Introduction
Osteosarcoma (OS) accounts for ~2.5% of all
malignancies in pediatric patients and ~ 20% of all primary bone
cancers (1), with a morphological
and malignant heterogeneity (2).
The majority of OS variant cells are extremely aggressive, with a
capability of rapid growth and early metastasis. Currently, >30%
of OS patients with localized disease eventually develop distant
metastases, mostly to the lungs and bones (3), even following chemotherapy and
surgical treatment. The outcome of OS patients has not
significantly improved over the last 20 years, and there has been
no significant advance in OS treatment, as the molecular mechanism
underlying the highly efficient proliferation and migration of OS
cells remains largely unknown. Thus, there is an urgency to
identify the details regarding tumor progression and to develop
novel therapy strategies for this disease.
microRNAs (miRNAs or miRs) are endogenous non-coding
RNAs with 18–24 nucleotides, which regulate gene expression
(4) by binding the target mRNA’s
3′ untranslated region (5), in a
wide range of organisms, and in a broad array of cell processes in
mammals (5–7). It is well known that cancer is driven
by the deregulation of a complexity of oncogenic and tumor
suppressive genes, and emerging evidence shows that miRNAs are
deregulated in various types of cancer (8–10),
and play oncogenic and tumor suppressive roles, contributing to
tumor formation and development (11–13).
Recently, various miRNAs have been confirmed to be deregulated in
OS (14,15). The oncogenic miRNA, miR-21,
which is aberrantly overexpressed in numerous types of tumor and
induces cancer cell growth, migration, invasion and metastasis
(16,17), has also been indicated to be
significantly overexpressed in OS tissues and induces invasion and
migration of the OS cell line, MG-63, by negatively regulating the
tumor suppressor gene, reversion-inducing-cysteine-rich protein
with kazal motifs (18). The
oncogenic miR-93 also induces proliferation and invasion in
OS (19), whereas miR-20a
promotes OS metastasis by regulating Fas expression
(20). By contrast, the tumor
suppressive miRNAs, including miR-199a-3p (21), miR-125b (22), miR-143 (23), miR-382 and miR-134
(24), are significantly
downregulated in OS cells and attenuate proliferation and
inhibition of migration, reduce cell viability and induce
apoptosis. miR-155 is well identified as an oncogenic miRNA
in leukemia (25,26) and breast cancer (14), contributing to tumorigenicity and
progression.
Neoadjuvant chemotherapy has improved the cure rate
of OS patients (27,28). However, patients that are not
sensitive to these drugs have a poor prognosis. In addition, the
frequent acquisition of drug-resistance is often associated with
chemotherapy and is a significant obstacle to achieving favorable
outcomes. Thus, exploring novel targets for therapy and developing
more effective treatment strategies for this disease is required.
Recently, Lauvrak et al (29) identified that miR-155
overexpression in OS cell lines was associated with aggressive
cancer phenotypes. In the present study, the aim was to evaluate
whether miR-155 is a sensitive target for therapy. The
regulatory role of miR-155 was determined in the
proliferation, invasion and migration of OS cells. Subsequently,
the miR-155 inhibitor was evaluated for its inhibition on
the OS cell proliferation and migration. The results demonstrated
that the miR-155 mimic significantly increased, whereas the
miR-155 inhibitor significantly reduced the proliferation
and migration of OS MG-63 cells. Therefore, the study revealed
miR-155 as a possible therapeutic target for OS.
Materials and methods
Reagents and cell culture
The human OS cell line, MG-63, was obtained from the
Cell Resource Center of the Chinese Academy of Medical Sciences
(Beijing, China). MG-63 cells were cultured in Eagle’s Minimum
Essential Medium (EMEM) (Invitrogen, Carlsbad, CA, USA),
supplemented with 2 mM glutamine, 1% non-essential amino acids and
10% fetal bovine serum (FBS) (Invitrogen). The cells were incubated
at 37°C with 5% CO2. The miR-155 mimic (Qiagen,
Valencia, CA, USA) or inhibitor (Qiagen) was used to elevate or
reduce the miR-155 level via lipofectamine 2000
(Invitrogen). miR-Con was used as a control.
