Introduction
Osteosarcoma is the most common type of primary bone
tumor and causes serious harm to the health of adolescents.
Osteosarcoma is highly invasive and is transferred by the blood in
the early stage, and progresses rapidly. It mainly occurs in
actively growing long bone metaphysis. This type of tumor has a
high degree of malignancy, recurrence and metastasis and the
prognosis is poor. The incidence was reported in 2009 as ~4 million
individuals per year (1).
Osteosarcoma cells have strong invasive ability, quick hematogenous
metastasis in early stage, rapid progression, and the five-year
survival rate was only 60% in 2008 (2). Although treatment of osteosarcoma has
been on the increase, the five-year survival rate remains low, and
the recurrence rate is high (3).
Therefore, investigation of the pathogenesis of osteosarcoma and
attempts to identify a novel approach to reduce the tumor
recurrence rate and improve the survival rate of the patients is of
significant importance in the clinic.
The development of osteosarcoma is complex, and the
molecular mechanism is not clear yet. Numerous studies have shown
that there are abnormal expression levels of the phosphate and
tension homolog (PTEN) gene in human osteosarcoma cells or tissues.
The PTEN gene deleted on chromosome 10, also known as
mutated in multiple advanced cancer 1 and TGF-β-regulated and
epithelial cell-enriched phosphatase, is located on chromosome
10q23.3. The gene consists of nine exons, encodes a protein which
is composed of 403 amino acids and has phosphatase enzyme activity
(4,5).
The PTEN gene was first identified in 1997
(6), and it is considered an
important tumor suppressor gene together with p53 and Rb. It is
also the first tumor suppressor gene with phosphatase activity to
be observed thus far.
The PTEN protein inhibits tumor occurrence and
development mainly through the following three pathways: i)
Inositol triphosphate kinase [phosphoinositide 3-kinase
(PI3K)/AKT]pathway. The protein encoded by PTEN has lipid
phosphatase activity, thus it competes with PI3K and causes
dephosphorylation of phosphatidylinositol (3,4,5)-triphosphate (PIP3), and this prevents
the growth factors’ signal transduction pathway regulated by PI3K.
The reduced PIP3 levels arrest the cell in G1 phase, thereby
inducing apoptosis of the tumor cells. ii) Mitogen-activated
protein kinase (MAPK) pathway. PTEN inhibits the upstream
extracellular signal-regulated kinase (ERK) of MAPK, the activation
of Ras and the phosphorylation of Shc. The PTEN gene also inhibits
the phosphorylation of MAPK kinase and blocks the cell in G1 phase,
thus inhibiting tumor growth (6–8).
iii) Focal adhesion kinase (FAK) pathway. FAK is an important
factor in the integrin-mediated signal transduction pathway.
Activated FAK activates several associated kinases and signaling
molecules that promote cell invasion and metastasis. PTEN inhibits
the activation of FAK by causing its dephosphorylation, thus
inhibiting the invasion and metastasis of tumor cells.
The abnormal expression levels of the PTEN gene play
an important role in tumor occurrence and development (9). A study concerning the expression
levels of PTEN in osteosarcoma tissues has demonstrated that there
is a significant reduction in the levels of PTEN protein expression
in osteosarcoma tissue. Through enhancement of the phosphorylation
levels of Akt, PTEN is inhibited and thus promotes the
proliferation of osteosarcoma cells (10). The reason for the expression levels
of the PTEN gene being abnormally low in osteosarcoma tissues
remains unclear.
Studies have confirmed that hypermethylation of
tumor suppressor genes is closely associated with the occurrence of
tumors and the methylation levels of the CpG islands in eukaryotic
DNA are closely associated with cell canceration (11–12).
If there is an unmethylated CpG island in a tumor suppressor gene,
this tumor suppressor gene easily becomes the attack target of DNA
methyltransferases (DNMTs). In cancer cells, the activation levels
of DNMTs are increased, tumor suppressor genes show
hypermethylation status of the CpG islands and cause
transcriptional inactivation. A number of studies have confirmed
that the abnormal methylation of the PTEN gene promoter leads to
abnormal gene expression levels (13–15)
and the methylation of the promoter enhancer region CpG islands
causes certain transcription factors to be unable to bind to DNA
and thus inhibits gene transcription.
Myc and Sp1 are the main transcription factors in
the PTEN promoter that regulate the transcription of PTEN (16,17).
To the best of our knowledge, it has not been reported whether DNA
demethylation influences the expression levels of the PTEN gene in
osteosarcoma cells and the methylation degree of the GC site that
binds to Myc and Sp1 in the PTEN promoter. To the best of our
knowledge, there are few studies concerning the epigenetic changes
of PTEN in osteosarcoma, i.e., whether the methylation status of
the PTEN gene promoter region affects the expression levels of the
PTEN protein, and if it does, the mechanisms by which this
happens.
