Introduction
The enzyme New Delhi metallo-β-lactamase-1 (NDM-1)
is expressed by certain bacteria that carry the
blaNDM-1 gene. The earliest report of NDM-1 was
by Yong et al (1) in
December 2009. Yong et al separated a strain of
Klebsiella pneumoniae that exhibited resistance to
antibacterial agents such as carbapenems from the urine specimen of
an Indian patient who was suffering from urinary tract infection in
Sweden, confirming that the resistance mechanism of Klebsiella
pneumoniae had led to the production of a new metalloenzyme. As
this patient had once received treatment in New Delhi, the enzyme
was designated NDM-1. An article published in The Lancet Infectious
Diseases in August 2010 (2)
reported that there had been 37 cases of Enterobacteriaceae
carrying the blaNDM-1 gene in England, 44 in
Chennai, India, 26 in Haryana, India, and 73 in other parts of
India and in Pakistan.
Since the NDM-1 drug resistance gene can reduce the
antibacterial activity of almost all β-lactam antibiotics,
including the most potent carbapenem antibiotics, its presence in
bacteria results in serious bacterial resistance. Drug resistance
genes can be transferred by plasmid; hence, these features of NDM-1
may cause global public health security issues. The initial article
in The Lancet Infectious Diseases drew worldwide attention. Shortly
thereafter, the United States, Canada, Sweden, Greece, Israel, the
Netherlands, Japan, Brazil and other countries (3), as well as Hong Kong (4) and Taiwan (5) in China began to carry out research
into blaNDM-1. The Chinese Ministry of Public
Health also made an announcement on October 25, 2010, requiring all
medical institutions to screen and monitor bacterial strains
containing NDM-1. To date, very few studies concerning the NDM-1
drug resistance gene have been published.
In 2011, clinical observations of Enterobacteriaceae
strains that had a sharp reduction of sensitivity to carbapenem
antibiotics and were suspected of carrying the
blaNDM-1 gene began in Hainan. The current study
investigated the existence of the blaNDM-1 gene
in 30 Enterobacteriaceae strains collected in Hainan, and
determined the species of these strains.
Materials and methods
Materials
The patients in this study attended the Hainan
Provincial Agricultural Reclamation General Hospital and
Traditional Chinese Medicine Hospital of Hainan Province (both
Haikou, China) from January to December in 2012. In total, 30
Enterobacteriaceae strains were isolated from the patients and
following the use of broth enrichment, were preserved at −70°C. The
present study was approved by the Medical Ethics Committee of
Hainan General Hospital (Haikou, China). Informed consent was
obtained from the patients or patients’ families prior to their
involvement in the present study.
Reagents and instruments
The API 20E biochemical identification system
(bioMerieux, Inc., Marcy l’Étoile, France) was used to identify the
bacteria. The VITEK2 automatic bacterial identification and
antibiotic susceptibility testing system (bioMerieux, Inc.) with
supporting reagents, and imipenem, meropenem, tigecycline
susceptibility and imipenem/imipenem inhibitor (IP/IPI) E-test
strips (AB Biodisk, bioMerieux Inc., Stockholm, Sweden) were used
to evaluate drug sensitivity and identify the metallo-β-lactamase
phenotype. An E-Cycle™ 96PCR cycler (CapitalBio Corporation,
Beijing, China), agarose gel electrophoresis (AGE) apparatus
(Beijing 61 Instrument Factory, Beijing, China), JS-380 automatic
gel imaging system used in AGE (Peiqing Inc., Shanghai, China) and
2X Power Taq PCR Master Mix (PR1700; BioTeke Corporation, Beijing,
China) kit were used in the polymerase chain reaction (PCR)
amplification process and a D2000 DNA Marker (MD114; Tiangen
Biotech Co., Ltd, Beijing, China) was used as an AGE marker.