RNA extraction and reverse transcription
quantitative polymerase chain reaction (RT-qPCR) miR-155 assay
The mirVana miRNA Isolation kit (Ambion, Austin, TX,
USA) was used to extract miRNAs from the MG-63 cells, and the
mirVana RT-qPCR miRNA Detection kit (Ambion) was used to quantify
the miR-155 expression, with the U6 small nuclear RNA as the
internal control. ΔΔCt method was used for relative quantification
(30). The RT-qPCR was performed
using SYBR Green with the LightCycle 2.0 (Roche Diagnostics GmbH,
Mannheim, Germany).
Cell viability assay and cell colony
formation assay
The MTT assay was adopted to determine the cell
viability. MG-63 cells were seeded in 96-well plates and
transfected with the miR-155 mimic, inhibitor or control,
with ~85% confluence. The cells were washed with warm PBS 6 h
post-tranfection and were replaced with RPMI-1640 medium containing
1% FBS, and were cultured for various time. Subsequently, the MTT
assay was conducted. Briefly, the incubation medium in the cell
wells was replaced with 50 μl 1× MTT solution, and the cells were
incubated for 2 h at 37°C. Post-incubation, the MTT solution was
discarded and 150 μl DMSO was added to dissolve the precipitate
completely at room temperature. The optical density was measured at
570 nm using a spectrophotometer, the cell viability was expressed
as relative viable cells (%) to the control MG-63 cells. For the
cell colony formation assay, 2×103 cells were incubated
in 6-well plates at 37°C containing 5% CO2. Ten days
post-incubation, the cells were stained with crystal violet
(0.005%) for 30 min and the colony numbers were recorded by Image J
software (National Institutes of Health, Bethesda, MD, USA). For
the proliferation assay, post-transfection with the miR-155
mimic, inhibitor or control, cells were incubated in cell counting
kit 8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan) for various
times. The 450 nm absorbance of each well was detected following
visual color occurrence.
Cell migration and invasion assay
The cell migration was determined by the scratch
assay. The cells were cultivated to 90% confluence on 12-well
plates and were transfected with the miR-155 mimic,
inhibitor or control. Subsequently, Cell Scrapers (Corning Inc.,
Corning, NY, USA) were utilized to scratch the confluent cells 24 h
post-transfection. The procedures of cellular growth were observed
at 0 and 96 h. All the experiments were repeated in triplicate. The
Transwell migration chambers were used to evaluate the MG-63 cell
invasion. The cells were first seeded at a density of
1×105 cells in serum-free media on the upper chamber
with the non-coated membrane (8 μm pore size; Millipore, Zug,
Switzerland). The lower chamber contained EMEM with 20% FBS as a
chemoattractant. The cells in the upper chamber were discarded
using cotton wool after 24 h and the migration cells in the lower
chamber were counted using a microscope (Olympus, Tokyo, Japan).
All the experiments were repeated in triplicate.
Statistical analysis
The results are expressed as mean ± standard error.
Student’s t-test was performed to compare the differences between
two groups. Statistical analysis was conducted by SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference; and in