The cultured MG-63 osteosarcoma cell line was used
for the present study. The growth inhibition and induced apoptosis
caused by different concentrations of 5-Zac added to MG-63 cells
were observed, and the changes in the PTEN gene mRNA and the
expression levels of the PTEN protein were detected. Bisulfite
sequencing was further used to detect the methylation status of the
CG site for binding to the transcription factor Myc in the PTEN
gene promoter, and the associations between them.
Materials and methods
Materials
Cell line
The MG-63 osteosarcoma cell line was provided by the
Cell Culture Centre of Xiangya School of Medicine, Central South
University (Changsha, China).
Reagents and instruments
5-Zac, formula
C8H12N4O5 and relative
molecular weight 244.205, was purchased from Sigma-Aldrich (St.
Louis, MO, USA). RPMI-1640 medium and fetal bovine serum Australia
origin were purchased from Gibco (Carlsbad, CA, USA). Rabbit
anti-human polyclonal and anti-PTEN antibodies were purchased from
Millipore (Billerica, MA, USA), and rabbit anti-human anti-β-actin
and peroxidase-conjugated goat anti-rabbit anti-IgG were purchased
from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). A DNA
extraction kit, Taq enzyme, dNTPs and a reverse transcription kit
were purchased from Promega GmbH (Madison, WI, USA). DNA reference
standards were purchased from Fermentas (Burlington, Canada).
TRIzol was purchased from Invitrogen (Carlsbad, CA, USA) and a
sodium bisulfite treatment kit was purchased from Chemicon American
Companies.
Methods
Cell culture. The MG-63 cells were cultured in
RPMI-1640 medium with 10% fetal bovine serum, and incubated at 37°C
in a humidified atmosphere with 5% CO2. After the cells
covered the bottom of a 9-cm petri dish, they were subcultured in a
6-cm dish and the medium was changed every 2–3 days. Experimental
intervention was exerted when the cells reached 60–70% fusion.
Methylation inhibitor 5-Zac
processing
There were three treatment groups in the study.
5-Zac with a final concentration of either 0, 5 or 10 μmol/l was
added to the medium of the cells, and then the cells were incubated
at 37°C in a humidified atmosphere with 5% CO2. The
culture medium and the same concentration of 5-Zac were changed
every 24 h, and after 72 h the treatment the cells were harvested
and tested. Each experiment was repeated three times.
Flow cytometry to detect the apoptotic
rate of the MG-63 cells
Following trypsin digestion, 1×106 MG-63
cells were harvested, washed twice with ice-cold phosphate-buffered
saline (PBS), fixed and permeabilized with 70% ethanol at −20°C for
24 h, and washed once with ice-cold PBS. After incubation with
propidium iodide (PI) staining buffer at 37°C for 1 h, the cells
were washed one more time with ice-cold PBS and DNA content
analysis was performed with a FASCalibur Flow Cytometer (Becton,
Dickinson and Company). The PI staining buffer contained 1X PBS,
100 μg/μl RNase and 40 μg/ml PI.
RNA extraction and reverse
transcription-polymerase chain reaction (RT-PCR)
The total RNA was extracted with TRIzol according to
the manufacturer’s instructions. Following quantification by UV
spectrophotometry, 1 μg of the total RNA was used for reverse
transcription reaction synthesis of cDNA with a reverse
transcription kit, according to the manufacturer’s instructions.
PCR was used to amplify the cDNA. The corresponding primer
sequences were as follows: forward, 5′-CCACCCATGGCAAATTCCATG-3′ and
reverse, 5′-TCTAGACGGCAGGTCAGGTCCACC-3′ for reference GAPDH; and
forward, 5′-TTGAAGACCATAACCCACCA-3′ and reverse,
5′-CACATAGCGCCTCTGACTG-3′ for PTEN.
Quantitative PCR was performed using a Rotor-Gene
3000 Real-Time PCR instrument (Corbett Research, Australia). PTEN
and β-actin mRNA were amplified by SYBR-Green real-time PCR using
the One Step PrimeScript RT-PCR kit (Takara Biotechnology Co.,
Ltd., Dalian, China). GAPDH mRNA was used as the internal control.
