Introduction
Parkinson’s disease (PD) is a common
neurodegenerative disease that is prevalent in 1–2% of individuals
aged ≥65 years. While the specific etiology of PD is yet to be
elucidated, genes involved in the development of the disease have
attracted much research attention. At present, three autosomal
recessive genes have been identified to be associated with PD,
including PTEN-induced putative kinase 1 (PINK1), parkin, and DJ-1
(1–3). The PINK1 gene encodes a cytosolic E3
ubiquitin ligase and a mitochondrial serine/threonine kinase. PINK1
mutations were initially observed in consanguineous families of
Italian and Spanish origin and are associated with slowly
progressive PD with an onset prior to 50 years of age (4,5).
PINK1 mutations have been reported in Filipino, Taiwanese, Israeli,
Japanese, Irish and North American populations, and also in Chinese
populations (6–11).
In order to investigate PINK1 mutations in PD and
analyze the distribution of PINK1 gene T313M polymorphisms in the
Uygur and Han populations of China, patients with PD and healthy
individuals were investigated. The significance of PINK1 T313M
polymorphisms in the pathogenesis of PD was also assessed.
Materials and methods
Diagnostic criteria
The present study was performed in accordance with
British Brain Bank diagnostic criteria (12), with partial diagnoses and complex
cases confirmed by a senior doctor from the Neurological Department
of the First Affiliated Hospital of Xinjiang Medical University
(Urumqi, China). Over the past 50 years, diagnostic criteria for
early onset and late onset PD have been defined by developments in
head magnetic resonance imaging and computed tomography
examinations. The present study excluded patients with secondary
PD, Parkinson-plus syndromes, nervous system disease,
hyperthyroidism and other genetic diseases. The present study was
conducted in accordance with the Declaration of Helsinki and was
conducted with approval from the Ethics Committee of the first
Affiliated Hospital of Xinjiang Medical University. Written
informed consent was obtained from all participants.
Patient data
In the present study, 364 patients with PD from the
Uygur and Han populations of Xinjiang, China were selected between
July 2010 and March 2011 as the case group. These patients included
175 individuals from the Uygur population, of which 99 were male
and 76 were female, aged between 31 and 95 years (mean, 62.63±12.71
years). The remaining 189 patients with PD were from the Han
population, of which 107 were male and 82 were female, aged between
25 and 85 years (mean, 61.76±12.31 years). The control group
comprised 346 healthy individuals without PD. These included 163
individuals from the Uygur population, of which 92 were male and 71
were female, aged between 33 and 90 years (mean, 62.78±12.50
years), and 183 individuals from the Han population, of which 101
were male and 82 were female, aged between 27 and 86 years (mean,
61.43±12.51 years). No significant differences in age or gender
were observed between the individuals in the PD group compared with
those in the control group (χ2gender=0.048,
P>0.05; tage=1.445, P>0.05).
DNA extraction
Subsequent to obtaining informed consent, 2 ml
venous blood was extracted from each patient. EDTA was used as an
anti-coagulant and a blood extraction kit (Tiangen Biotech Co.,
Ltd., Beijing, China) was used to extract the DNA.
Primers
Primer3 software was used to design the primer
sequences required for polymerase chain reaction (PCR) analysis.
The primer sequences for exon 4 of the PINK1 gene were as follows:
5′-GAATGTCAGTGC CAGTGTTGG-3′ (forward) and 5′-AGATATGTTCCCTTT
GCATGGC-3′ (reverse). The length of amplified fragment was 429 bp
and the primers were synthesized by Huada Gene Company (Beijing,
China).
Restriction fragment length polymorphism
analysis
Restriction fragment length polymorphism analysis
was performed using the HgaI endonuclease (New England
Biolabs, Inc., Ipswich, MA, USA) in a restriction enzyme reaction
system with a total reaction volume of 20 μl. The reaction
consisted of 10 μl PCR product, 2 μl NEBuffer 1.1 (pH=8.0), 7.7 μl
deionized double-distilled water and 0.3 μl HgaI, at 37°C
overnight.