particular, the results are shown as no significance,
*P<0.05, **P<0.01 or
***P<0.001.
Results
miR-155 inhibitor reduces the viability
and proliferation of MG-63 cells
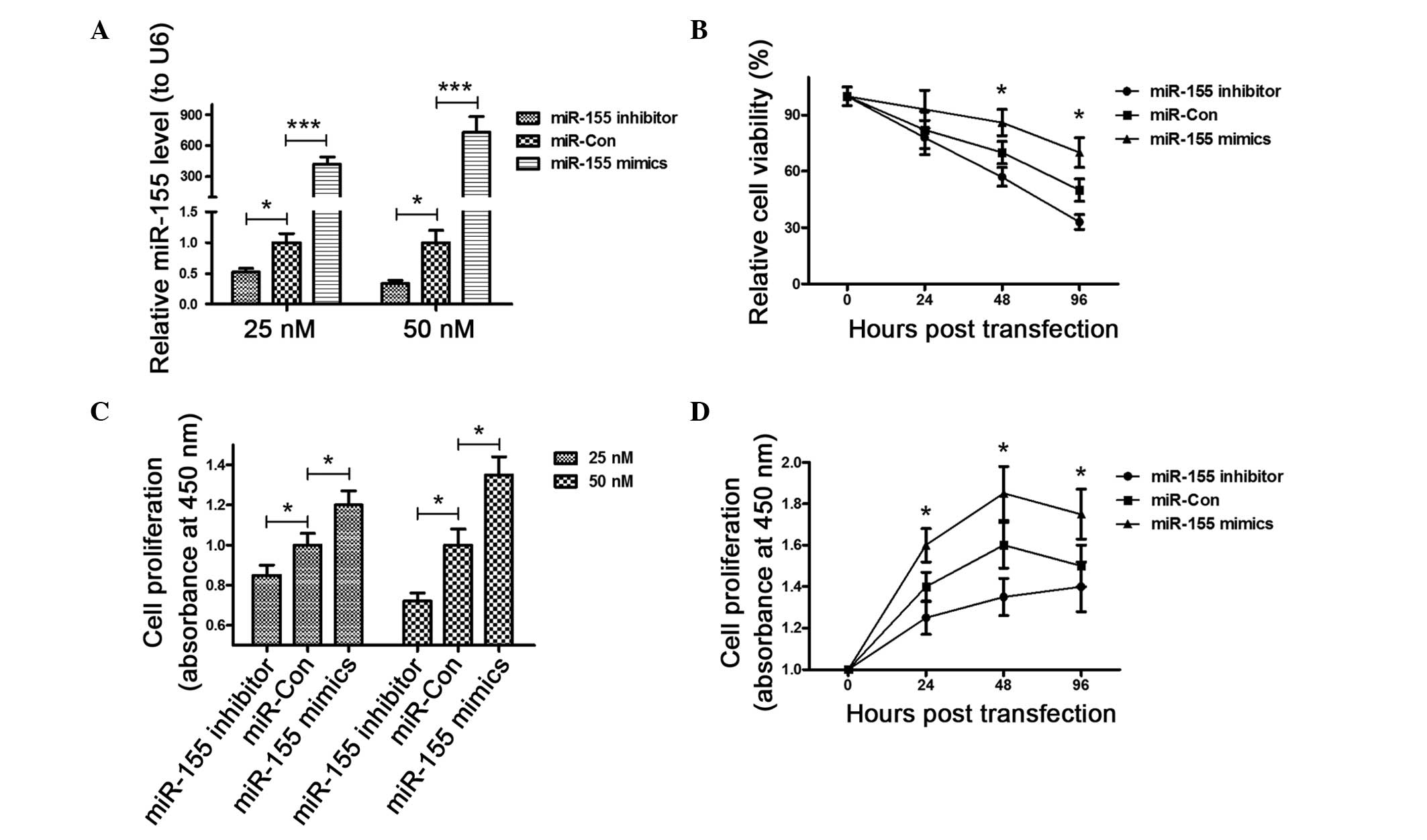
To confirm the promotion of miR-155 to the OS
cell proliferation, the miR-155 expression level was
manipulated in MG-63 cells, via transfection with the
miR-155 mimic or inhibitor. The miR-155 in
mimic-transfected cells was significantly higher than that of the
control cells (P<0.001) 48 h post transfection, whereas the
miR-155 level in the miR-155 inhibitor-transfected
cells was significantly lower than in the control cells (P<0.05)
(Fig. 1A). Subsequently, the
influence of the miR-155 mimic, inhibitor or control on the
cell viability was examined. The MTT assay results (Fig. 1B) demonstrated that the viability
of the MG-63 cells 48 h post-transfection decreased significantly
following the transfection of the miR-155 inhibitor compared
to the transfection of miR-Con (P<0.05); whereas the
transfection of the miR-155 mimic ameliorated the viability
reduction of MG-63 cells (P<0.05). Finally, the proliferation of
MG-63 cells was determined post-transfection for 24 h with the
miR-155 mimic, inhibitor or control in a 25 or 50 nM
concentration by the CCK-8 assay. Fig.