Strain identification and drug
sensitivity test
The API 20E biochemical identification system was
used to identify bacteria, using the specific methods described by
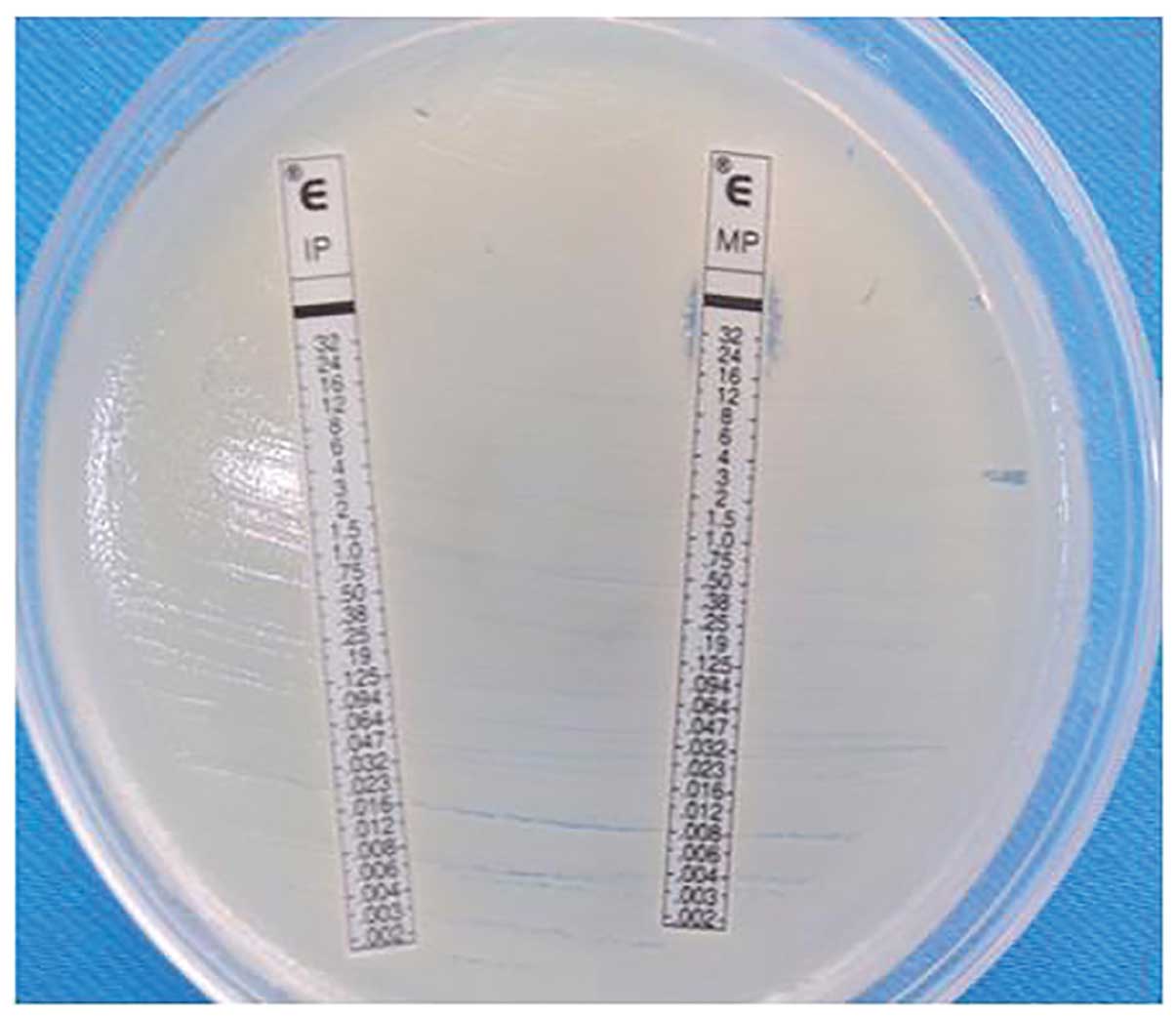
the manufacturer. The concentration gradient-based E-test strip
method was used to measure the minimum inhibitory concentration
(MIC) values (Fig. 1) in imipenem
and meropenem in vitro susceptibility tests; the 2010
Clinical Laboratory Standards Institute (CLSI) threshold (6) was used to judge the sensitivity,
medium and drug resistance. The E-test strip method was also used
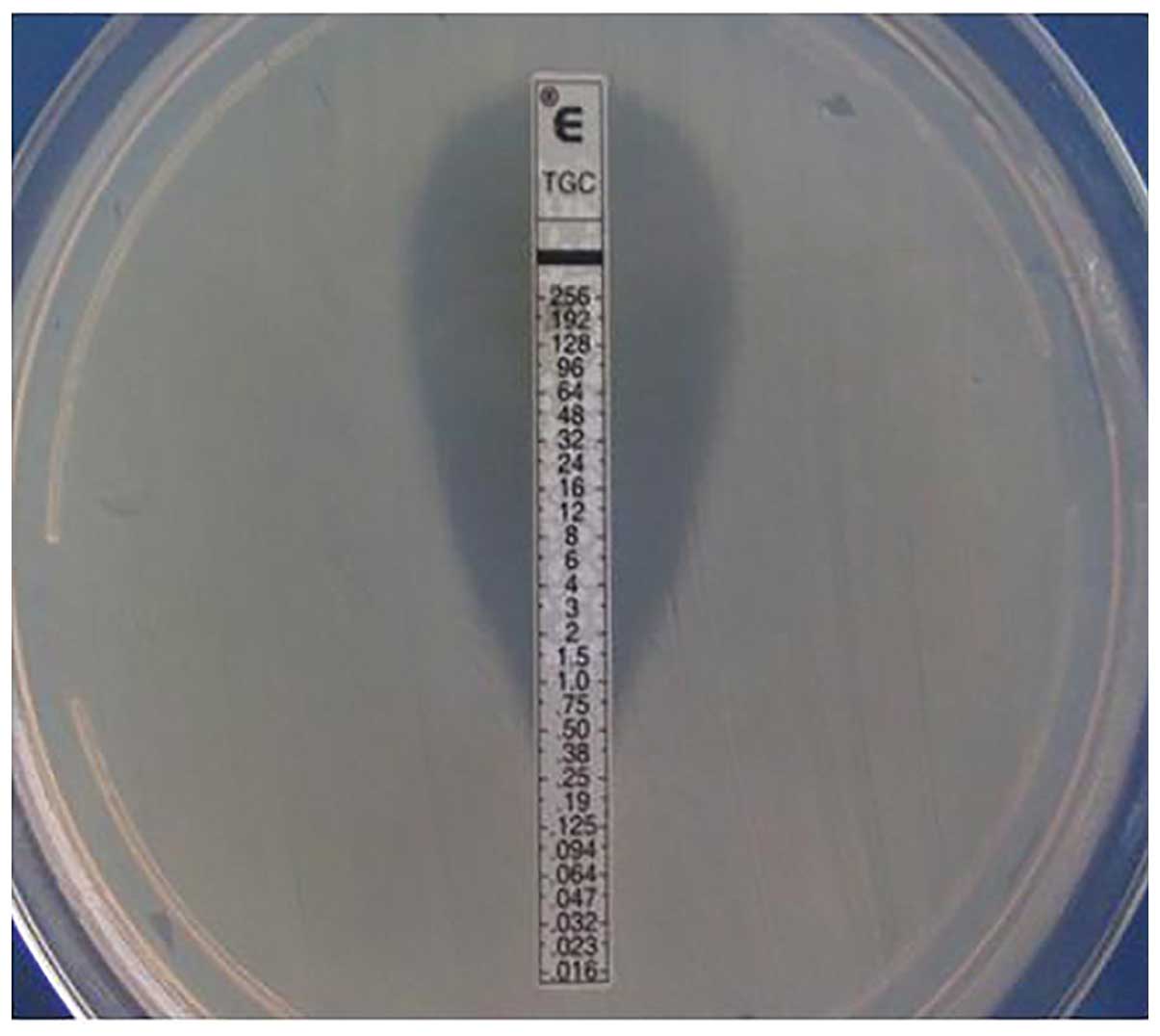
to test in vitro susceptibility to tigecycline; the results
were determined in accordance with the US Food and Drug
Administration Enterobacteriaceae criteria (an MIC ≤2 μg/ml is
defined as sensitive; Fig. 2)
(7). The VITEK2 automated
bacterial identification system and supporting reagents were used
to determine susceptibility (MIC values) to other antibacterial
agents; the 2010 CLSI threshold (6) was used to judge the sensitivity,
medium and drug resistance.
Confirmation of metallo-β-lactamase
phenotype
IP/IPI E-test papers were used to test the target
bacterial strains as follows: The strains were cultured overnight.
Sterile saline was used to dilute the suspension to 0.5 McFarland
turbidity. The bacteria were coated onto the Müller-Hinton (MH)
agar plate, the plate was allowed to dry for 10 min, the IP/IPI
E-test strip was placed on the surface of the agar plate, and the
plate was incubated for 16–18 h at 35°C. An IP/IPI ratio >8
indicated a positive result (Fig.
3).
Extraction of bacterial DNA
The fresh culture was put into 100–200 μl sterile
deionized water, boiled for 10 min and then subjected to
centrifugation at 14,800 × g for 5 min. The supernatant fluid was
extracted and preserved at −20°C.
