Introduction
Hypertension has an increased prevalence in cold
regions or during winter. Cold winters increase the severity of
hypertension and trigger myocardial infarction and stroke in
hypertensive patients, leading to high mortality and morbidity
rates from cardiovascular complications (1–6).
Therefore, it is important to further investigate the mechanisms
underlying cold-induced hypertension (CIH). Previous studies that
have focused on clinical epidemiology in humans have indicated that
cold exposure is a risk factor of hypertension (6–8).
However, the cause of CIH remains unknown. The
thermoregulatory-vascular remodeling hypothesis indicates that
avoiding the ingestion of sodium chloride is key to preventing
hypertension (9). By contrast,
advances in mechanistic studies of CIH in animals indicate that the
activated sympathetic nervous system (10,11)
initiates CIH via the renin-angiotensin system (12,13).
Additional studies have also revealed that cold exposure suppresses
the expression of endothelial nitric oxide synthase and thus the
formation of nitric oxide (14),
as well as increasing the production of endothelin-1 (15). The hypothesis that hypertension is
caused by increased oxidative stress remains controversial
(16–20).
In the present study, gene expression data from
cold-exposed mice, generated by an Affymetrix microarray, were
selected from the Gene Expression Omnibus (GEO) database. Gene
expression analyses were performed with the aim of identifying the
expression patterns of CIH-associated genes. Integrating the
results of the present study with previous related studies allowed
a more comprehensive interpretation of the molecular mechanism
underlying CIH to be constructed at the gene expression level.
Materials and methods
Microarray data
The GSE13432 dataset was downloaded from the GEO
database (http://www.ncbi.nlm.nih.gov/geo/). Sample information
of the 12 C57/Bl6 mice that were maintained at 30 or 4°C for 1 or 5
weeks is listed in Table I. Data
from the three mice in each group were used for gene expression
analysis using the Mouse Genome 430 2.0 Array system (Affymetrix,
Inc., Santa Clara, CA, USA). The robust multichip analysis
algorithm was used to polish the microarray data prior to further
analysis (21).
 | Table ICharacteristics of the sample
information. |
Table I
Characteristics of the sample
information.
| GEO
identification | Sample
description | Sample name | Raw file data |
|---|
| GSM338983 | 1 week 30°C rep
1 | C1-1 | GSM338983.cel.gz |
| GSM338984 | 1 week 30°C rep
2 | C1–2 | GSM338984.cel.gz |
| GSM338985 | 1 week 30°C rep
3 | C1-3 | GSM338985.cel.gz |
| GSM338986 | 1 week 4°C rep 1 | D1-1 | GSM338986.cel.gz |
| GSM338987 | 1 week 4°C rep 2 | D1-2 | GSM338987.cel.gz |
| GSM338988 | 1 week 4°C rep 3 | D1-3 | GSM338988.cel.gz |
| GSM338989 | 5 weeks 30°C rep
1 | C5-1 | GSM338989.cel.gz |
| GSM338990 | 5 weeks 30°C rep
2 | C5-2 | GSM338990.cel.gz |
| GSM338991 | 5 weeks 30°C rep
3 | C5-3 | GSM338991.cel.gz |
| GSM338992 | 5 weeks 4°C rep
1 | D5-1 | GSM338992.cel.gz |
| GSM338993 | 5 weeks 4°C rep
2 | D5-2 | GSM338993.cel.gz |
| GSM338994 | 5 weeks 4°C rep
3 | D5-3 | GSM338994.cel.gz |
Differentially expressed gene (DEG)
detection
Based on the Bayesian statistical model, DEGs were
identified between the 4 and 30°C groups at the various timescales
(4°C 1 week/30°C 1 week, 4°C 5 weeks/30°C 5 weeks). Statistical
t-tests with multiple test correction using the Benjamini and
Hochberg procedure (22) were
performed with the threshold of significantly expressed genes set
at a false discovery rate (FDR) of 0.01. Up- or downregulation of
DEGs was determined as fold change.