1C shows that in either concentration, the miR-155 mimic
group exhibited a higher proliferation than miR-155 control,
whereas the miR-155 inhibitor group reduced proliferation
(P<0.05). In addition, the time-dependent promoting or reducing
effect in cell proliferation of the miR-155 mimic or
inhibitor was indicated under the condition of enhanced or reduced
miR-155 levels in the MG-63 cells (P<0.05) (Fig. 1D).
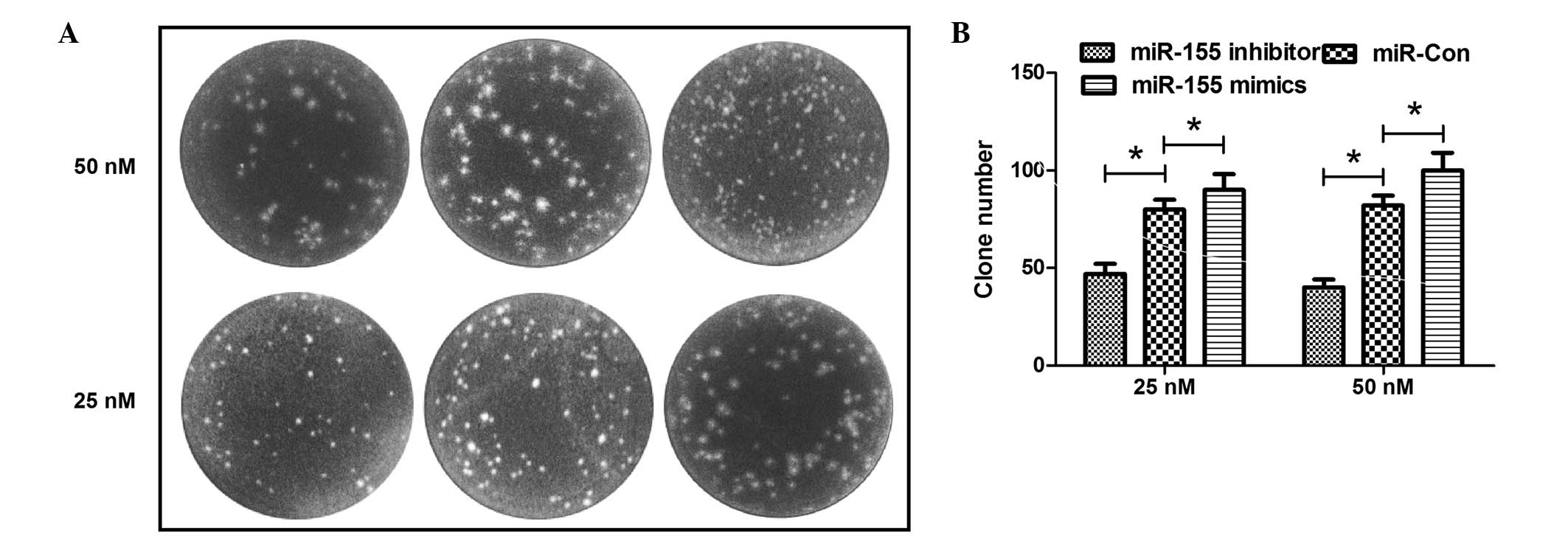
miR-155 inhibitor reduces clone formation
of MG-63 cells
The difference in colony formation was also detected
for the MG-63 cells transfected with the miR-155 mimic,
inhibitor or control in the 25 or 50 nM concentration. The image of
the colonies is shown in Fig. 2A,
and the MG-63 cells that were transfected with the miR-155
mimic in a 25 or 50 nM concentration formed more colonies than the
miR-control-transfected cells, whereas the miR-155
inhibitor reduced the colony formation of MG-63 cells (P<0.05)
(Fig. 2B). All these findings
indicate that the miR-155 inhibitor reduced the clonegenesis
of MG-63 cells, while the upregulated miR-155 in the cells
had a significant role in enhancing the proliferative capability
and colony formation of the MG-63 cells.