The reactions used the following cycling conditions: 94°C initial
denaturation for 3 min, 94°C denaturation for 30 sec, 60°C
annealing for 30 sec and 72°C extension for 30 sec for a total of
35 cycles, and a final extension at 72°C for 7 min. Relative PTEN
mRNA expression levels normalized to those of β-actin mRNA were
calculated using the equation: 2−ΔΔCt, where
ΔΔCt(relative quantification) = (CTPTEN -
CTβ-actin)CHB patient - (CTPTEN -
CTβ-actin)Normal control
Western blot analysis of the
expression levels of PTEN protein in MG-63 cells
The MG-63 cells treated with different
concentrations of 5-Zac were cultured and collected, and the
protein samples were extracted by radioimmunoprecipitation assay
lysate. The bicinchoninic acid (BCA; Keygen Biotech, Nanjing,
China) method, using a microplate reader (Thermo, Waltham, MA, USA)
at 570 nm wavelength, was used to detect the total protein
concentration. Protein (20 μg) was collected from each group,
electrophoresized in 12% SDS-PAGE and a wet electrostatic transfer
method was used to transfer the protein to a nitrocellulose
membrane. Non-fat milk (5%) was used to block the membrane at room
temperature for 1 h, then anti-PTEN (working concentration, 1:500)
and internal reference anti-β-actin (1:2,000) were added to the
membrane and it was incubated at 4°C overnight. The membrane was
washed in PBS three times, each time for 10 min, and
peroxidase-labeled anti-IgG was added as the secondary antibody
(working concentration, 1:1,000). The membrane was incubated for 1
h at room temperature and washed with PBS three times, each time
for 10 min. An ECL chemiluminescence kit (Thermo) was used to
develop the membrane. The experiment was repeated three times.
DNA extraction and bisulfate
sequencing to detect the methylation status of the PTEN gene
fragment
The genomic DNA of the cells was extracted using a
DNA extraction kit according to the manufacturer’s instructions.
Following identification and quantification by UV
spectrophotometry, 1 μg DNA was collected to perform the bisulfate
conversion with a CpGenome™ DNA Modification kit (Millipore),
according to the manufacturer’s instructions. PCR was used to
amplify 287 bp from the binding region of the PTEN promoter region
and the transcription factor Myc in the bisulfate-converted DNA.
The amplification primer sequences were:
5′-TATTTATAAGGTGGAAGTTTTGAGG-3′ and
5′-ATAAAAAATAAACTCAACCCCACTC-3′. The PCR amplification conditions
were 94°C for 3 min; 35 cycles of 94°C for 30 sec, 55°C for 30 sec
and 72°C for 30 sec; and a final extension at 72°C for 7 min. The
PCR products were cloned into a T-vector and transformed into
Escherichia coli (E. coli) cells (DH5α).
Subsequently, the E. coli were inoculated in
Ampicillin+ (100 μg/ml) LB agar plates, incubated at
37°C for 12–16 h and then five independent clones were sequenced
for the amplified fragment. The demethylation rate of the CpG pairs
in the MG-63 cells treated with or without different concentrations
of 5-Zac was calculated from the sequencing results.
Statistical analysis
SPSS software, version 16.0 for Windows (SPSS, Inc.,
Chicago, IL, USA) was used to store and analyze the data. The
routine test of homogeneity of variance and the normality test were
performed. Measurement and experimental data are expressed as the
mean ± standard deviation. Multiple sets of data were compared
using the F-test (one way analysis of variance). P<0.05 was
considered to indicate a statistically significant difference.
Results
MG-63 cell growth is inhibited by
5-Zac
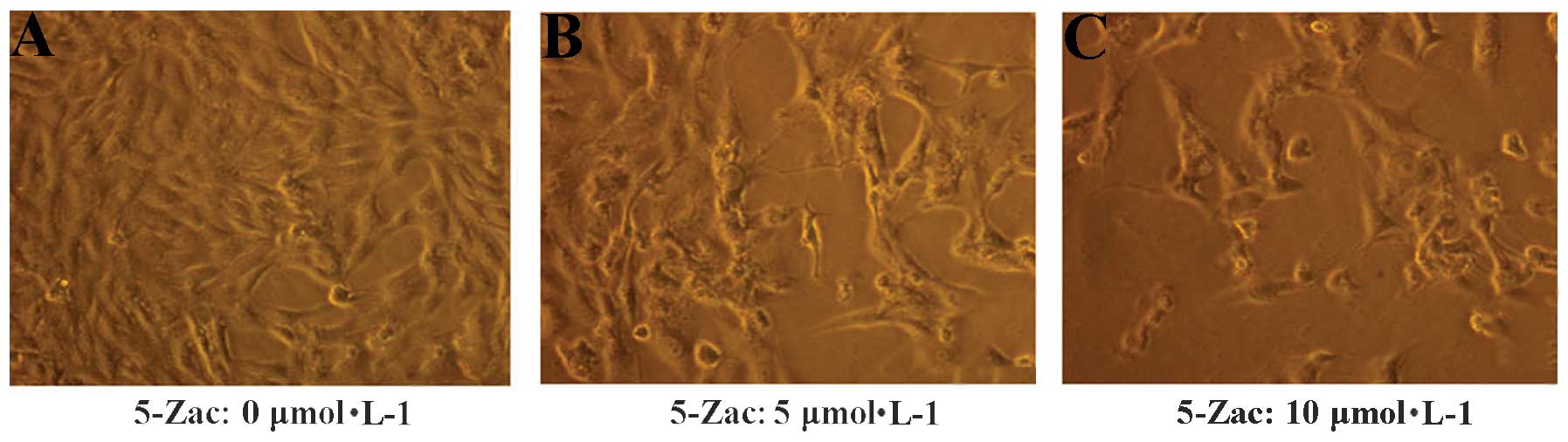
The morphology of the MG-63 cells was altered
following the addition of 5-Zac (Fig.