Digested products confirmation
Digested products were subjected to agarose gel
electrophoresis and the enzymes were analyzed using UV gel
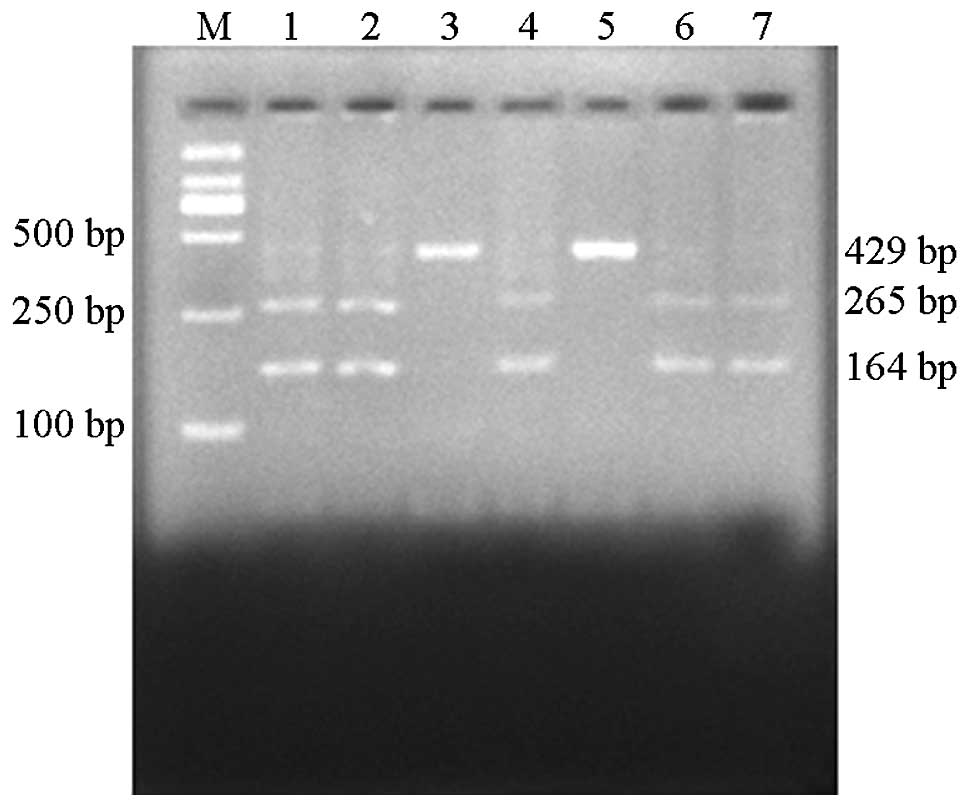
electrophoresis. In brief, the gene amplified by PCR was a 429-bp
sequence of the fourth exon of the PINK1 gene fragment, which
contained a HgaI enzyme restriction site. Upon HgaI
restriction digestion, homozygous wild-type sequences were digested
into two fragments of 265 bp and 164 bp. This genotype was type
C/C. At the 938 site, a C to T mutation occurs, resulting in loss
of the HgaI restriction site. Thus, mutant homozygote
genotypes are not digested and remain as 429 bp fragments, type
T/T. Upon HgaI restriction digestion, heterozygous sequences
are digested into fragments of 429, 265 and 164 bp. This
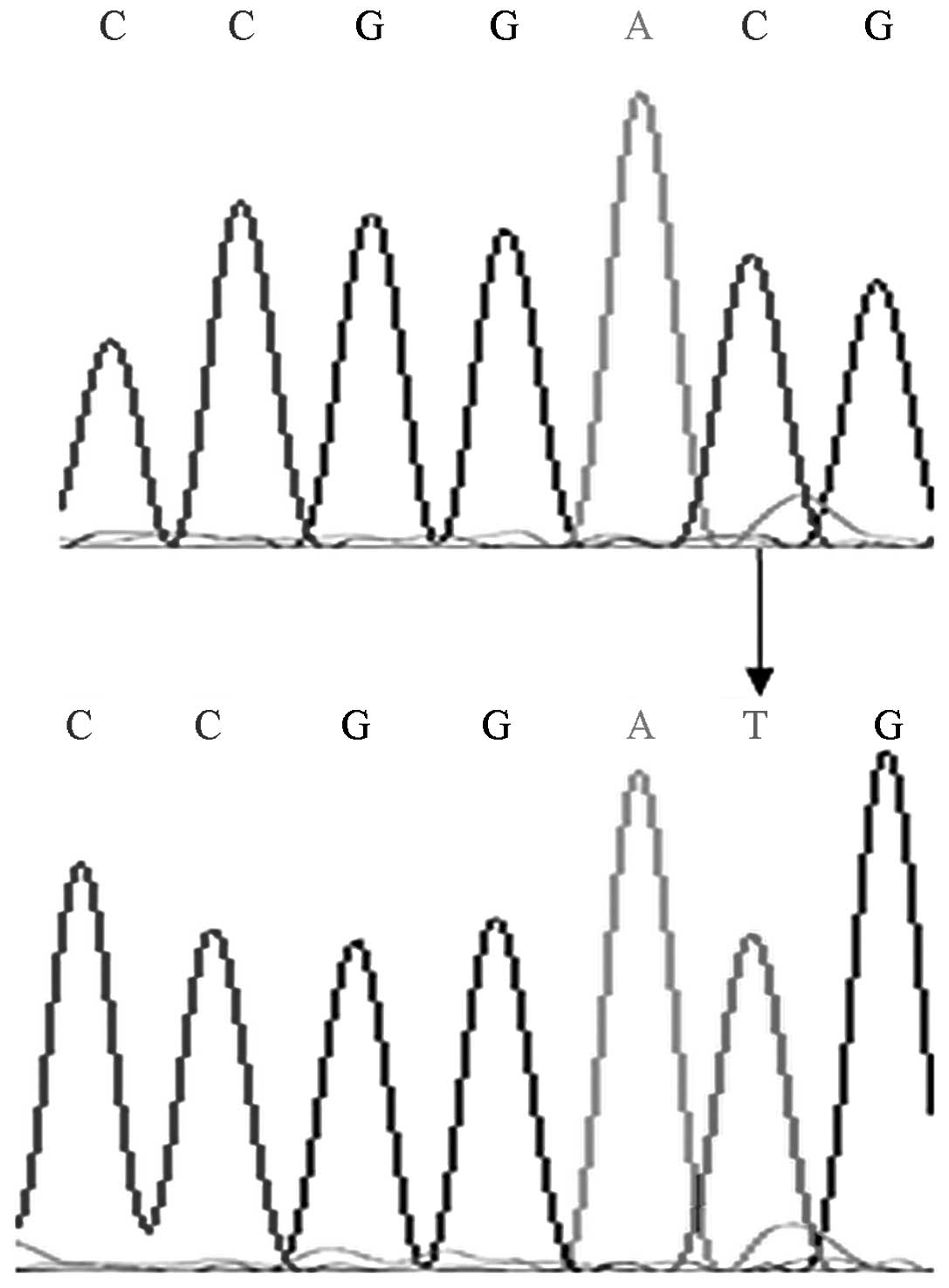
heterozygous genotype is type C/T (Fig. 1). The mutant genotype was sequenced
which confirmed that the digestion results were accurate (Fig. 2).
Statistical analysis
Genotypes and allele frequencies were calculated
using a direct counting method. Genotype data were subjected to
Hardy-Weinberg equilibrium tests and alleles and genotypes were
compared using the χ2 test. The constituent ratio of
each group was analyzed using the χ2 test. All
statistical analyses were performed using SPSS software, version
17.0 (SPSS, Inc., Chicago, IL, USA).
Results
Hardy-Weinberg equilibrium test
To show that the PINK1 genotype frequencies were
equal in the case and control groups, the genotype frequencies of
the case and control groups were assessed using alignment
inspection. The PINK1 genotype distributions in the case and
control groups were in Hardy-Weinberg equilibrium and exhibited
good consistency (P>0.05).
