Introduction
Coronary heart disease (CHD) is a major cause of
mortality and disability worldwide (1). The global number of CHD-induced
mortalities is increasing rapidly and is hypothesized to reach 23.3
million by 2030 (2). CHD is
induced by the accumulation of plaques on vascular endothelial
surfaces (3,4). As a complex disease, CHD is
influenced by lifestyle, environmental and genetic factors
(1,5). Twin studies have provided evidence
that genetic and environmental factors play key roles in the
occurrence and development of CHD (6–8).
The IKZF2 gene is located on chromosome 2q13
and encodes the zinc-finger protein, Helios, which is a member of
the Ikaros family of zinc-finger proteins. IKZF2 protein is a
functional component in lymphocyte differentiation (9) and plays a key role in the growth of
the early hematopoietic system (10). Hematopoietic progenitors can
develop into neutrophils, eosinophils, dendritic cells, Langerhans
cells, macrophages and osteoclasts (11). Infiltration of eosinophils has been
found in the myocardial tissue of patients with hypereosinophilic
syndrome (12). The rs12619285
polymorphism of the IKZF2 gene has been associated with a
variation in the blood eosinophil count (13). Furthermore, genes that are involved
in the regulation of eosinophil numbers have been shown to be
involved in the inflammatory regulation and immune responses that
occur during the development of CHD (14–16).
Eosinophils, as a type of white blood cell, exert multidimensional
functions in the occurrence and development of autoimmune diseases
(17), particularly in the
pathogenesis of CHD (18,19) by promoting thrombus formation
(20).
In addition, a previous study found that the
IKZF2 rs12619285 (G/A) polymorphism in European populations
(G allele frequency, 26%; P=5.4×10−10) and East Asian
populations (G allele frequency, 64%; P=0.017) was significantly
associated with CHD (21),
although there were large allele differences between the European
and East Asian populations. Previously, preliminary results
indicated that there was an association between IKZF2
rs12619285 and CHD in the Chinese Han population [G allele
frequency, 62.8%; P=0.07; odds ratio (OR), 1.38, 95% confidence
interval (CI), 0.97–1.98], with no departure from the
Hardy-Weinberg equilibrium (HWE) in the controls (22). Thus, an association was observed
between IKZF2 rs12619285 and CHD (162 cases and 113
controls); however, the statistical power was only 45.2% (22). The small sample size used in this
preliminary study may have been unable to indicate the authentic
association between rs12619285 and CHD (22). Thus, the aim of the present study
was to investigate the association between the rs12619285
polymorphism of the IKZF2 gene in CHD patients and non-CHD
controls using an increased sample size.
Materials and methods
Sample collection
In total, 1,352 samples were collected between
September 2011 and July 2013 in Ningbo Lihuili Hospital (Ningbo,
China) and Ningbo Yinzhou People’s Hospital (Ningbo, China). CHD
cases were confirmed with angiographic evidence that showed
vascular stenosis of >50% in at least one major coronary artery.
In addition, participants with a history of angioplasty or coronary
artery bypass surgeries were classified as CHD cases. Control
samples comprised patients whose vascular stenosis was <50% in
each of the major coronary arteries (23). The severity of CHD was classified
according to the number of major coronary arteries affected by
>50% stenosis (single, double and triple). All the participants
were unrelated and of Han Chinese origin, habituating in Zhejiang
province. Patients were excluded from the study if they suffered
from congenital heart disease, cancer and severe liver or kidney
diseases. All the blood samples were collected by the same
investigator. This study was approved by the Ethical Committees of
Ningbo Lihuili Hospital and Ningbo Yinzhou People’s Hospital. All
the individuals provided written informed consent.
Single nucleotide polymorphism
genotyping
Genomic DNA from the peripheral blood was extracted
using a nucleic acid automatic extractor (Lab-Aid 820; Zeesan
Biotech Co., Ltd., Xiamen, China) and all the DNA samples were
stored in Tris-EDTA buffer. Genotyping was performed using the
melting temperature-shift polymerase chain reaction (PCR) method
(24,25). The Tm-shift PCR approach was used
to differentiate the two allele-specific PCR products that were
amplified using two forward primers and one common reverse primer.