Gene Ontology (GO) and pathway analysis
of DEGs
All cold-induced DEGs were mapped according to the
GO (http://www.geneontology.org) and Kyoto
Encyclopedia of Genes and Genomes pathway terms (http://www.genome.jp/kegg/). Multiple models for
pathway enrichment evaluation were utilized (23,24).
Blood pressure-associated DEGs were defined by blood
pressure-associated GO function terms, while
hypertension-associated DEGs were selected from
hypertension-associated pathways identified by known hypertension
genes in the Online Mendelian Inheritance in Man (OMIM) database
and the Mammalian Phenotype Ontology in the Mouse Genome
Informatics (MGI) database (http://www.omim.org and http://www.informatics.jax.org/). Odds ratios and FDRs
were calculated with a threshold FDR value of 0.05 to identify
statistical significance of pathway enrichment. Kernel principal
component analysis was used to evaluate the overall gene expression
differences between samples. All these procedures were performed
with R statistical software (v 2.14.1; http://www.r-project.org/), and Bioconductor limma
packages (3.12.1) and libraries (25) (http://www.bioconductor.org/packages/devel/bioc/html/limma.html).
Protein-protein interaction (PPI) network
analysis
Network inference was performed using Cytoscape
(26) (v 2.8.3; http://www.cytoscape.org/). The interaction dataset
between genes was downloaded from the Biomolecular Interaction
Network Database (http://bind.ca) and the MIPS Mammalian
Protein-Protein Interaction Database (MPPI; http://mips.helmholtz-muenchen.de/proj/ppi/). The
network was analyzed for cold-induced DEGs associated with blood
pressure regulation and hypertension in two case/control groups
(4°C for 1 week vs. 30°C for 1 week and 4°C for 5 weeks vs. 30°C
for 5 weeks).
Results
Cold-induced DEG profiling
The downloaded GSE13432 dataset contained the gene
expression data of four groups of mice (three replicates in each)