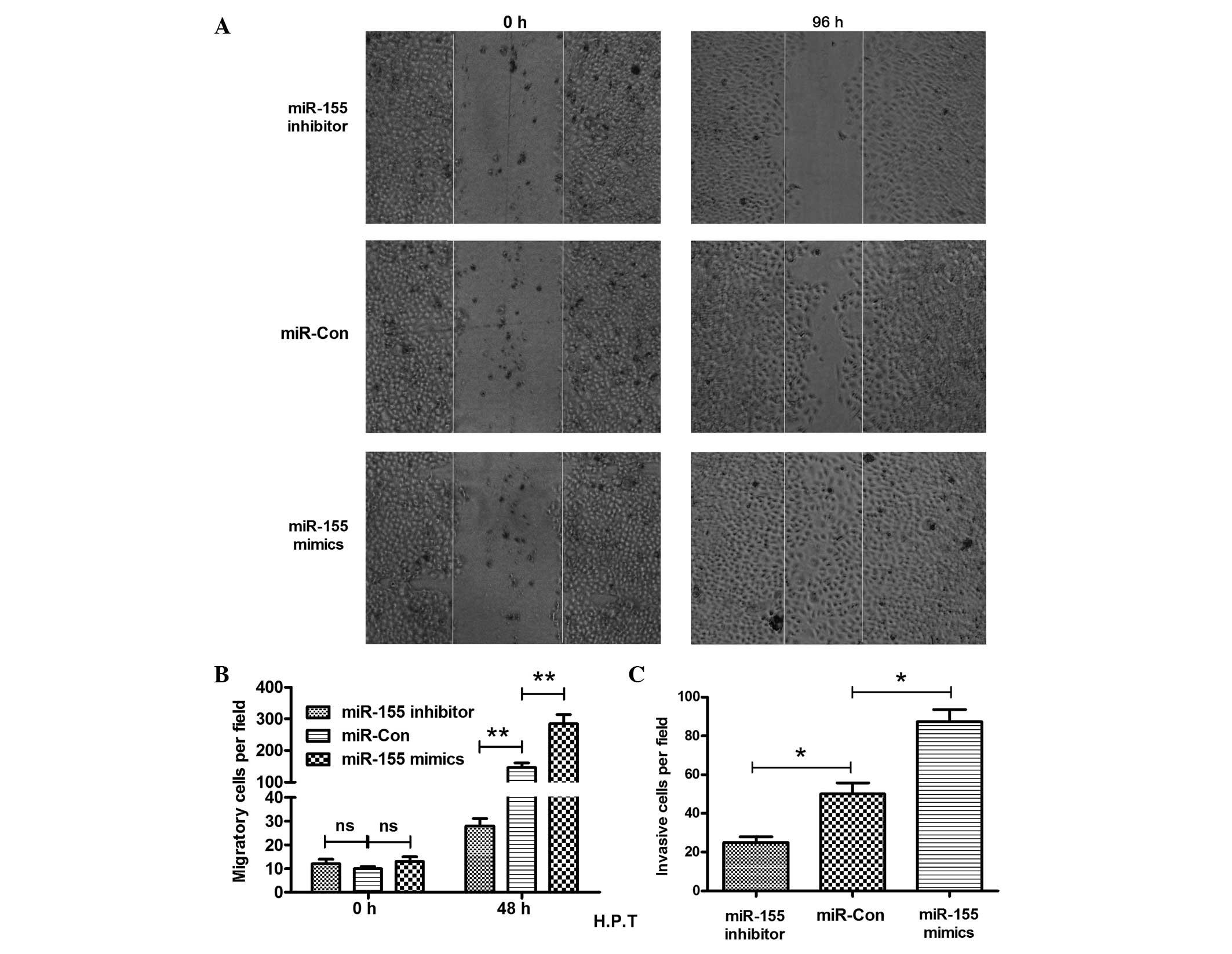
miR-155 inhibitor reduces the migration
and invasion of MG-63 cells
Cell migration is known to contribute to tumor
metastasis (31). The migration of
the MG-63 cells was determined post-transfection of the
miR-155 mimic, inhibitor or control by the scratch assay.