The two forward primers comprised one long and one short primer to
generate two PCR products with different Tm values. The primers
used were as follows: IKZF2-g forward, 5′-GCGGGCAGGGCGGCA
CCAAGGAAAATGGAGCTTCTG-3′; IKZF2-a forward,
5′-GATTACCGACCAAGGAAAATGGAGCTTCTA-3′); and IKZF2 reverse,
5′-GCCTCTTTAGGTAGGGAAGAG AGAACACA-3′. The PCR amplification program
consisted of an initial denaturation at 95°C for 30 sec, followed
by denaturation at 95°C for 30 sec, annealing at 59°C for 30 sec
and extension at 72°C for 30 sec for 40 cycles, and a final
extension at 72°C for 30 sec. PCR was performed using the ABI
GeneAmp® PCR System 9700 with the 96-Well Sample Block
Module (Applied Biosystems Life Technologies, Foster City, CA,
USA). Subsequently, melting curve analysis was performed using a
Roche LightCycler 480® fluorescence quantitative PCR
instrument (Roche Diagnostics, Basel, Switzerland). The melting
curve analysis program was 95°C for 15 sec, 60°C for 30 sec,
followed by increasing the temperature by 0.11°C per sec, until a
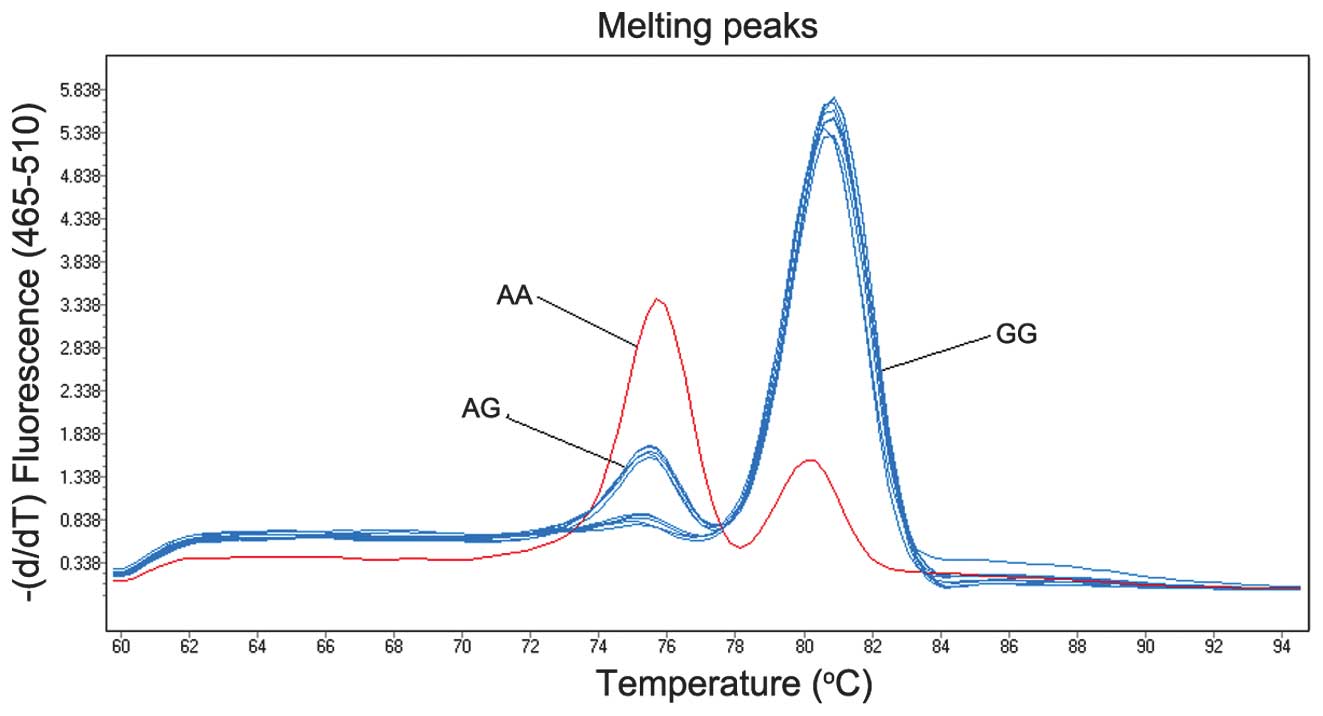
temperature of 95°C was reached. As shown in Fig. 1, the data of the melting curve
analysis was obtained by clustering the fluorescence intensity
analysis (25).
Statistical analysis
HWE analysis was performed using Arlequin software
(version 3.5; Zoological Institute, University of Bern, Bern,
Switzerland) (26). Differences in
the genotype and allele frequencies between the case and control
groups were calculated using CLUMP 22 software (Institute of
Psychiatry, Denmark Hill, London, UK) with 10,000 Monte Carlo
simulations (27). OR and 95% CI
values were determined using an online program (http://faculty.vassar.edu/lowry/odds2x2.html). The
Cochran-Mantel-Haenszel (CMH) test was performed using SAS 9.2
software (SAS Institute, Marlow, UK), while power analysis was
conducted using the software of Power and Sample Size Calculation
(version 3.0.43; Department of Biostatistics, Vanderbilt
University, Nashville, TN, USA) (28). A two-tailed P-value of <0.05 was
considered to indicate a statistically significant difference.