exposed to four conditions: 4°C for 1 and 5 weeks and their
respective controls at 30°C for 1 and 5 weeks (27). In order to demonstrate the gene
expression level changes that resulted from cold exposure, analysis
of DEGs was performed between the cold exposure mice and the
control groups for the two timescales. Multiple testing with FDR
control was conducted to improve the accuracy of the results.
In total, 5,183 DEGs were identified in the 4°C 1
week vs. 30°C 1 week group, while 5,580 DEGs were identified in the
4°C 5 week vs. 30°C 5 week group (FDR, <0.01). Among these DEGs,
2,800 genes maintained increased or decreased expression levels at
week 1 and 5, while >2,000 genes were detected only at week 1 or
5, as shown in Fig. 1.
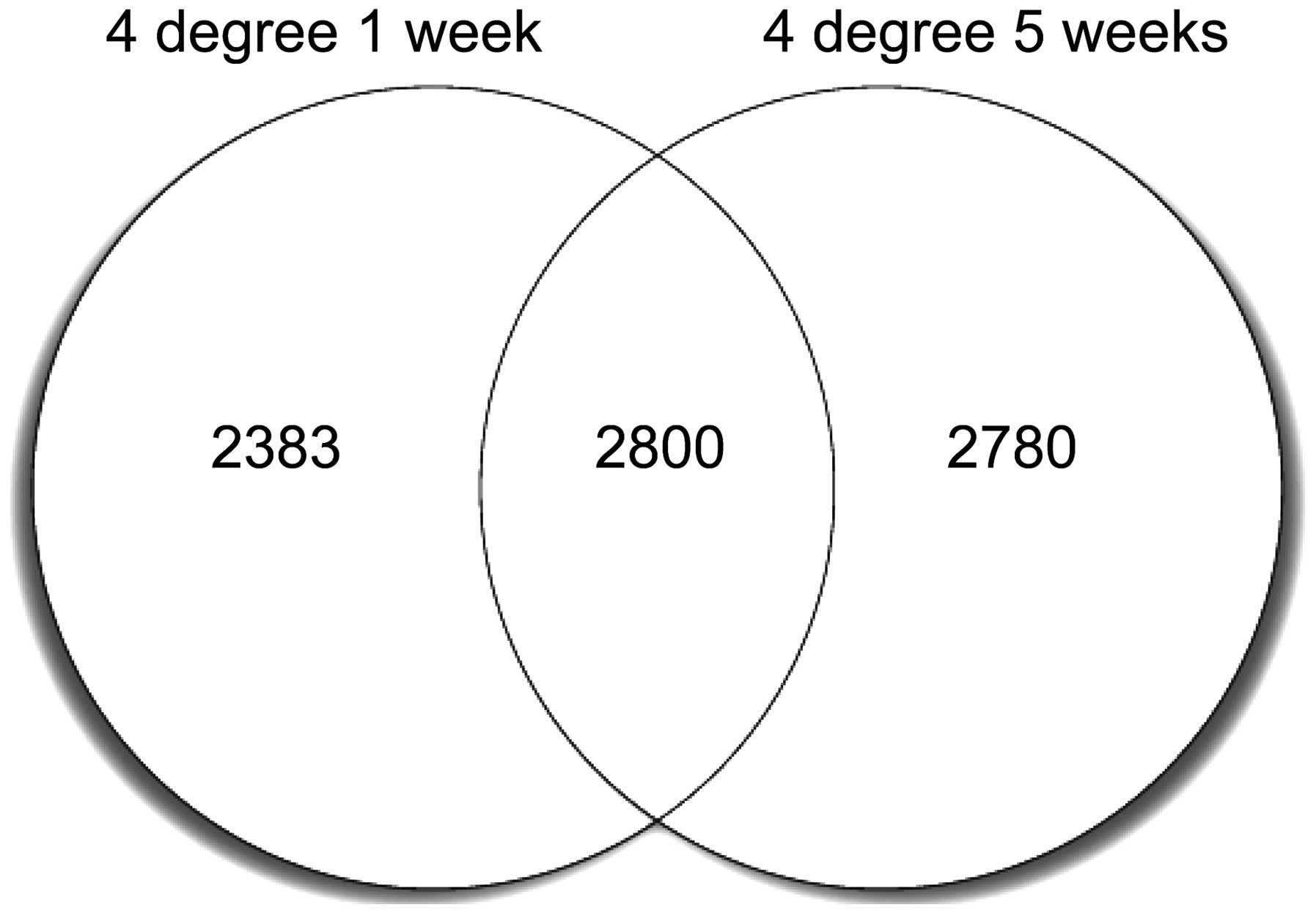 | Figure 1Venn plot showing the identified DEGs
in the two cold-exposure groups. A total of 5,183 DEGs were
identified in the 4°C 1 week vs. 30°C 1 week group, while 5,580
DEGs were identified in the 4°C 5 week vs. 30°C 5 week group (FDR,
<0.01). Among the DEGs, 2,800 genes maintained increased or
decreased expression levels after week 1 and 5, while >2,000
genes were detected only at week 1 or 5. DEGs, differentially
expressed genes; FDR, false discovery rate. |
With regard to the most significant DEGs, the
expression levels of uncoupling protein 1 (UCP1), a
mitochondrial proton carrier, and a series of other fatty acid
metabolic genes, including ELOVL3, FABP3 and
CPT1B, were markedly elevated in the two cold exposure
groups. In addition, energy metabolism genes associated with
oxidative phosphorylation, the citrate cycle and mitochondrial
function were also shown to be upregulated, including the
ubiquinol-cytochrome c reductase (UQCR) and
cytochrome c oxidase (COX) gene families. Peroxisome
genes were also activated in the two groups. Notably, following
cold exposure, the number of downregulated genes was nearly double
the number of upregulated genes.