The results shown in Fig. 3A
indicate that more inoculation occurred 96 h post-scratch. The
MG-63 cells post miR-155 mimic-transfection migrated
significantly faster than the miR-Con-transfected MG-63
cells, as there were more cells crossing the base line (P<0.01)
(Fig. 3B). In addition, the
miR-155 inhibitor reduced the migration of MG-63 cells
significantly, as less cells crossed the base line in this group
than in the control group (P<0.01) (Fig. 3B). The miR-155 inhibitor
clearly reduced the MG-63 cell migration. The blockage of the
miR-155 inhibitor to the cell invasion was also
demonstrated. The Transwell invasion chamber assay demonstrated
clearly that there was a significant difference in the cell
invasion between the miR-155 mimic and control groups, or
between the miR-155 inhibitor and control groups. The number
of invasive cells was 50±10 cells in the control group, whereas the
invasive cell number in the miR-155 mimic or inhibitor group
was 88±12 and 25±4 cells, respectively (Fig. 3C) (P<0.05, respectively). All
the results indicated that overexpression of miR-155
stimulated the migration and invasion of OS cells, and the
miR-155 inhibitor reduced the migration and invasion of the
MG-63 cells.
Discussion
As the most common malignant primary bone tumor in
childhood (32), OS maintains a
high recurrence of 30–40%, and 80% of OS patients with metastatic
disease at diagnosis will relapse (27,33,34),
regardless of the significant improvements in the overall survival
rate of high-grade OS patients during the past decades. Failure of
standard multimodal therapy for the disease is associated with an
extremely poor prognosis, and therefore, novel drugs or combination
therapies are required for patients with recurrent or refractory
high-grade OS. Several clinical studies have been conducted to
evaluate the efficiency of a combined therapy with gemcitabine and
docetaxel in recurrent or refractory OS, and the effect of the
gemcitabine-docetaxel combination regimen in recurrent or
refractory OS patients remains controversial (35–37).
Extensive studies have been conducted to identify
the oncogenes that are suitable to become targets of monoclonal
antibodies and small inhibitors. Antibodies or inhibitors were used
to knockdown the tyrosine kinase receptors, KIT, platelet-derived
growth factor receptors and vascular endothelial growth factor
receptors (38–41), however, their inhibition lacked
antitumor activity. The monoclonal antibody anti-insulin-like
growth factor receptor-I was also promising preclinically, but was
not confirmed to be effective in the clinical setting (42). Recently, several studies have
focused on the signal transduction pathways of phosphatidylinositol
3′-kinase/mammalian target of rapamycin (43) and mitogen-activated protein
kinases. Their inhibition proved highly effective in OS preclinical
models (44).
Previously, various miRNAs have been confirmed to be
deregulated in OS (14,15). Several oncogenic miRNAs, including
miR-21, miR-93 and miR-29, have been indicated
to be overexpressed and to induce cancer cell growth, migration,
invasion and metastasis (16–19,45).
Recently, the miR-155 dysregulation in OS was discovered by
microarray analysis (29). In the
present study, the regulation of miR-155 was explored on the
OS cell proliferation, migration and invasion on the MG-63 cell
in vitro. The miR-155 mimic was shown to promote the
cell proliferation, colony formation, migration and invasion
significantly, compared to the control miRNA. An miR-155
inhibitor was also used to evaluate whether miR-155 could
serve as a therapeutic target for OS. The results demonstrated that
the miR-155 inhibitor significantly reduced the
proliferation, colony formation, migration and invasion of MG-63 OS
cells.
In conclusion, the present study confirmed that the
oncogenic regulation on the OS progression of miR-155 could
serve as a therapeutic target with an miR-155 inhibitor.
References
|
1
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer treatment and research. 152:3–13. 2009.