1). The MG-63 cells in the standard RPMI-1640 complete medium
were adherent and exhibited adequate growth (Fig. A), with the
cells arranged similar to epithelia. The nuclear shape was round,
the cell membrane was integrated, the cytoplasm presented a high
degree of uniformity, and the cells possessed a high quality of
refraction. Subsequently, 5-Zac (5 or 10 μmol/l) was added to the
cells. After 72 h, cell proliferation was stagnated, numerous cells
were crimpled, the vacuole was evident, the number of granules was
increased in the cytoplasm, and high levels of impurity including
cell fragments were identifiable (Fig.
1B and C).
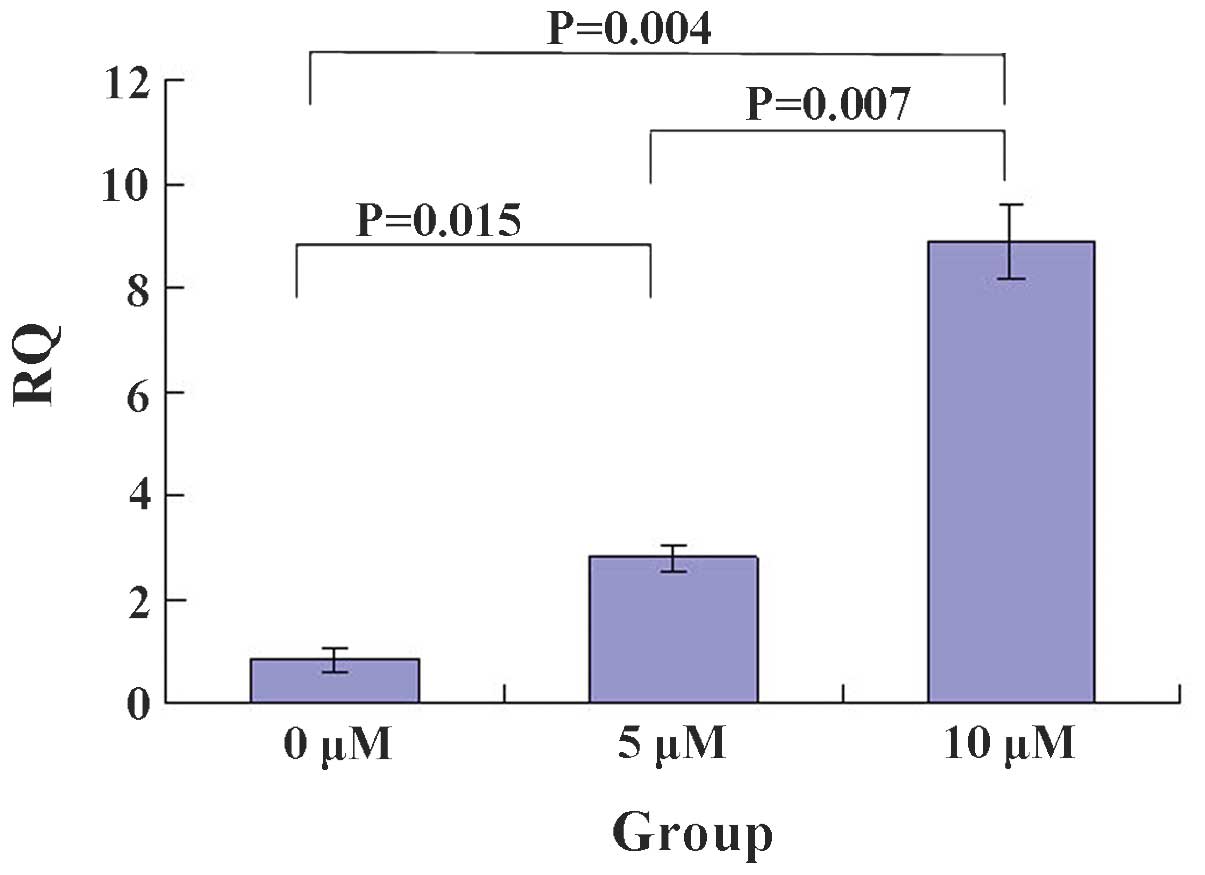
5-Zac enhances PTEN mRNA expression
levels in MG-63 cell
The RT-PCR showed that 5-Zac increases the PTEN mRNA
levels in a concentration-dependent manner (Figs. 2 and 3). The relative quantification (RQ) value
of each group was 0.80±0.02 for the control group (0 μmol/l),
0.90±0.02 for the 5 μmol/l 5-Zac group, and 0.95±0.01 for the 10
μmol/l 5-Zac group. The statistical significance was calculated by
comparing each combination of two groups: P<0.05 for the
comparison between the 0 and the 5 and 10 μmol/l groups; and
P=0.007 for the comparison between the 5 and 10 μmol/l groups.