Results
Distribution of genotypes and alleles in
the case and control groups
In total, 721 CHD cases and 631 controls were
recruited for the study in order to evaluate the contribution of
the IKZF2 rs12619285 polymorphism to CHD. Genotype
distributions of the IKZF2 rs12619285 polymorphism were
shown to not deviate from the HWE in the CHD cases, non-CHD
controls and additional subgroups divided by gender or age
(P>0.05; Tables I, II and III). The results did not reveal a
statistically significant association between the rs12619285
polymorphism and CHD in the case-control study (P=0.17; OR, 1.139,
95% CI, 0.972–1.334; Table I).
 | Table IDistribution of genotypes and alleles
in the case and control subjects. |
Table I
Distribution of genotypes and alleles
in the case and control subjects.
| Genotype | Controls (n=631) | Single vessel
(n=352) | Double vessels
(n=168) | Triple vessels
(n=201) | Total cases
(n=721) | χ2 | P-value (df=2) | P-value (df=1) | OR (95% CI) |
|---|
| AA | 84 | 45 | 11 | 17 | 73 | | | | |
| AG | 291 | 156 | 79 | 101 | 336 | | | | |
| GG | 256 | 151 | 78 | 83 | 312 | 3.546 | 0.17 | 0.109 | 1.139
(0.972–1.334) |
 | Table IIDistribution of genotypes and alleles
according to gender. |
Table II
Distribution of genotypes and alleles
according to gender.
| Gender | Genotype (n) | χ2 | P-value (df=2) | HWE | Allele (n) | χ2 | P-value (df=1) | OR (95% CI) |
|---|
|
|
|---|
| GG | AG | AA | G | A |
|---|
| Male |
| Cases (n=516) | 217 | 242 | 57 | | | 0.441 | 676 | 356 | | | |
| Controls
(n=345) | 140 | 157 | 48 | 1.588 | 0.452 | 0.730 | 437 | 253 | 0.852 | 0.356 | 1.099
(0.899–1.344) |
| Female |
| Cases (n=205) | 95 | 94 | 16 | | | 0.326 | 284 | 126 | | | |
| Controls
(n=286) | 116 | 134 | 36 | 3.534 | 0.171 | 0.898 | 366 | 206 | 2.978 | 0.084 | 1.269
(0.968–1.663) |
 | Table IIIDistribution of genotypes and alleles
according to age. |
Table III
Distribution of genotypes and alleles
according to age.
| Genotype (n) | | | | Allele (n) | | | |
|---|
|
| | | |
| | | |
|---|
| Age (years) | GG | AG | AA | χ2 | P-value (df=2) | HWE | G | A | χ2 | P-value (df=1) | OR (95% CI) |
|---|
| ≤55 |
| Cases (n=166) | 70 | 79 | 17 | | | 0.494 | 219 | 113 | | | |
| Controls
(n=224) | 87 | 108 | 29 | 0.862 | 0.650 | 0.667 | 282 | 166 | 0.756 | 0.385 | 1.141
(0.848–1.536) |
| 55–65 |
| Cases (n=244) | 99 | 113 | 32 | | | 1.000 | 311 | 177 | | | |
| Controls
(n=243) | 95 | 113 | 35 | 0.215 | 0.898 | 0.892 | 303 | 183 | 0.200 | 0.655 | 1.061
(0.818–1.377) |
| ≥65 |
| Cases (n=311) | 143 | 144 | 24 | | | 0.148 | 430 | 192 | | | |
| Controls
(n=164) | 74 | 70 | 20 | 2.654 | 0.265 | 0.601 | 218 | 110 | 0.705 | 0.401 | 1.130
(0.849–1.503) |
Associations with age and gender
Since the development of CHD may be involved with
the interaction between genovariation and the environment (29), the samples were divided into
subgroups according to age and gender (30,31),
from which subgroup association tests were performed. Subgroup
analysis by gender did not identify an association between the
rs12619285 polymorphism and CHD in males (P=0.356; OR, 1.099, 95%
CI, 0.899–1.344; Table II) or
females (P=0.084; OR, 1.269, 95% CI, 0.968–1.663). In addition, an
association was not identified between rs12619285 and CHD in the
breakdown analysis by age (P>0.05; Table III).