PCR amplification and sequencing
In the screening for the NDM-1-encoding gene, the
primer sequences used and the PCR product details are shown in
Table I. The PCR reaction
conditions were initial denaturation at 94°C for 5 min;
degeneration at 94°C for 15 sec, annealing at 55°C for 30 sec and
72°C for 30 sec, for a total of 25 cycles; followed by extension at
72°C for 5 min. Following the PCR amplification, 1.5% AGE of the
products was conducted at 120 V for 40 min and the JS-380 imaging
system was used to observe the results and capture photographic
images. The products were submitted to Shanghai Biological
Engineering Co., Ltd. (Shanghai, China) to conduct the sequencing.
For full-length blaNDM-1 gene detection, the
primer was designed with reference to FN396876.1 in Genbank, with
details as shown in Table I. The
acquired sequences along with the nucleotide sequence of FN396876.1
were entered into DNAStar software (DNASTAR Inc., Madison, WI, USA)
and sequence alignment was performed by the ClustalW method. The
PCR reaction conditions and processing for the full-length gene
were the same as those described for the screening PCR.
 | Table IPCR primers. |
Table I
PCR primers.
| Primer | Details | Primer sequences
(5′-3′) | Product length
(bp) | Amplified fragment,
location of CDS |
|---|
| Screening | Forward |
CAGCACACTTCCTATCTC | 292 | Reverse,
2751–2993 |
| Reverse |
CCGCAACCATCCCCTCTT | | |
| Full-length | Upstream |
TCGCATAAAACGCCTCTG | 1001 | Forward,
2564–3444 |
| Downstream |
GAAACTGTCGCACCTCAT | | |
Quality control strains
The quality control strain for bacterial
identification and drug susceptibility testing was ATCC25922
Escherichia coli, acquired from the Clinical Inspection
Center of the Ministry of Health (Beijing, China).
Results
Results of screening for metalloenzyme
production
When the samples were analyzed using the IP/IPI
E-test, 21 of the 30 collected Enterobacteriaceae strains were
found to be positive, confirming that 70% of the bacterial strains
were producing carbapenemases of the metallo-β-lactamase
phenotype.
Results of blaNDM-1 gene
screening by PCR amplification and sequencing
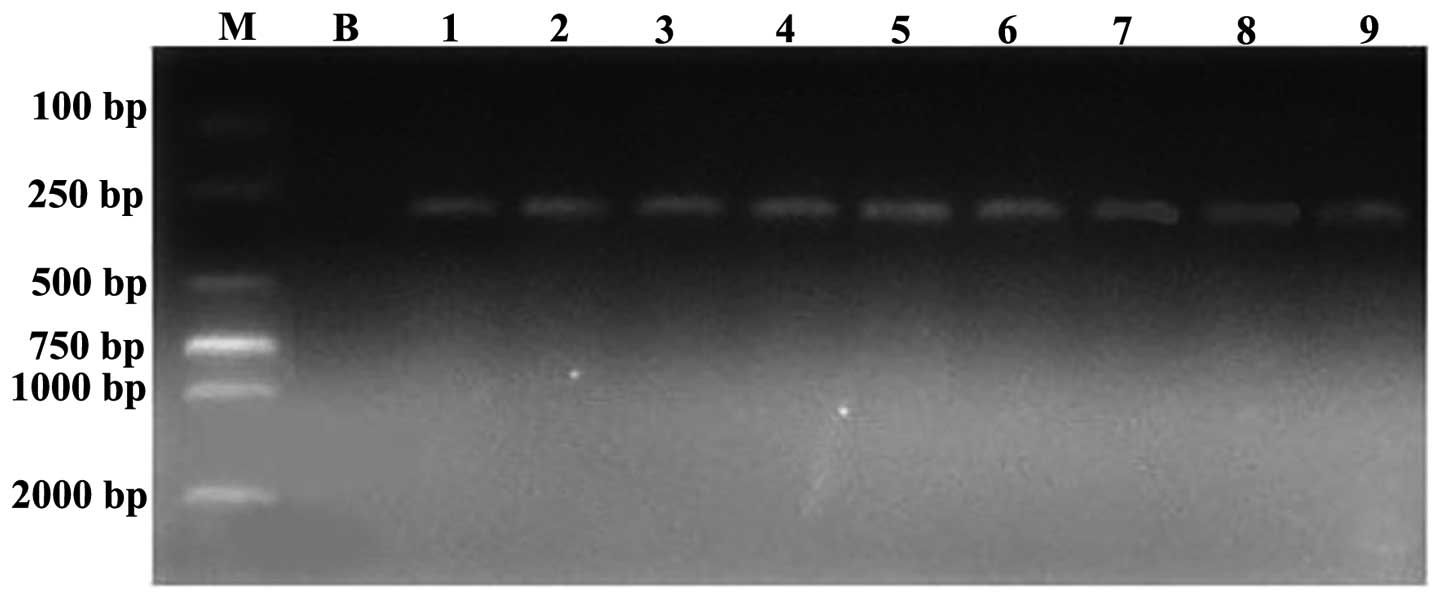
It was found that nine of the 21 strains that
produced metallo-β-lactamase also presented positive segments at
292 bp (Fig. 4). Through DNA
sequencing and DNAStar analysis, compared with the FN396876.1
sequence in GenBank, the homology comparison result was 100% in
each case (reverse, 2751–2993), confirming that nine of the strains
carried the blaNDM-1 gene.