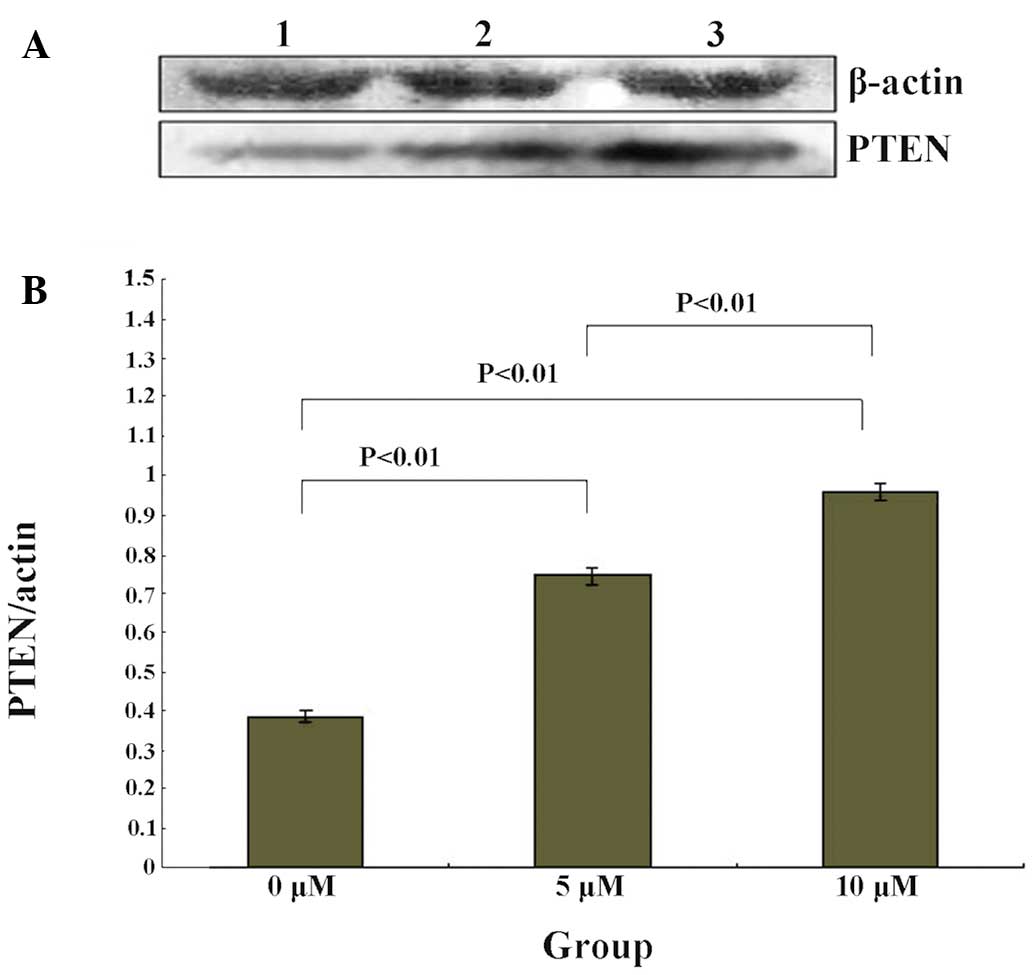
PTEN expression levels are upregulated by
5-Zac in MG-63 cells
The PTEN protein expression levels of the MG-63
cells were measured following treatment with different
concentrations of 5-Zac. The results are shown in Fig. 3, which demonstrate that the PTEN
protein expression levels increase with the increasing
concentration of 5-Zac, and are concentration-dependent. The ratio
between the PTEN protein and internal reference β-actin levels was
0.39±0.01 for the control group (0 μmol/l), 0.75±0.02 for the 5
μmol/l 5-Zac group, and 0.96±0.01 for the 10 μmol/l 5-Zac group.
