction and
probe synthesis
The cDNA of 48 hpf embryos was used as a template to
amplify the mpo gemonic fragment by PCR. The PCR product was
verified by 1% agarose gel electrophoresis and the single band at
2400 bp was observed as expected. The PCR product was cloned into
the pBK-CML carrier and the mpo/pBK-CML recombinant plasmid
was constructed. The plasmid was linearized by SalI and the
single band at 7300 bp was observed. The plasmid was digested with
two enzymes (XhoI and EcoRI), producing two bands at
5200 and 2100 bp, respectively, which were the pBK-CML vector and
mpo gene segment. This demonstrated that plasmid
construction was successful (Fig.
1A). The DIG-labeled antisense mpo mRNA probe was
generated by T7 in vitro transcription and confirmed in the
electrophoresis tank soaked with DEPC H2O overnight.
There was a single band near 2100 bp (Fig. 1B).
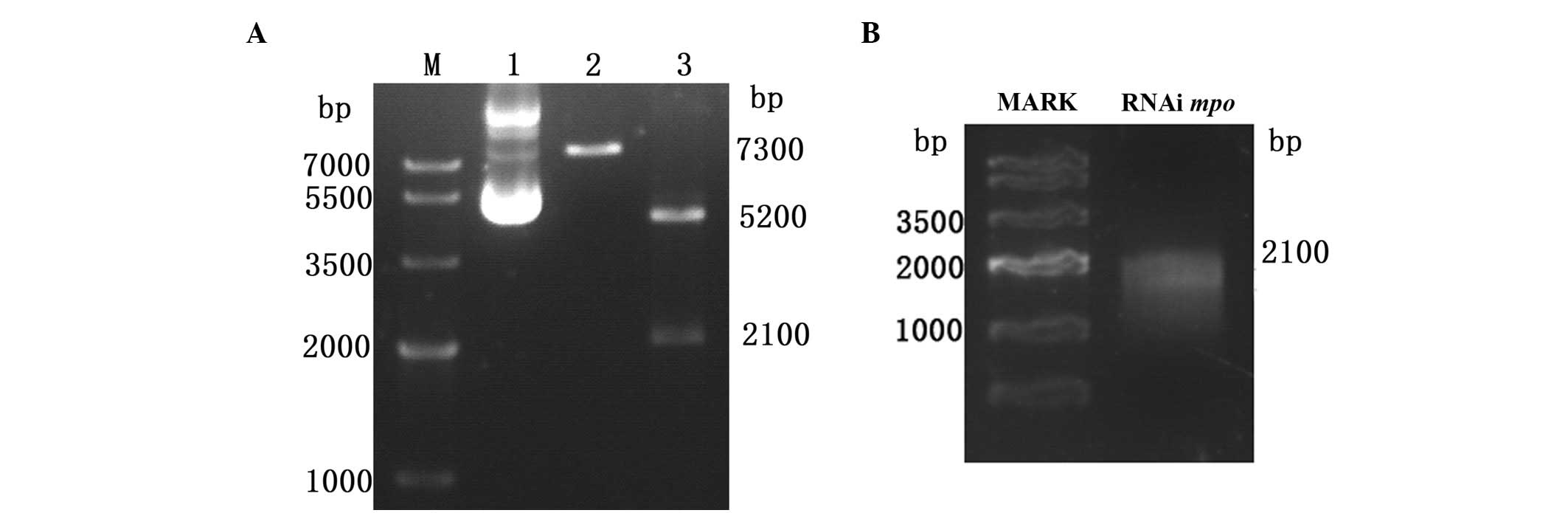 | Figure 1Electrophoresis map of recombinant
plasmid and mpo probe. (A) Electrophoresis map of the
mpo/pBK-CML recombinant plasmid. M, Tiangen marker IV; lane
1, plasmid crude extract. Three bands were observed; relaxed DNA,
open circle plasmid and superhelix plasmid; lane 2, the plasmid was
linearized by SalI enzyme, which obtained the single band at
7300 bp; lane 3, two enzymes, XhoI and EcoRI,
obtained two bands at 5200 and 2100 bp, respectively, which are the
pBK-CML vector and the mpo gene segment. (B) Electrophoresis
map of the antisense mpo probe. The band was at 2100 bp.