View Article : Google Scholar
|
|
2
|
Dorfman HD and Czerniak B: Bone cancers.
Cancer. 75:203–210. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meyers PA, Heller G, Healey J, et al:
Chemotherapy for nonmetastatic osteogenic sarcoma: the Memorial
Sloan-Kettering experience. J Clin Oncol. 10:5–15. 1992.PubMed/NCBI
|
|
4
|
Ambros V: MicroRNA pathways in flies and
worms: growth, death, fat, stress, and timing. Cell. 113:673–676.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reinhart BJ, Slack FJ, Basson M, et al:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nature Revs Cancer.
6:259–269. 2006. View
Article : Google Scholar
|
|
9
|
Wang D, Qiu C, Zhang H, Wang J, Cui Q and
Yin Y: Human microRNA oncogenes and tumor suppressors show
significantly different biological patterns: from functions to
targets. PLoS One. 5:e130672010. View Article : Google Scholar
|
|
10
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ventura A and Jacks T: MicroRNAs and
cancer: short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Spizzo R, Nicoloso MS, Croce CM and Calin
GA: SnapShot: MicroRNAs in Cancer. Cell. 137:586–586 e581. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou G, Shi X, Zhang J, Wu S and Zhao J:
MicroRNAs in osteosarcoma: from biological players to clinical
contributors, a review. J Int Med Res. 41:1–12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Y, Zhang J, Zhang L, Si M, Yin H and Li
J: Diallyl trisulfide inhibits proliferation, invasion and
angiogenesis of osteosarcoma cells by switching on suppressor
microRNAs and inactivating of Notch-1 signaling. Carcinogenesis.
34:1601–1610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Asangani IA, Rasheed SA, Nikolova DA, et
al: MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor
suppressor Pdcd4 and stimulates invasion, intravasation and
metastasis in colorectal cancer. Oncogene. 27:2128–2136. 2008.
View Article : Google Scholar
|
|
17
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ziyan W, Shuhua Y, Xiufang W and Xiaoyun
L: MicroRNA-21 is involved in osteosarcoma cell invasion and
migration. Med Oncol. 28:1469–1474. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Montanini L, Lasagna L, Barili V, et al:
MicroRNA cloning and sequencing in osteosarcoma cell lines:
differential role of miR-93. Cell Oncol (Dordr). 35:29–41. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang G, Nishimoto K, Zhou Z, Hughes D and
Kleinerman ES: miR-20a encoded by the miR-17-92 cluster increases
the metastatic potential of osteosarcoma cells by regulating Fas
expression. Cancer Res. 72:908–916. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duan Z, Choy E, Harmon D, et al:
MicroRNA-199a-3p is downregulated in human osteosarcoma and
regulates cell proliferation and migration. Mol Cancer Ther.
10:1337–1345. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu LH, Li H, Li JP, et al: miR-125b
suppresses the proliferation and migration of osteosarcoma cells
through down-regulation of STAT3. Biochem Biophys Res Commun.
416:31–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang H, Cai X, Wang Y, Tang H, Tong D and
Ji F: microRNA-143, down-regulated in osteosarcoma, promotes
apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol
Rep. 24:1363–1369. 2010.PubMed/NCBI
|
|
24
|
Thayanithy V, Sarver AL, Kartha RV, et al:
Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma.
Bone. 50:171–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eis PS, Tam W, Sun L, et al: Accumulation
of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad
Sci USA. 102:3627–3632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kluiver J, Poppema S, de Jong D, et al:
BIC and miR-155 are highly expressed in Hodgkin, primary
mediastinal and diffuse large B cell lymphomas. J Pathol.