Also, significant statistical differences are shown among the three
treatment groups, all P<0.01.
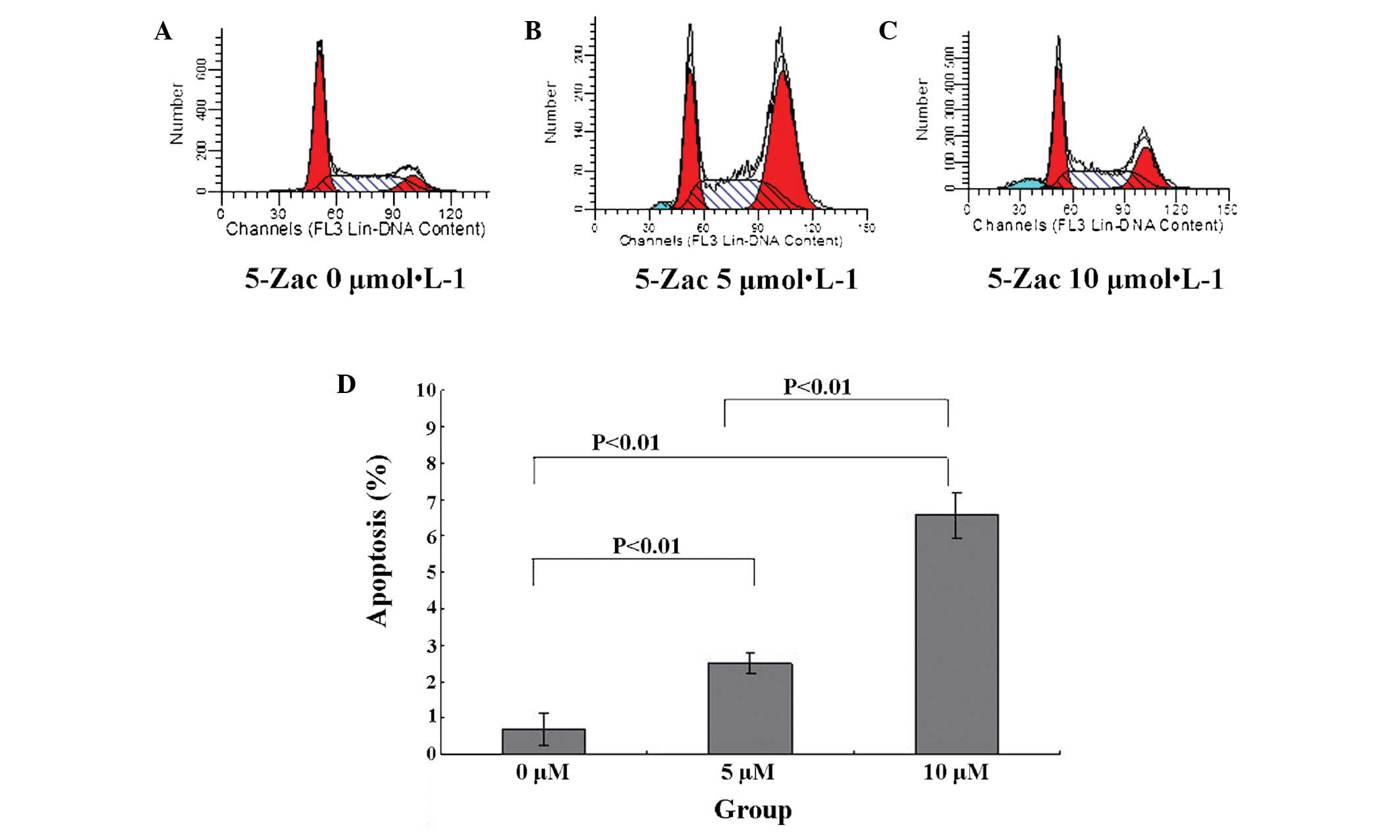
5-Zac induces apoptosis of MG-63 human
osteosarcoma cells
The MG-63 cell apoptotic rate is 0.69±0.42% in the
absence of 5-Zac. When 5-Zac is added to the cells and they are
cultured for 72 h, the MG-63 cell apoptotic rate increases
gradually, as Fig. 4 presents. It
is observable that the cell apoptotic rate increases by 2.50±0.30%
for the 5 μmol/l group and 6.59±0.62% for the 10 μmol/l group.
Comparing the three groups, P<0.01 is obtained, and the
differences are evidently statistically significant. Furthermore,
the PTEN expression rate is higher in the cells treated with a
higher concentration of 5-Zac.