T313M allele frequency distribution
Differences in polymorphic T313M alleles and
genotype frequencies in the PD group compared with the control
group were not observed to be statistically significant (P>0.05;
Table I). Furthermore, in the
Uygur population, the genotype and allele frequencies in the PD
group were not observed to be significantly different from those in
the control group (P>0.05). Moreover, in the Han population, the
genotype frequencies of the PD group showed no significant
difference from those in the control group (P>0.05), whereas
allele frequencies were observed to be significantly different
between the PD and control groups (χ2=6.247, P<0.05;
Table II).
 | Table IAllele and genotype frequencies of the
PINK1 gene T313M polymorphism in patients with PD and healthy
control individuals. |
Table I
Allele and genotype frequencies of the
PINK1 gene T313M polymorphism in patients with PD and healthy
control individuals.
| Genotype
frequency | Allele frequency |
|---|
|
|
|
|---|
| Groups | No. of cases | T/T, n (%) | C/T, n (%) | C/C, n (%) | T type, n (%) | C type, n (%) |
|---|
| PD | 364 | 5 (1.4) | 0 (0.0) | 359 (98.6) | 10 (1.4) | 718 (98.6) |
| Control | 346 | 5 (1.4) | 0 (0.0) | 341 (98.6) | 10 (1.4) | 682 (98.6) |
 | Table IIAllele and genotype frequencies of the
PINK1 gene T313M polymorphism in patients with PD from Uygur and
Han populations compared with healthy control individuals. |
Table II
Allele and genotype frequencies of the
PINK1 gene T313M polymorphism in patients with PD from Uygur and
Han populations compared with healthy control individuals.
| | Genotype
frequency | Allele frequency |
|---|
| |
|
|
|---|
| Groups | No. of cases | T/T, n (%) | C/T, n (%) | C/C, n (%) | T type, n (%) | C type, n (%) |
|---|
| Uygur population |
| PD | 175 | 5 (2.9) | 0 (0.0) | 170 (97.1) | 10 (2.9) | 340 (97.1) |
| Control | 163 | 2 (1.2) | 0 (0.0) | 161 (98.8) | 4 (1.2) | 322 (98.8) |
| Han population |
| PD | 189 | 0 (0.0) | 0 (0.0) | 189 (100.0) | 0 (0.0) | 378 (100.0) |
| Control | 183 | 3 (1.6) | 0 (0.0) | 180 (98.4) | 6 (1.6) | 360 (98.4) |
When the Uygur and Han patients with PD were
compared, the genotype and allele frequencies were observed to
differ significantly between the two groups (χ2=5.475,
χ2=10.950, P<0.05; Table III). When the study subjects were
grouped according to age, (≤50 years and >50 years), the T313M
polymorphisms in the PD group compared with the control group
showed no significant difference (P>0.05; Table IV). Furthermore, when the study
subjects were grouped according to gender, T313M polymorphisms in
the PD group compared with the control group also showed no
significant difference (P>0.05, Table V).
 | Table IIIAllele and genotype frequencies of the
PINK1 gene T313M polymorphism in patients with PD from Uygur and
Han populations. |
Table III
Allele and genotype frequencies of the
PINK1 gene T313M polymorphism in patients with PD from Uygur and
Han populations.