Cold exposure regulates the expression
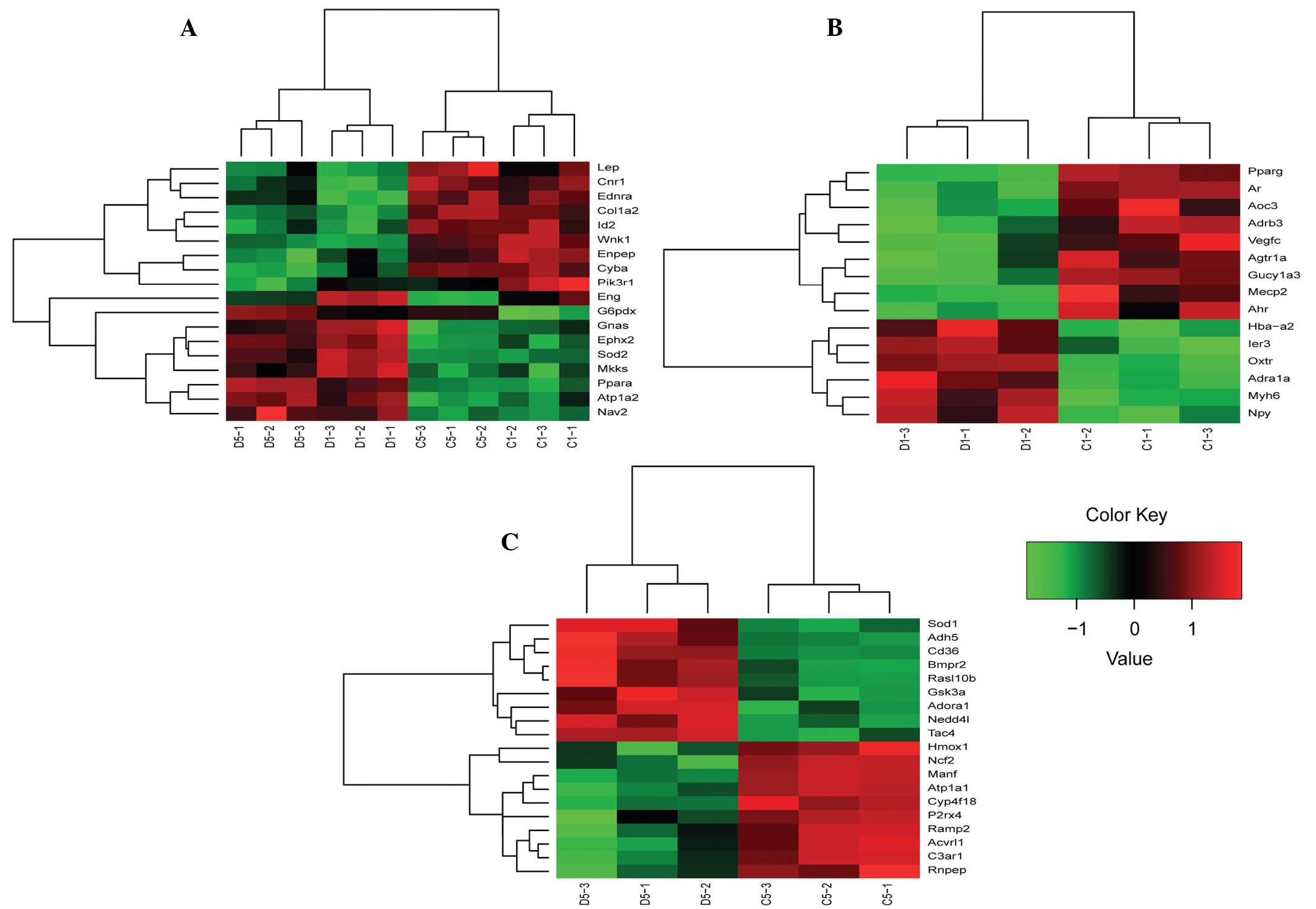
levels of blood pressure-associated genes
A total of 52 DEGs associated with blood pressure
regulation were selected according to the GO terms and
descriptions. Expression patterns were constructed with clustering
analysis and are shown in Fig. 2.
In total, 18 DEGs were observed that were differentially expressed
after 1 and 5 weeks of exposure. In addition, six genes were
identified to have a larger difference at the expression level with
long term exposure, including superoxide dismutase 2 (SOD2),
glucose-6-phosphate dehydrogenase X-linked (G6pdx), WNK
lysine deficient protein kinase 1 (Wnk1), peroxisome
proliferator activated receptor α (Ppara), cytochrome b-245,
α polypeptide (Cyba) and ATPase,
Na+/K+ transporting, α 2 polypeptide
(Atp1a2). Furthermore, there were a number of 1 week
specific DEGs that exhibited a transient expression pattern
following 1 week exposure, but returned to the initial values by
week 5, including angiotensin II receptor type 1a (Agtr1a)
and adrenergic receptor α 1a (Adra1a). There were also
specific genes considered to be slow-response genes that were only
observed after 5 weeks of cold exposure.