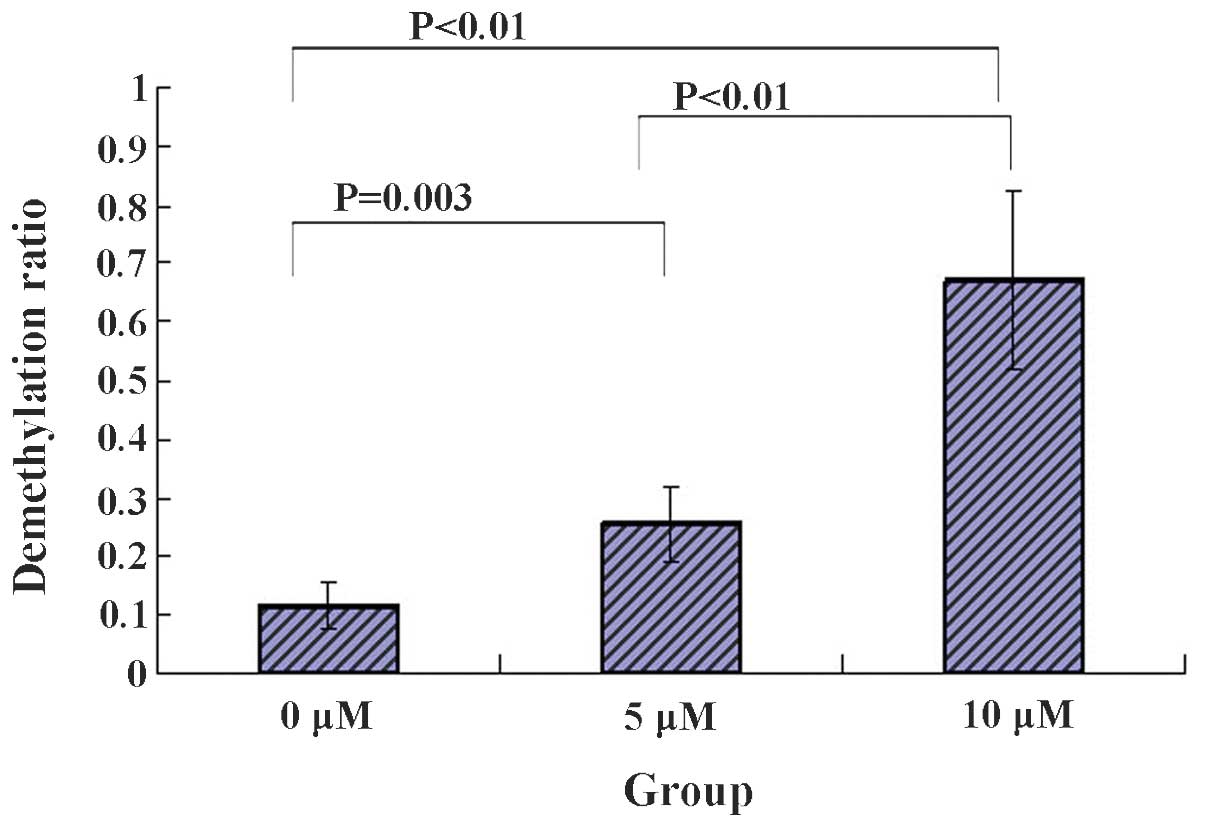
5-Zac reduces the methylation of the PTEN
promoter in MG-63 cells
The degree of methylation of 22 CpG points between
−263 and 0 bp in the PTEN gene promoter was detected in each group
of MG-63 cells following treatment with 5-Zac. Five clones from
each group were selected for sequencing. The average methylation
levels were produced following the sequencing. The results are
shown in Fig. 5, where 1
represents full demethylation and 0, full methylation. It was found
that the demethylation levels were largely increased for the CG
points that bind to the transcription regulation factors Myc and
Sp1 in the PTEN promoter; a large demethylation difference was
evident for the 5 and 10 μmol/l groups by comparing with that of
the 0 μmol/l group; the demethylation levels increase with the
increasing concentration of 5-Zac; and the differences were evident
by comparing the methylation among the three groups. Therefore, it
is suggested that the methylation inhibitor 5-Zac affects the
expression levels of PTEN in MG-63 cells possibly via the
demethylation of the GC site that binds to the transcription
factors Myc and Sp1 in the PTEN promoter.
Discussion
Osteosarcoma is the most common type of primary
malignant tumor in bone. The pathogenesis of osteosarcoma is
complex and the precise molecular mechanisms have yet to be
determined.
The present study cultured the MG-63 osteosarcoma
cell line and treated the cells with different concentrations of
the methylation inhibitor 5-Zac to detect the expression levels of
the PTEN protein, the mRNA transcription levels of the PTEN gene,
and the influence of the methylation status of the GC site that
binds to the transcription factors Myc and Sp1 in the PTEN
promoter.