mpo, myeloperoxidase. |
Temporal and spatial expression patterns
of mpo
The expression patterns of mpo were
investigated in zebrafish embryos from 12 to 72 hpf by in
situ hybridization using the DIG-labeled antisense RNA probe.
As shown in Fig. 2, the earliest
expression of zebrafish mpo was detected in cells of the ICM
at 18 hpf and 1 to 2 h later, it was detected in cells in the
rostral blood island (RBI). Strong signals were observed in the
anterior ICM, then it spread over the yolk sac. By 72 hpf the
mpo-expressing cells were in the circulation and distributed
throughout the embryo. In our previous study, we established the
transgenic enhanced green fluorescent protein
[Tg(zlyz:EGFP)] zebrafish line (7), which expresses EGFP in primitive
neutrophils (mpo+). The EGFP distribution
coincided with the result of in situ hybridization of
mpo (Fig. 2).
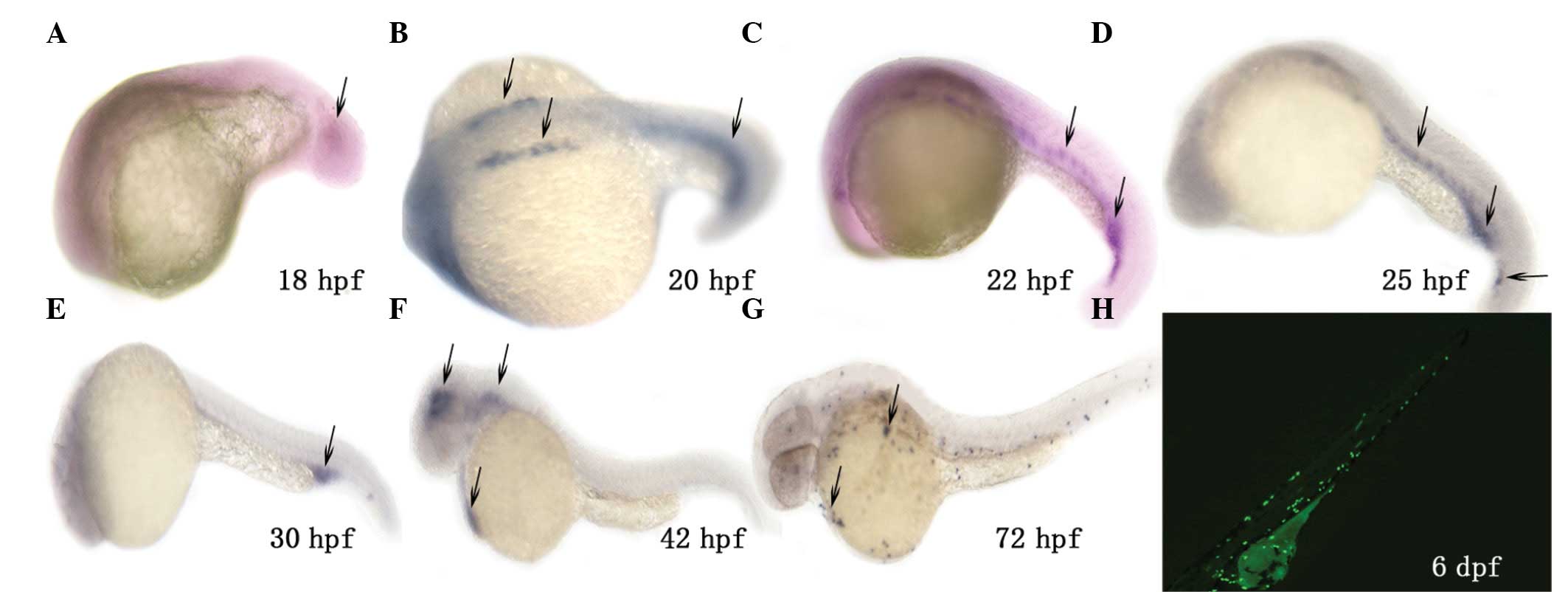 | Figure 2Spatio-temporal spectrum of mpo
expression in the AB zebrafish strain. In situ hybridization
with a digoxigenin-labeled RNA probe was used to detect zebrafish
mpo expression in embryos at 18 hpf (A), 20 hpf (B), 22 hpf
(C), 25 hpf (D), 30 hpf (E), 42 hpf (F) and 72 hpf (G). Black
arrows indicate mpo expression in the intermediate cell mass
(ICM) (A-E), rostral blood island (RBI) and pericardial cavity (F).