| | Genotype
frequency | Allele frequency |
|---|
| |
|
|
|---|
| Groups | No. of cases | T/T, n (%) | C/T, n (%) | C/C, n (%) | T type, n (%) | C type, n (%) |
|---|
| Uygur with PD | 175 | 5 (2.9) | 0 (0.0) | 170 (97.1) | 10 (2.9) | 340 (97.1) |
| Han with PD | 189 | 0 (0.0) | 0 (0.0) | 189 (100.0) | 0 (0.0) | 378 (100.0) |
 | Table IVAssociation between age and allele and
genotype frequencies of the PINK1 gene T313M polymorphism in
patients with PD and healthy individuals |
Table IV
Association between age and allele and
genotype frequencies of the PINK1 gene T313M polymorphism in
patients with PD and healthy individuals
| | | Genotype
frequency | Allele frequency |
|---|
| | |
|
|
|---|
| Age | Group | No. of cases | T/T, n (%) | C/T, n (%) | C/C, n (%) | T type, n (%) | C type, n (%) |
|---|
| ≤50 years | PD | 80 | 1 (1.3) | 0 (0.0) | 79 (98.8) | 2 (1.3) | 158 (98.8) |
| Control | 74 | 0 (0.0) | 0 (0.0) | 74 (100.0) | 0 (0.0) | 148 (100.0) |
| >50 years | PD | 284 | 4 (1.4) | 0 (0.0) | 280 (98.6) | 8 (1.4) | 560 (98.6) |
| Control | 285 | 5 (1.8) | 0 (0.0) | 280 (98.2) | 10 (1.8) | 560 (98.2) |
 | Table VAssociation between gender and allele
and genotype frequencies of the PINK1 gene T313M polymorphism in
patients with PD and healthy individuals. |
Table V
Association between gender and allele
and genotype frequencies of the PINK1 gene T313M polymorphism in
patients with PD and healthy individuals.
| | | Genotype
frequency | Allele
frequency |
|---|
| | |
|
|
|---|
| Gender | Group | No. of cases | T/T, n (%) | C/T, n (%) | C/C, n (%) | T type, n (%) | C type, n (%) |
|---|
| Male | PD | 206 | 3 (1.5) | 0 (0.0) | 203 (98.5) | 6 (1.5) | 406 (98.5) |
| Control | 193 | 1 (0.5) | 0 (0.0) | 192 (99.5) | 2 (0.5) | 384 (99.5) |
| Female | PD | 158 | 2 (1.3) | 0 (0.0) | 156 (98.7) | 4 (1.3) | 312 (98.7) |
| Control | 153 | 4 (2.6) | 0 (0.0) | 149 (97.4) | 8 (2.6) | 298 (97.4) |
Discussion
Mutations in the PINK1 gene on chromosome 1p36 have
been reported in ~5% of patients with autosomal recessive PD
(13). PINK1 mutations have been
reported in patients with autosomal recessive early-onset PD (AREP)
in Italy (14,15). The frequency of PINK1 mutations in
patients with AREP is between 2.9 and 29%, with the mutation
frequency varying greatly depending on ethnicity (16–20).
In patients with sporadic early-onset PD (EOP), the pathogenic role
of PINK1 mutation is particularly important (13,21).
The mutation frequency range is wide. PINK1 gene mutation has not
been observed in patients with sporadic PD in the USA (22). Furthermore, among a Chinese
population in Taiwan, only one heterozygous PINK1 gene mutation was
found in 73 patients with sporadic EOP (22). In Italy, patients with sporadic EOP
have been reported to have relatively high mutation rates (14). Klein et al (15) observed that PINK1 gene mutations in
patients with sporadic EOP were similar to those in the parkin gene
(15). At present, few reports on
minority PD genes are available, particularly concerning the PINK1
T313M mutation in populations in Xinjiang, China.
The present study showed that genotype frequencies
and allele frequency distributions were not significantly different
between patients with sporadic PD and healthy control individuals
in the total study population. However, in patients with PD, the
distribution of T313M polymorphisms was significantly different
between patients from the Uygur and Han populations, and all were
homozygous mutants. These findings suggest that differences in gene
polymorphisms may exist between patients with PD from Uygur and Han
populations. These findings are inconsistent with those of Guo
et al (23) and Zhang et al
(24), who identified that in 120
patients with sporadic PD, as well as in patients with pedigree
mutations, mutations may not only exist in patients with familial
PD, but also those with sporadic PD in the Han population. However,
the sample size should be further expanded and uneven sampling may
be associated with the results observed. In the present study, in
the Uygur populations, the allele frequencies of the T313M
polymorphism in the PD group were not found to be significantly
different from those in the control group. However, in the Han
population, the allele frequencies were observed to be
significantly different in the PD group compared with the control
group. These findings differ from those obtained in other regions
of mainland China (25). When
grouped according to age and gender, the PD and control groups
showed no statistically significant difference. Further expansion
of the sample size of the patients with early-onset PD is necessary
to verify these findings. It has previously been reported that
homozygous or compound heterozygous PINK1 mutations are more common
in patients with AREP and single heterozygous mutations are found
in patients with sporadic EOP (19,26,27).