Results of full-length
blaNDM-1 gene screening by PCR amplification and
sequencing
Nine Enterobacteriaceae strains were carriers of the
NDM-1-encoding gene; following full-length
blaNDM-1 gene amplification, all nine strains
exhibited positive results with bands at 1,001 bp (Fig. 5). Through sequencing and DNASTAR
analysis, compared with the FN396876.1 sequence in GenBank, the
homology was 100%. The gene sequences of certain strains were
registered in GenBank; the accession numbers were KC573881.1,
KC573880.1 and KC573878.1.
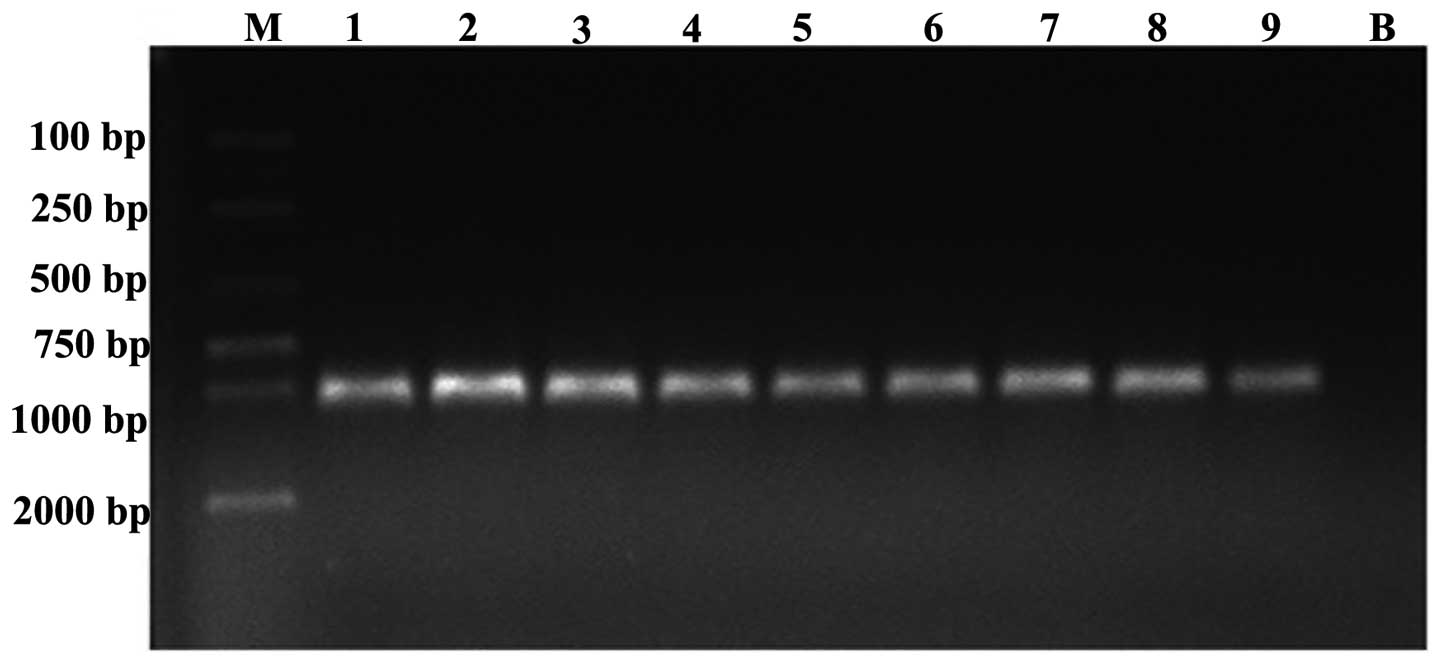 | Figure 5Electrophoresis results for the
full-length PCR products of nine strains carrying the
blaNDM-1 resistance gene (DNA fragment, 1,001
bp). Lane M, marker (D2000); lanes 1, 5, 6 and 8, Klebsiella
pneumoniae; lanes 4 and 9, Escherichia coli; lanes 2 and
3, Enterobacter cloacae; lane 7, Enterobacter
aerogenes; lane B, blank control. |
Results of strain identification and
susceptibility tests
Following testing with the API 20E biochemical
identification system, four strains were identified to be
Klebsiella pneumoniae (Nos. 1, 5, 6 and 8), two strains were
Escherichia coli (Nos. 4 and 9), two strains were
Enterobacter cloacae (Nos. 2 and 3) and one strain was
Enterobacter aerogenes (No. 7). Specific information
concerning the types of bacteria and drug susceptibility results
are shown in Table II. The
sensitivity rate of the blaNDM-1-positive strains
in vitro was 77.8% to amikacin, 44.4% to ciprofloxacin and
33.3% to gentamicin. The sensitivity rates to polymyxin and
tigecycline were 100 and 88.9%, respectively.