At 72 hpf these mpo-expressing cells were in the circulation
and distributed throughout the embryo (G). (H) Distribution of
fluorescent neutrophils (mpo+) in our previously
established Tg(zlyz:EGFP) embryo at 6 dpf. mpo,
myeloperoxidase; EGFP, enhanced green fluorescent protein; hpf,
hours post-fertilization; dpf, days post-fertilization. |
In situ hybridization and cytological
analyses
To test whether mpo expression detected by
in situ hybridization at an early stage could predict the
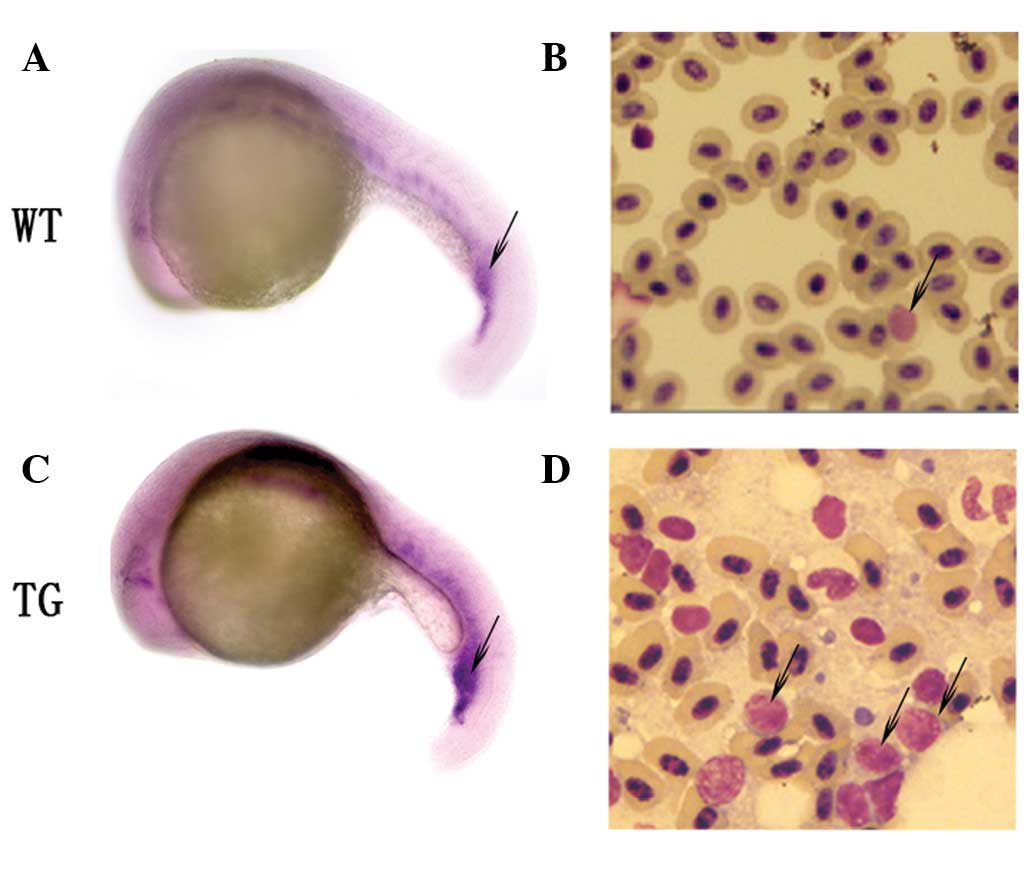
myelopoiesis in zebrafish development, we compared the peripheral
blood smear between wild type (WT) and Tg zebrafish. In our
previous study, we established the Tg(MYCN:HSE:EGFP)
zebrafish line, which suggested that MYCN converted
erythropoiesis to myelopoiesis (Shen et al, the influence of
MYCN gene on the transcriptional regulation of hematopoiesis. abs.