The present study showed that in the Xinjiang area, T313M
polymorphism of the PINK1 gene in the PD and control groups was a
homozygous mutation. Significant differences were found between the
Uygur and Han populations; the T allele frequency was 2.9% in the
Ugyur patients with PD, whereas the T allele was absent from the
Han patients with PD, suggesting that T313M gene polymorphism was
related to nationality. The Uygur population may be expected to
have an increased prevalence of PD due to the incidence of PINK1
mutations. However, no significant difference was observed between
the PD group and the control group, and the frequency of the type T
allele (1.4%) was not found to increase PD prevalence. In the
present study, no significant difference was observed in the allele
and genotype frequencies of the PINK1 gene T313M polymorphism in
patients aged ≤50 years compared with those aged >50 years. This
finding suggests that T313M may not be a predisposing factor of
early-onset PD.
The present study identified that differences in
T313M polymorphisms exist between the Uygur and Han populations.
The PINK1 gene may be associated with genetic susceptibility to PD,
particularly in the Han population, which may be associated with
the particular area of Xinjiang, living environment and genetic
background. Furthermore, sampling may be not uniform. Further
investigations, including a larger sample size, are required to
validate these results.
References
|
1
|
Koziorowski D, Hoffman-Zacharska D, Sławek
J, et al: Incidence of mutations in the PARK2, PINK1, PARK7 genes
in Polish early-onset Parkinson disease patients. Neurol Neurochir
Pol. 47:319–324. 2013.PubMed/NCBI
|
|
2
|
Vincow ES, Merrihew G, Thomas RE, et al:
The PINK1-Parkin pathway promotes both mitophagy and selective
respiratory chain turnover in vivo. Proc Natl Acad Sci USA.
110:6400–6405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Billia F, Hauck L, Grothe D, et al:
Parkinson-susceptibility gene DJ-1/PARK7 protects the murine heart
from oxidative damage in vivo. Proc Natl Acad Sci USA.
110:6085–6090. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Valente EM, Abou-Sleiman PM, Caputo V, et
al: Hereditary early-onset Parkinson’s disease caused by mutations
in PINK1. Science. 304:1158–1160. 2004.
|
|
5
|
Valente EM, Bentivoglio AR, Dixon PH, et
al: Localization of a novel locus for autosomal recessive
early-onset parkinsonism, PARK6, on human chromosome 1p35–p36. Am J
Hum Genet. 68:895–900. 2001.PubMed/NCBI
|
|
6
|
Rogaeva E, Johnson J, Lang AE, et al:
Analysis of the PINK1 gene in a large cohort of cases with
Parkinson disease. Arch Neurol. 61:1898–1904. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Weng YH, Chou YH, Wu WS, et al: PINK1
mutation in Taiwanese early-onset parkinsonism : clinical, genetic,
and dopamine transporter studies. J Neurol. 254:1347–1355. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ephraty L, Porat O, Israeli D, et al:
Neuropsychiatric and cognitive features in autosomal-recessive
early parkinsonism due to PINK1 mutations. Mov Disord. 22:566–569.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kumazawa R, Tomiyama H, Li Y, et al:
Mutation analysis of the PINK1 gene in 391 patients with Parkinson
disease. Arch Neurol. 65:802–808. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deng H, Le W, Shahed J, et al: Mutation
analysis of the parkin and PINK1 genes in American Caucasian
early-onset Parkinson disease families. Neurosci Lett. 430:18–22.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang BR, Hu ZX, Yin XZ, et al: Mutation
analysis of parkin and PINK1 genes in early-onset Parkinson’s
disease in China. Neurosci Lett. 477:19–22. 2010.