Hypertension-associated pathway
analysis
In order to exclude unassociated pathways that were
caused by cold acclimation, the OMIM and MGI databases were
searched and 51 genes were identified as known
hypertension-associated genes. In order to identify how these genes
were affected by cold exposure, enrichment analysis was performed
in the associated pathways. Nervous system-associated pathways,
including Huntington’s and Alzheimer’s diseases, were found to be
significantly enriched (odds ratio, >2; FDR,
<10−6) in the two cold exposure groups. In addition,
immune system pathways were specifically enriched in the 5 week
exposure group, including the chemokine signaling pathway, B cell
receptor signaling pathway and Fc γ R-mediated phagocytosis
(Table II).
 | Table IIKEGG enriched pathways of
hypertension-associated genes in the OMIM and MGI databases. |
Table II
KEGG enriched pathways of
hypertension-associated genes in the OMIM and MGI databases.
| Pathways | Odds ratio | P-value | FDR | Subclass |
|---|
| 4°C 1 week group |
| Huntington’s
disease | 3.087 | 7.465E-13 | 4.852E-11 | Neurodegenerative
disease |
| Alzheimer’s
disease | 2.924 | 1.590E-11 | 8.270E-10 | Neurodegenerative
disease |
| Peroxisome | 2.824 | 3.937E-06 | 1.462E-04 | Transport and
catabolism |
| Fatty acid
metabolism | 3.635 | 2.425E-05 | 6.736E-04 | Lipid metabolism |
| 4°C 5 week group |
| Huntington’s
disease | 2.631 | 6.559E-10 | 4.263E-08 | Neurodegenerative
disease |
| Alzheimer’s
disease | 2.433 | 1.903E-08 | 8.246E-07 | Neurodegenerative
disease |
| Phagosome | 2.364 | 7.954E-08 | 2.954E-06 | Transport and
catabolism |
| Epstein-Barr virus
infection | 1.988 | 1.306E-06 | 3.525E-05 | Infectious
disease |
| Toxoplasmosis | 2.483 | 1.746E-06 | 4.127E-05 | Infectious
disease |
| Chemokine
signaling | 1.976 | 4.920E-06 | 9.841E-05 | Immune system |
| Osteoclast
differentiation | 1.989 | 1.207E-04 | 1.847E-03 | Development |
| B cell receptor
signaling | 2.330 | 1.886E-04 | 2.725E-03 | Immune system |
| Tuberculosis | 1.744 | 2.129E-04 | 2.913E-03 | Infectious
disease |
| Hepatitis B | 1.807 | 3.237E-04 | 4.208E-03 | Infectious
disease |
| Viral
carcinogenesis | 1.618 | 4.905E-04 | 5.545E-03 | Cancer |
| Fc γ R-mediated
phagocytosis | 2.038 | 7.372E-04 | 7.986E-03 | Immune system |
| Leishmaniasis | 2.108 | 1.980E-03 | 1.716E-02 | Infectious
disease |
| Pertussis | 2.013 | 2.506E-03 | 1.975E-02 | Infectious
disease |
| Acute myeloid
leukemia | 2.112 | 3.787E-03 | 2.825E-02 | Cancer |
| Hepatitis C | 1.605 | 5.119E-03 | 3.597E-02 | Infectious
disease |
| Peroxisome | 1.825 | 5.296E-03 | 3.624E-02 | Transport and
catabolism |
| Estrogen
signaling | 1.723 | 5.638E-03 | 3.758E-02 | Endocrine
system |
| Fatty acid
metabolism | 2.168 | 8.170E-03 | 4.968E-02 | Lipid
metabolism |
| T cell receptor
signaling | 1.623 | 8.245E-03 | 4.968E-02 | Immune system |
| Leukocyte
transendothelial migration | 1.597 | 8.364E-03 | 4.968E-02 | Immune system |
PPI network
According to the MPPI database, numerous genes (4°C
1 week group, 327; 4°C 5 week group, 515) were identified for
construction of the network. It was possible to organize the
majority of the genes into a single network. Proteins associated
with signal transduction were observed in the PPI network of the
4°C 1 week group, including certain proteins associated with the
hypoxia-inducible factor (HIF-1) and mitogen-activated protein
kinase (MAPK) signaling pathways. A number of additional proteins
associated with the immune system were identified in the 4°C 5-week
mice, including mammalian target of rapamycin (mTOR) and B and T
cell receptor signaling pathways.