751, 9–12 June, 2011). As demonstrated by in situ
hybridization, the MYCN gene increases the expression of
mpo in embryos at 22 hpf (Fig.
3A and C). For cytological analyses, blood cells collected from
the zebrafish at 60 dpf were transferred onto glass slides using
Cytospin, stained with Wright-Giemsa stain and examined under oil
immersion by light microscopy (8).
This revealed that the blood cells from WT fish were predominantly
erythrocytes, with myeloid cells occasionally observed. By
contrast, erythrocytes were significantly inhibited in Tg fish,
which were enriched with granulocytes (Fig. 3B and D).
Modified WISH
In order to avoid background staining,
unincorporated nucleotides were removed from the probe preparation.
We routinely used Ambion NucAway Spin Columns to purify the RNA
probe according to the manufacturer’s instructions (Cat. AM10070).
Following the final precipitation, the hydrolyzed probe was placed
in HYB+ (final concentration, ∼1 ng/μl) and
stored at −20°C. Each time the probe was used, it was degenerated
in advance by placing it at 95°C for 5 min then on ice for 5 min to
eliminate single RNA spontaneous formation of hairpin structures.
Probes were reused for 4–5 times. When 24 hpf embryos were
collected, the conventional decolorization method was used and they
were placed in 0.03% phenylthiourea (PTU) in fish water at 12–36
hpf. We used the improved decolorizing liquid of 5% hydrogen
peroxide and 5% potassium peroxide in fish water. As a result, the
embryos only required soaking at 24 hpf for 10–15 min. The effect
of decolorization was good without damaging the integrity of the
embryo. Prior to hybridization, the embryos were permeabilized by
digesting with proteinase K (10 μg/ml in PBST) at RT for
5–30 min, depending on the developmental stage. The permeability of
the probe was appropriate and the staining background was low. In
addition, the traditional permeabilization method included
digesting the embryos at RT for 5–30 min, depending on the
developmental stage. This is difficult to control. We improved the
digestion temperature to 37°C and the process was shorted to 5
min.
Discussion
The zebrafish system has a number of unique
advantages compared to other vertebrate model organisms. The
embryos are externally fertilized and transparent, enabling in
vivo visualization of early embryonic processes ranging from
birth of HSCs in the mesoderm to migration of blood cells. In
addition, large production of embryos makes phenotype-based forward
genetics feasible (9). A steadily
increasing number of hematopoietic-specific genes have been cloned
in zebrafish, providing molecular reagents and markers for specific
stages of hematopoietic differentiation and specific cell types. We
constructed the DIG-labeled mpo RNA probe to investigate
mpo gene expression in zebrafish embryos. The earliest
mpo expression was detected in cells of the ICM at 18 hpf
and 1–2 h later, it was detected in cells in the RBI. Strong
signals were observed in the anterior ICM, then it spread over the
yolk sac. By 72 hpf the mpo-expressing cells were in the
circulation and distributed throughout the embryo, with a tendency
for a subpopulation of mpo-expressing cells to be aggregated
in the ventral vein region. Later-stage expression was difficult to
distinguish by WISH.