|
|
12
|
Hughes AJ, Daniel SE, Kilford L and Lees
AJ: Accuracy of clinical diagnosis of idiopathic Parkinson’s
disease: a clinico-pathological study of 100 cases. J Neurol
Neurosurg Psychiatry. 55:181–184. 1992.
|
|
13
|
Hilker R, Pilatus U, Eggers C, et al: The
bioenergetic status relates to dopamine neuron loss in familial PD
with PINK1 mutations. PLoS One. 7:e513082012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Scornaienchi V, Civitelli D, De Marco EV,
et al: Mutation analysis of the PINK1 gene in Southern Italian
patients with early- and late-onset parkinsonism. Parkinsonism
Relat Disord. 18:651–653. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Klein C, Djarmati A, Hedrich K, et al:
PINK1, Parkin, and DJ-1 mutations in Italian patients with
early-onset parkinsonism. Eur J Hum Genet. 13:1086–1093. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Moura KC, Campos Junior M, de Rosso AL, et
al: Genetic analysis of PARK2 and PINK1 genes in Brazilian patients
with early-onset Parkinson’s disease. Dis Markers. 35:181–185.
2013.
|
|
17
|
Lohmann E, Dursun B, Lesage S, et al:
Genetic bases and phenotypes of autosomal recessive Parkinson
disease in a Turkish population. Eur J Neurol. 19:769–775. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yonova-Doing E, Atadzhanov M, Quadri M, et
al: Analysis of LRRK2, SNCA, Parkin, PINK1, and DJ-1 in Zambian
patients with Parkinson’s disease. Parkinsonism Relat Disord.
18:567–571. 2012.PubMed/NCBI
|
|
19
|
Zhang X, Zhang H, Liao B, Guo J, Xia K and
Tang B: Mutation analysis of PINK1 gene in patients with
early-onset Parkinsonism. Zhong Nan Da Xue Xue Bao Yi Xue Ban.
36:490–497. 2011.PubMed/NCBI
|
|
20
|
Keyser RJ, Lesage S, Brice A, Carr J and
Bardien S: Assessing the prevalence of PINK1 genetic variants in
South African patients diagnosed with early- and late-onset
Parkinson’s disease. Biochem Biophys Res Commun. 398:125–129.
2010.PubMed/NCBI
|
|
21
|
Kondapalli C, Kazlauskaite A, Zhang N, et
al: PINK1 is activated by mitochondrial membrane potential
depolarization and stimulates Parkin E3 ligase activity by
phosphorylating serine 65. Open Biol. 2:1200802012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fung HC, Chen CM, Hardy J, et al: Analysis
of the PINK1 gene in a cohort of patients with sporadic early-onset
parkinsonism in Taiwan. Neurosci Lett. 394:33–36. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo JF, Xiao B, Liao B, et al: Mutation
analysis of Parkin, PINK1, DJ-1 and ATP13A2 genes in Chinese
patients with autosomal recessive early-onset Parkinsonism. Mov
Disord. 23:2074–2079. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang YH, Tang BS, Guo JF, et al: Mutation
analysis of PINK1 gene in Chinese patients with autosomal recessive
early-onset parkinsonism type 6. Zhonghua Yi Xue Za Zhi.
85:1538–1541. 2005.(In Chinese).
|
|
25
|
Guo JF, Zhang XW, Nie LL, et al: Mutation
analysis of Parkin, PINK1 and DJ-1 genes in Chinese patients with
sporadic early onset parkinsonism. J Neurol. 257:1170–1175. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bonifati V, Rohé CF, Breedveld GJ, et al:
Early-onset parkinsonism associated with PINK1 mutations:
frequency, genotypes, and phenotypes. Neurology. 65:87–95. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moura KC, Junior MC, de Rosso AL, et al:
Exon dosage variations in Brazilian patients with Parkinson’s
disease: analysis of SNCA, PARKIN, PINK1 and DJ-1 genes. Dis
Markers. 32:173–178. 2012.PubMed/NCBI
|