Discussion
Cold exposure is hypothesized to be an important
factor contributing to hypertension. However, the pathogenic
mechanism of CIH is not fully understood. Although hypertension is
defined as a condition of high blood pressure, the disorder is
complex with numerous phenotypes and a number of causative factors,
including genetics and environmental factors (8). Thus, mice models that were only
exposed to cold were selected for the present study. The results of
the present study indicate that a myriad of genes were expressed
differentially when adapting to cold acclimation. Genes associated
with thermogenesis (UCP1) and energy metabolism (UQCR
and COX families) were activated as more heat and energy
were required to maintain temperature homeostasis (9). Thus, the compensatory increase in the
metabolic rate causes tissue hypoxia, as previously demonstrated
(27). The expression of
endothelial PAS domain-containing protein 1 (Epas1) was
upregulated, indicating that the HIF signaling pathway was
activated. HIF-associated genes also exhibited regulated expression
levels and were observed in the PPI networks. Genes associated with
oxidative stress, including SOD2 and epoxide hydrolase 2
(EPHX2), were also shown to be significantly upregulated
with DEG and pathway analyses of blood pressure-associated genes.
In addition, hypertension-associated gene analysis revealed that
pathways associated with oxidative stress were markedly enriched in
the initial exposure group (4°C for 1 week). Huntington’s and
Alzheimer’s disease pathways are associated with mitochondrial
dysfunction and the peroxisome pathway plays a crucial role in free
radical detoxification. Furthermore, a number of additional immune
system pathways were enriched in the chronic exposure group (4°C
for 5 week), which may further induce inflammatory reactions and
increase oxidative stress (28–30).
Therefore, the results of the present study indicate that an
imbalance in reactive oxygen species and superoxide caused by
hypoxia and oxidative stress may contribute to the initiation of
hypertension, while chronic cold exposure may accumulate the effect
of oxidative stress in the process of hypertension.
References
|
1
|
Aubinière-Robb L, Jeemon P, Hastie CE, et
al: Blood pressure response to patterns of weather fluctuations and
effect on mortality. Hypertension. 62:190–196. 2013.PubMed/NCBI
|
|
2
|
Fu S, Cao Y and Li Y: Epidemiological
study of hypertension in Heilongjiang province. Zhonghua Nei Ke Za
Zhi. 41:114–116. 2002.(In Chinese).
|
|
3
|
Gyllerup S, Lanke J, Lindholm LH and
Scherstén B: High coronary mortality in cold regions of Sweden. J
Intern Med. 230:479–485. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jakovljević D, Salomaa V, Sivenius J, et
al: Seasonal variation in the occurrence of stroke in a Finnish
adult population. The FINMONICA Stroke Register Finnish Monitoring
Trends and Determinants in Cardiovascular Disease. Stroke.
27:1774–1779. 1996.
|
|
5
|
Seretakis D, Lagiou P, Lipworth L,
Signorello LB, Rothman KJ and Trichopoulos D: Changing seasonality
of mortality from coronary heart disease. JAMA. 278:1012–1014.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun Z: Cardiovascular responses to cold
exposure. Front Biosci (Elite Ed). 2:495–503. 2010. View Article : Google Scholar
|
|
7
|
Kim JY, Jung KY, Hong YS, Kim JI, Jang TW
and Kim JM: The relationship between cold exposure and
hypertension. J Occup Health. 45:300–306. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brook RD, Weder AB and Rajagopalan S:
‘Environmental hypertensionology’ the effects of environmental
factors on blood pressure in clinical practice and research. J Clin
Hypertens (Greenwich). 13:836–842. 2011.
|
|
9
|
Blankfield RP: The
thermoregulatory-vascular remodeling hypothesis: an explanation for
essential hypertension. Med Hypotheses. 66:1174–1178. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun Z, Cade R and Morales C: Role of
central angiotensin II receptors in cold-induced hypertension. Am J
Hypertens. 15:85–92. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Patel D, Böhlke M, Phattanarudee S, Kabadi
S, Maher TJ and Ally A: Cardiovascular responses and
neurotransmitter changes during blockade of angiotensin II
receptors within the ventrolateral medulla. Neurosci Res.