We identified that the level of mpo
expression detected by WISH at an early stage was consistent with
the result of cytological analyses of adult fish (Fig. 3). This method enabled us to track
the gene changes that took place before morphological phenotypes
were detected, as well as investigate the hematopoietic cell fate
in mutational or transgenic models in vivo. Given the
considerable morphological and functional parallels between
zebrafish and mammalian myeloid cells, it is not surprising that
zebrafish show conservation of the molecular regulation of
myelopoiesis and the molecular tools for myeloid cell function
(3).
In this study, we modified several steps of WISH. We
carried out several pretreatment steps to ensure the purity and
concentration of the probe, as well as shorten the digestion time
by proteinase K at higher temperatures. In the decolorization step,
we avoided using PTU, which simplified the zebrafish embryo culture
steps and enhanced the environmental protection. Finally, we
selected BM purple dye to simplify the staining step. The improved
hybridization results demonstrated high specificity, distinct
coloration and low background figures.
Using WISH in zebrafish, we have the ability to
identify and study the lesions of myelopoiesis. Therefore, the
powerful genetic approaches applicable in this model, the genomic
resources being collected by the international zebrafish and
genomic communities and the ability to study myeloid development in
this model organism provide new insights into the myeloid arm of
developmental hematology.
Acknowledgements
The authors thank all members of the
Shanghai Research Center for Biomodel Organisms and the Shanghai
Institute of Hematology for excellent technical support. This study
was supported in part by the National Natural Science Foundation of
China Grant (30900636), the Science and Technology Commission of
Shanghai Municipality Grant (08JCl414900), the Science and
Technology Commission of Shanghai Fund Project of the TCM Guide
Project (12401906700) and the Science and Technology Fund Project
of Shanghai Jiaotong University School of Medicine (09XJ21066).
References
|
1
|
Paik EJ and Zon LI: Hematopoietic
development in the zebrafish. Int J Dev Biol. 54:1127–1137. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu J, Du L and Wen Z: Myelopoiesis during
zebrafish early development. J Genet Genomics. 39:435–442. 2012.
View Article : Google Scholar
|
|
3
|
Forrester AM, Berman JN and Payne EM:
Myelopoiesis and myeloid leukaemogenesis in the zebrafish. Adv
Hematol. 2012:3585182012.PubMed/NCBI
|
|
4
|
Tautz D and Pfeifle C: A non-radioactive
in situ hybridization method for the localization of specific RNAs
in Drosophila embryos reveals translational control of the
segmentation gene hunchback. Chromosoma. 98:81–85. 1989. View Article : Google Scholar
|
|
5
|
Westerfield M: The Zebrafish Book: A Guide
for the Laboratory Use of Zebrafish (Danio rerio).
University of Oregon Press; Eugene: pp. 1.1–1.27. 1995
|
|
6
|
Kimmel CB, Ballard WW, Kimmel SR, Ullmann
B and Schilling TF: Stages of embryonic development of the
zebrafish. Dev Dyn. 203:253–310. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Bai XT, Zhu KY, et al: In vivo
interstitial migration of primitive macrophages mediated by
JNK-matrix metalloproteinase 13 signaling in response to acute
injury. J Immunol. 181:2155–2164. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yeh JR, Munson KM, Chao YL, Peterson QP,
Macrae CA and Peterson RT: AML1-ETO reprograms hematopoietic cell
fate by downregulating scl expression. Development. 135:401–410.
2008.PubMed/NCBI
|
|
9
|
Shafizadeh E and Paw BH: Zebrafish as a
model of human hematologic disorders. Curr Opin Hematol.
11:255–261. 2004. View Article : Google Scholar : PubMed/NCBI
|