 | Table IIBacterial susceptibility results
(μg/ml). |
Table II
Bacterial susceptibility results
(μg/ml).
| Variable | Bacterial strain
no. | Sensitivity rate
(%) |
|---|
|
|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|
| Strain type | Kpn | Ecl | Ecl | Eco | Kpn | Kpn | Eae | Kpn | Eco | |
| Antibacterial
agent |
| Polymyxin B | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | ≤2 | 100 |
| Tigecycline
(E-test strip) | 1 | 0.75 | 1 | 1.5 | 3 | 1 | 1 | 1 | 0.25 | 88.9 |
| Amikacin | 8 | ≥64 | ≥64 | 8 | 8 | 8 | ≤2 | ≤2 | ≤2 | 77.8 |
| Ciprofloxacin | ≤0.25 | ≥4 | ≥4 | ≥4 | ≤0.25 | ≤0.25 | ≤0.25 | 2 | ≥4 | 44.4 |
| Gentamicin | ≥16 | ≥16 | ≥16 | ≤1 | ≤1 | ≥16 | ≤1 | ≥16 | ≥16 | 33.3 |
| Cotrimoxazole | ≥320 | ≥320 | ≥320 | ≥320 | ≤20 | ≥320 | ≥320 | ≥320 | ≥320 | 11.1 |
| Aztreonam | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | 16 | 0 |
| Ceftazidime | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | 0 |
| Ceftriaxone | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | 0 |
| Cefepime | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | 32 | 0 |
| Cefotetan | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | ≥64 | 0 |
|
Piperacillin/tazobactam | ≥128 | ≥128 | ≥128 | ≥128 | ≥128 | ≥128 | ≥128 | ≥128 | ≥128 | 0 |
| Imipenem (E-test
strip) | 8 | >32 | >32 | 3 | 32 | 3 | 24 | 4 | 8 | 0 |
| Meropenem (E-test
strip) | >32 | >32 | >32 | 4 | 16 | 6 | >32 | >32 | >32 | 0 |
Discussion
Carbapenem antibiotics are atypical β-lactam
antibiotics with a broad spectrum and strong antibacterial
activity. In the past 20 years, along with the increasing clinical
use of cephalosporins, the rate of multi-resistant bacteria that
produce high levels of AmpC enzymes and extended-spectrum
β-lactamases (ESBLs) has also increased sharply (8) Thus, carbapenems have been used to
treat illness caused by drugs that induce the production of AmpC
enzymes and ESBLs. However, with the worldwide use of carbapenems
as antibacterial agents in AmpC- and ESBL-producing bacteria and
other multi-drug resistant bacteria, carbapenem-resistant
Enterobacteriaceae (CRE) such as Pseudomonas aeruginosa and
Acinetobacter have become prevalent in the clinic (9). In recent years, the clinical
treatment and control of CRE infection has faced great challenges.
In 2009, guidelines for CRE infection control in hospitals were
published (10). This indicates
that CRE had already begun to attract worldwide attention. The
resistance mechanism of CRE is mainly due to the production of
carbapenemases (11–13).
Carbapenemases are β-lactamases that are able to
hydrolyze carbapenem antibiotics, including those of the A, B and D
types according to the Ambler molecular classification.