It was demonstrated that the PTEN expression and
mRNA transcription levels in the MG-63 cells gradually increased
along with the increasing 5-Zac concentration, and the apoptotic
rate of the MG-63 cells was also positively correlated to the 5-Zac
concentration.
In a further experiment, it was revealed that
following treatment with 5-Zac, the methylation status of the
transcription factor binding fragment of the PTEN promoter had
significantly changed.
These results suggest that following treatment with
the methylation inhibitor 5-Zac, the PTEN expression and
transcription levels in MG-63 osteosarcoma cells were significantly
increased, and the number of apoptotic cells was increased. The
gene transcription levels may be affected by methylation
regulation. This study provides a novel perspective for future
studies concerning the regulation mechanism of the PTEN gene, which
is closely associated with osteosarcoma, and presents a novel
theory that change in the methylation status of PTEN may be
effective as a treatment for osteosarcoma.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (81272947).
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bielack SS and Carrle D: State-of-the-art
approach in selective curable tumors: bone sarcoma. Ann Oncol.
19(Suppl 7): vii155–vii160. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Picci P, Mercuri M, Ferrari S, et al:
Survival in high-grade osteosarcoma: improvement over 21 years at a
single institution. Ann Oncol. 21:1366–1373. 2010.PubMed/NCBI
|
|
4
|
Yim EK, Peng G, Dai H, et al: Rak
functions as a tumor suppressor by regulating PTEN protein
stability and function. Cancer Cell. 15:304–314. 2009. View Article : Google Scholar
|
|
5
|
Song MS, Carracedo A, Salmena L, et al:
Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a
phosphatase-independent manner. Cell. 144:187–199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li L, Ernsting BR, Wishart MJ, Lohse DL
and Dixon JE: A family of putative tumor suppressors is
structurally and functionally conserved in humans and yeast. J Biol
Chem. 272:29403–29406. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu J, Zhang SS, Saito K, et al: PTEN
regulation by Akt-EGR1-ARF-PTEN axis. EMBO J. 28:21–33. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tamura M, Gu J, Danen EH, et al: PTEN
interactions with focal adhesion kinase and suppression of the
extracellular matrix-dependent phosphatidylinositol 3-kinase/Akt
cell survival pathway. J Biol Chem. 274:20693–20703. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chung JH, Ostrowski MC, Romigh T, et al:
The ERK1/2 pathway modulates nuclear PTEN-mediated cell cycle
arrest by cyclin D1 transcriptional regulation. Hum Mol Genet.
15:2553–2559. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang T, Sun J, Lu G, et al: Expression
and significance of PTEN in osteosarcoma. J Chin Med Univ.
33:245–246. 2004.
|
|
11
|
Nielsen-Preiss SM, Silva SR and Gillette
JM: Role of PTEN and Akt in the regulation of growth and apoptosis
in human osteoblastic cells. J Cell Biochem. 90:964–975. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsumura A, Hayakawa T, Kumaki Y, et al:
Maintenance of self-renewal ability of mouse embryonic stem cells
in the absence of DNA methyltransferases Dnmtl, Dnmt3a and Dnmt3b.
Genes Cells. 11:805–814. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Polanowska J, Martin JS, Garcia-Muse T, et
al: A conserved pathway to activate BRCA1-dependent ubiquitylation
at DNA damage sites. EMBO J. 25:2178–2188. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hashemi M, Rezaei H, Eskandari-Nasab E,
Kaykhaei MA and Taheri M: Association of promoter methylation and
32-bp deletion of the PTEN gene with susceptibility to metabolic
syndrome. Mol Med Rep. 7:342–346. 2013.PubMed/NCBI
|
|
15
|
Lubecka-Pietruszewska K, Kaufman-Szymczyk
A, Stefanska B and Fabianowska-Majewska K: Folic acid enforces DNA
methylation-mediated transcriptional silencing of PTEN, APC and
RARbeta2 tumour suppressor genes in breast cancer. Biochem Biophys
Res Commun. 430:623–628. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bian EB, Huang C, Ma TT, Tao H, Zhang H,
Cheng C, Lv XW and Li J: DNMT1-mediated PTEN hypermethylation
confers hepatic stellate cell activation and liver fibrogenesis in
rats. Toxicol Appl Pharmacol. 264:13–22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kou XX, Hao T, Meng Z, Zhou YH and Gan YH:
Acetylated Sp1 inhibits PTEN expression through binding to PTEN
core promoter and recruitment of HDAC1 and promotes cancer cell
migration and invasion. Carcinogenesis. 34:58–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peyrou M, Bourgoin L and Foti M: PTEN in
liver diseases and cancer. World J Gastroenterol. 16:4627–4633.
2010. View Article : Google Scholar : PubMed/NCBI
|