60:340–348. 2008. View Article : Google Scholar
|
|
12
|
Sun Z, Fregly MJ and Cade JR: Effect of
renal denervation on elevation of blood pressure in cold-exposed
rats. Can J Physiol Pharmacol. 73:72–78. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang X, Sun Z and Cade R: Prolonged
attenuation of cold-induced hypertension by adenoviral delivery of
renin antisense. Kidney Int. 68:680–687. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun Z, Wang X, Wood CE and Cade JR:
Genetic AT1A receptor deficiency attenuates cold-induced
hypertension. Am J Physiol Regul Integr Comp Physiol.
288:R433–R439. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen GF and Sun Z: Effects of chronic cold
exposure on the endothelin system. J Appl Physiol (1985).
100:1719–1726. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grossman E: Does increased oxidative
stress cause hypertension? Diabetes Care. 31(Suppl 2): S185–S189.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nishiyama A, Yao L, Nagai Y, et al:
Possible contributions of reactive oxygen species and
mitogen-activated protein kinase to renal injury in
aldosterone/salt-induced hypertensive rats. Hypertension.
43:841–848. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Park JB, Touyz RM, Chen X and Schiffrin
EL: Chronic treatment with a superoxide dismutase mimetic prevents
vascular remodeling and progression of hypertension in salt-loaded
stroke-prone spontaneously hypertensive rats. Am J Hypertens.
15:78–84. 2002. View Article : Google Scholar
|
|
19
|
Rodriguez-Iturbe B, Zhan CD, Quiroz Y,
Sindhu RK and Vaziri ND: Antioxidant-rich diet relieves
hypertension and reduces renal immune infiltration in spontaneously
hypertensive rats. Hypertension. 41:341–346. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tanito M, Nakamura H, Kwon YW, et al:
Enhanced oxidative stress and impaired thioredoxin expression in
spontaneously hypertensive rats. Antioxid Redox Signal. 6:89–97.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Giorgi FM, Bolger AM, Lohse M and Usadel
B: Algorithm-driven artifacts in median polish summarization of
microarray data. BMC Bioinformatics. 11:5532010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. Journal of the Royal Statistical Society, Series
B. 57:289–300. 1995.
|
|
23
|
Kirik U, Cifani P, Albrekt AS, Lindstedt
M, Heyden A and Levander F: Multimodel pathway enrichment methods
for functional evaluation of expression regulation. J Proteome Res.
11:2955–2967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shen K and Tseng GC: Meta-analysis for
pathway enrichment analysis when combining multiple genomic
studies. Bioinformatics. 26:1316–1323. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Smyth GK, Michaud J and Scott HS: Use of
within-array replicate spots for assessing differential expression
in microarray experiments. Bioinformatics. 21:2067–2075. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Saito R, Smoot ME, Ono K, et al: A travel
guide to Cytoscape plugins. Nat Methods. 9:1069–1076. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xue Y, Petrovic N, Cao R, et al:
Hypoxia-independent angiogenesis in adipose tissues during cold
acclimation. Cell Metab. 9:99–109. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pruijm M, Hofmann L, Vogt B, et al: Renal
tissue oxygenation in essential hypertension and chronic kidney
disease. Int J Hypertens. 2013:6965982013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nangaku M and Eckardt KU: Hypoxia and the
HIF system in kidney disease. J Mol Med (Berl). 85:1325–1330. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Prchal JT: Delivery on demand - a new era
of gene therapy? N Engl J Med. 348:1282–1283. 2003. View Article : Google Scholar : PubMed/NCBI
|