Carbapenemases may be of metal-free or metal-containing types. The
former type includes NmcA and SME-1, while the latter includes L1,
IMP-1 and VIM-1. NDM-1 is a gene that encodes a class B
carbapenemase which hydrolyzes and inactivates the overwhelming
majority of carbapenem antibiotics. Unlike the L1, IMP-1 and VIM-1
metallo-β-lactamase genes, the blaNDM-1 gene is
located on a 140-kb plasmid (2),
which can be transferred at a horizontal level between bacteria. It
has been confirmed that strains with the blaNDM-1
gene can lead to the emergence and spread of multi-resistant
bacteria among different species of bacteria (14). Hence, it is suggested that the
monitoring and research of strains carrying the
blaNDM-1 gene may be useful in preventing and
controlling new types of multi-resistant bacteria.
Among the nine blaNDM-1-expressing
Enterobacteriaceae samples isolated from various parts of the body
of different patients in the present study, five were isolated from
sputum, two were separated from the blood, and the other two were
isolated from urine. The older patients had underlying diseases,
including chronic obstructive pulmonary disease, cerebral
infarction, hypertension and coronary heart disease, and had no
medical history from other provinces or countries. Four patients
were in an intensive care unit while the other five were from
intensive medicine and geriatric, neurology, neurosurgery and
endocrinology departments, respectively. Considering the history of
the patients, it appears that the nine strains were local
bacteria.
At present, there are very few reports from China
concerning bacteria carrying the blaNDM-1 gene,
and even fewer on Enterobacteriaceae carrying
blaNDM-1. On October 26, 2010, the Chinese
Military Academy of Medical Sciences and the Chinese Center for
Disease Control and Prevention announced that three strains
carrying the blaNDM-1 gene had been found in
mainland China, two of which were enterococci from the feces of
infants with diarrhea from Ningxia province (15) and the third was Acinetobacter
baumannii in a sputum specimen from a lung cancer patient in
Fujian province (16). A study
reported in August 2011 that the gene had been identified in three
strains of Acinetobacter and a case of Klebsiella
ozaenae in Guangzhou (17). A
strain of a Klebsiella oxytoca carrying the gene in Yunnan
was reported in 2012 (18). A
study from Hunan published in August 2012 reported that
Klebsiella pneumoniae carrying blaNDM-1
had been isolated from the sputum specimen of a 8-month-old infant
(19). In comparison with these
other provinces, Hainan has a greater quantity of the
blaNDM-1-carrying bacteria, which are from
different bacterial genera. Among these bacteria, four are
Klebsiella pneumoniae, two are Escherichia coli, two
are Enterobacter cloacae and one is Enterobacter
aerogenes.
The sensitivity rate of the
blaNDM-1-positive strains in vitro was
77.8% to amikacin, 44.4% to ciprofloxacin and 33.3% to gentamicin.
The strains were highly sensitive to amikacin but only moderately
sensitive to ciprofloxacin and gentamicin. In a previous study by
Kumarasamy et al (2), the
sensitivity rates of 180 NDM-1-producing Enterobacteriaceae strains
to gentamicin and ciprofloxacin were only 5–10%. The sensitivity
rates to polymyxin (100%) and tigecycline (88.9%) in the present
study are similar to those in the study by Kumarasamy et al
(2), which were 80–90 and 90%,
respectively. Resistance to broad-spectrum antibiotic plus
β-lactamase inhibitor, cephalosporins and carbapenems has reached
100%, which is in agreement with the study by Kumarasamy et
al (2).
Therefore, it appears that the isolated
Enterobacteriaceae strains carrying the blaNDM-1
gene have different biological behaviors, which requires further
in-depth study.
Acknowledgements
This study was supported by the Health Department
and Science Foundation of Hainan Province (No: 2010-9).
References
|
1
|
Yong D, Toleman MA, Giske CG, Cho HS,
Sundman K, Lee K and Walsh TR: Characterization of a new
metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin
esterase gene carried on aunique genetic structure in Klebsiella
pneumoniae sequence type 14 from India. Antimicrob Agents
Chemother. 53:5046–5054. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kumarasamy KK, Toleman MA, Walsh TR, et
al: Emergence of a new antibiotic resistance mechanism in India,
Pakistan, and the UK: a molecular, biological, and epidemiological
study. Lancet Infect Dis. 10:597–602. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huang LY: About NDM-1 superbug: Some
thoughts. Jie Fang Jun Yi Xue Za Zhi. 35:1409–1411. 2010.(In
Chinese).
|
|
4
|
Ho PL, Lo WU, Yeung MK, et al: Complete
sequencing of pNDM-HK encoding NDM-1 carbapenemase from a
multidrug-resistant Escherichia coli strain isolated in Hong Kong.