207:243–249. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Provisor AJ, Ettinger LJ, Nachman JB, et
al: Treatment of nonmetastatic osteosarcoma of the extremity with
preoperative and postoperative chemotherapy: a report from the
Children’s Cancer Group. J Clin Oncol. 15:76–84. 1997.
|
|
28
|
Goorin AM, Schwartzentruber DJ, Devidas M,
et al; Pediatric Oncology Group. Presurgical chemotherapy compared
with immediate surgery and adjuvant chemotherapy for nonmetastatic
osteosarcoma: Pediatric Oncology Group Study POG-8651. J Clin
Oncol. 21:1574–1580. 2003. View Article : Google Scholar
|
|
29
|
Lauvrak SU, Munthe E, Kresse SH, et al:
Functional characterisation of osteosarcoma cell lines and
identification of mRNAs and miRNAs associated with aggressive
cancer phenotypes. Br J Cancer. 109:2228–2236. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
32
|
Nagarajan R, Weigel BJ, Thompson RC and
Perentesis JP: Osteosarcoma in the first decade of life. Med
Pediatr Oncol. 41:480–483. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bramwell VH, Burgers M, Sneath R, et al: A
comparison of two short intensive adjuvant chemotherapy regimens in
operable osteosarcoma of limbs in children and young adults: the
first study of the European Osteosarcoma Intergroup. J Clin Oncol.
10:1579–1591. 1992.
|
|
34
|
Bacci G, Picci P, Ferrari S, et al:
Primary chemotherapy and delayed surgery for nonmetastatic
osteosarcoma of the extremities. Results in 164 patients
preoperatively treated with high doses of methotrexate followed by
cisplatin and doxorubicin. Cancer. 72:3227–3238. 1993. View Article : Google Scholar
|
|
35
|
Mora J, Cruz CO, Parareda A and de Torres
C: Treatment of relapsed/refractory pediatric sarcomas with
gemcitabine and docetaxel. J Pediatr Hematol Oncol. 31:723–729.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
McTiernan A and Whelan JS: A Phase II
Study of Docetaxel for the Treatment of Recurrent Osteosarcoma.
Sarcoma. 8:71–76. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Navid F, Willert JR, McCarville MB, et al:
Combination of gemcitabine and docetaxel in the treatment of
children and young adults with refractory bone sarcoma. Cancer.
113:419–425. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
McGary EC, Weber K, Mills L, et al:
Inhibition of platelet-derived growth factor-mediated proliferation
of osteosarcoma cells by the novel tyrosine kinase inhibitor
STI571. Clin Cancer Res. 8:3584–3591. 2002.PubMed/NCBI
|
|
39
|
Sulzbacher I, Birner P, Trieb K, Traxler
M, Lang S and Chott A: Expression of platelet-derived growth
factor-AA is associated with tumor progression in osteosarcoma. Mod
Pathol. 16:66–71. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kubo T, Piperdi S, Rosenblum J, et al:
Platelet-derived growth factor receptor as a prognostic marker and
a therapeutic target for imatinib mesylate therapy in osteosarcoma.
Cancer. 112:2119–2129. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kaya M, Wada T, Akatsuka T, et al:
Vascular endothelial growth factor expression in untreated
osteosarcoma is predictive of pulmonary metastasis and poor
prognosis. Clin Cancer Res. 6:572–577. 2000.PubMed/NCBI
|
|
42
|
Kolb EA, Kamara D, Zhang W, et al: R1507,
a fully human monoclonal antibody targeting IGF-1R, is effective
alone and in combination with rapamycin in inhibiting growth of
osteosarcoma xenografts. Pediatr Blood Cancer. 55:67–75.
2010.PubMed/NCBI
|
|
43
|
Manara MC, Nicoletti G, Zambelli D, et al:
NVP-BEZ235 as a new therapeutic option for sarcomas. Clin Cancer
Res. 16:530–540. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pignochino Y, Grignani G, Cavalloni G, et
al: Sorafenib blocks tumour growth, angiogenesis and metastatic
potential in preclinical models of osteosarcoma through a mechanism
potentially involving the inhibition of ERK1/2, MCL-1 and ezrin
pathways. Mol Cancer. 8:1182009. View Article : Google Scholar
|
|
45
|
Zhang W, Qian JX, Yi HL, et al: The
microRNA-29 plays a central role in osteosarcoma pathogenesis and
progression. Mol Biol (Mosk). 46:622–627. 2012. View Article : Google Scholar : PubMed/NCBI
|