PLoS One. 6:e179892011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu HS, Chen TL, Chen IC, Huang MS, Wang
FD, Fung CP and Lee SD: First identification of a patient colonized
with Klebsiella pneumoniae carrying blaNDM-1 in Taiwan.
J Chin Med Assoc. 73:596–598. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clinical and Laboratory Standards
Institute (CLSI). Performance Standards for Antimicrobial
Susceptibility Testing; Twentieth Informational Supplement (June
2010 Update) (M100-S20-U). CLSI; Wayne, PA, USA: 2010
|
|
7
|
Wang H, Ni YX, Chen MJ, et al: Operating
regulations of drug sensitive test in vitro with Tigecyclin, a new
antibacterial agent of glycyleycline. Zhonghua Jianyan Yixue Zazhi.
32:1208–1213. 2009.(In Chinese).
|
|
8
|
Zhang LL, Ji DM and Jin HY:
Pharmacological features and clinical application of
carbapenem-resistant. Zhongguo Linchuang Yanjiu. 4:335–336.
2010.(In Chinese).
|
|
9
|
Zhu BQ, Shen P, Yu YS and Zhang XG: Study
on the homology and carbapenemases genotypes of imipenem-resistant
Acinetobacter baumannii strains. Zhe Jiang Yi Xue. 30:459–462.
2008.(In Chinese).
|
|
10
|
Centers for Disease Control and Prevention
(CDC). Guidance for control of infections with carbapenem-resistant
or carbapenemase-producing Enterobacteriaceae in acute care
facilities. JAMA. 301:1979–1982. 2009.
|
|
11
|
Nordmann P, Naas T and Poirel L: Global
spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect
Dis. 17:1791–1798. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng X, Wang P, Wang Y, Zhang H, Tao C,
Yang W, Liu M and Jia W: Identification and distribution of the
clinical isolates of imipenem-resistant Pseudomonas aeruginosa
carrying metallo-beta-lactamase and/or class 1 integron genes. J
Huazhong Univ Sci Technolog Med Sci. 28:235–238. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qiu LP, Chang YZ, Sun J, Wei ZG, Zhou H
and Yu YS: Study on the antibiotic resistance mechanism and the
molecular epidemiology of Klebsiella pneumoniae with reduced
susceptibility to carbapenems. Zhongguo Wei Sheng Jian Yan Za Zhi.
22:1420–1422. 2012.(In chinese).
|
|
14
|
Lv JY and Qu F: Multi-drug Resistant
Microorganisms and Countermeasures. 1st edition. People’s Medical
Publishing; Beijing: pp. 202–208. 2011
|
|
15
|
Hao Q, Liu X, Guo BC, Xie MY, Zuo LQ, Wang
Z and Jing HZ: A preliminary study on the bacteria strain genes
with NDM-1 in Ningxia. Ningxia Yi Xue Za Zhi. 34:289–291. 2012.(In
Chinese).
|
|
16
|
Chen S, Qiu SF, Xia LL, et al: Sequence
and expression of Acinetobacter baumannii blaNDM-1 gene. Zhong Guo
Ren Shou Gong Huan Bing Xue Bao. 28:471–473. 2012.(In Chinese).
|
|
17
|
Yang YM, Ye HF, Zhang WH, Chen HL and Zhou
XM: Detecttion of New Delhi metallo-β-lactamase 1 gene in
Klebsiella ozaenae and Acinetobacter baumannii. Guo Ji Jian Yan Yi
Xue Za Zhi. 32:1407–1409. 2011.(In Chinese).
|
|
18
|
Yin JW, Xu W, Gu WP, Zhou YM, Li CQ, Yang
JB and Fu XQ: Detection of a strain of Klebsiella oxytoca carrying
blaNDM-1 in Yunnan province. Ji Bing Jian Ce. 27:211–217. 2012.(In
Chinese).
|
|
19
|
Zou MX, Wu JM, Li J, Dou QY, Zhou RR,
Huang Y and Liu WE: NDM-1-producing Klebsiella pneumoniae in
mainland China. Zhongguo Dang Dai Er Ke Za Zhi. 14:616–621.
2012.(In Chinese). PubMed/NCBI
|