Associations with genetic models and the
severity of CHD
Association tests were also performed using dominant
and recessive genetic models. However, no statistically significant
difference in the distribution of rs12619285 genotypes or alleles
were identified between the case and control subjects (P>0.05;
Table IV). In addition, CMH
statistical analysis was performed to investigate the association
between rs12619285 and the number of arteries with stenosis in the
CHD patients. Similarly, no statistically significant association
was identified between the rs12619285 polymorphism and the severity
of CHD (P>0.05; data not shown). In addition, the G allele
frequency was found to be 63.6% in the Han Chinese population,
which was similar to the previous preliminary study where the G
allele frequency was 62.8% (22).
The power of the present case-control study was 63.3%.
 | Table IVGenotyping under dominant and
recessive models. |
Table IV
Genotyping under dominant and
recessive models.
| rs12619285 | Dominant
(GG/AG+AA) | χ2 | P-value (df=1) | OR (95% CI) | Recessive
(GG+AG/AA) | χ2 | P-value (df=1) | OR (95% CI) |
|---|
| Total cases | 312/409 | | | | 648/73 | | | |
| Total controls | 256/375 | 1.009 | 0.315 | 1.117
(0.900–1.388) | 547/84 | 3.330 | 0.068 | 1.363
(0.977–1.903) |
| Male cases | 217/299 | | | | 459/57 | | | |
| Male controls | 140/205 | 0.185 | 0.667 | 1.063
(0.806–1.402) | 297/48 | 1.587 | 0.208 | 1.301
(0.863–1.963) |
| Female cases | 95/110 | | | | 189/16 | | | |
| Female
controls | 116/170 | 1.629 | 0.202 | 1.266
(0.881–1.818) | 250/36 | 2.884 | 0.090 | 1.701
(0.916–3.157) |
Discussion
In the present case-control study, a statistically
significant association between IKZF2 rs12619285 and CHD was
not identified, although there was a borderline statistical
difference between the CHD cases and non-CHD controls in the female
subgroup (P=0.08). The case-control study included 1,352
individuals that comprised 721 CHD cases and 631 controls. However,
the current study was relatively small compared with a previous
genome-wide association study that included 12,118 European and
5,212 East Asian individuals (21). Insufficient sample size may explain
the negative association observed in the present study (statistical
power, 63.3%).
Further analysis indicated that there was an ethnic
difference in the allele frequency of the IKZF2 rs12619285
polymorphism. The G allele frequency of rs12619285 in the HapMap
European population was found to be 24.3%, which was much smaller
compared with that of the HapMap Han Chinese in Beijing (CHB)
population (59.3%). In addition, the G allele frequency was 63.6%
in the controls of the present study, which was similar to that of
the HapMap-CHB (59.3%) population (32). The discrepancy in the allele
frequency may help to explain why the present study failed to
confirm the previously identified positive association between the
IKZF2 rs12619285 polymorphism and CHD in the Han Chinese
population.
Genetic heterogeneity may be an additional
explanation for the negative results. A total of 3,355
polymorphisms have been identified in the IKZF2 gene. The
current study only focused on one polymorphism of the IKZF2
gene; thus, the function of the IKZF2 gene may not be fully
demonstrated. Future studies should investigate a greater number of
polymorphisms in order to improve the understanding of the role of
the IKZF2 gene in the susceptibility of CHD.
IKZF2 protein has been identified at an early phase
of development in thymocytes; thus, IKZF2 has been regarded as a
key regulator in lymphocyte differentiation (9). In addition, IKZF2 has been
demonstrated to be involved in the earliest hematopoietic
differentiation of human embryonic stem cells (33), and neutrophils and eosinophils are
developed from hematopoietic progenitors (11). Eosinophils participate in the
production and reproduction of inflammation (21), which may promote the development of
CHD (34,35). Thus, it was hypothesized that the
IKZF2 gene may play a role in the pathogenesis of CHD.
In conclusion, the present case-control study
demonstrated a lack of association between the IKZF2
rs12619285 polymorphism and CHD in the Han Chinese population. This
observation indicated that other IKZF2 polymorphisms or
different genes can affect the variation in blood eosinophil
numbers, which may further the understanding into the contribution
of inflammatory pathways in CHD.
Acknowledgements
The study was supported by grants from the National
Natural Science Foundation of China (nos. 31100919 and 81371469),
the Natural Science Foundation of Zhejiang Province (no.
LR13H020003), the K.C. Wong Magna Fund in Ningbo University and the
Ningbo Social Development Research Projects (no. 2012C50032).
References
|
1
|
Lopez AD, Mathers CD, Ezzati M, Jamison DT
and Murray CJ: Global and regional burden of disease and risk
factors, 2001: systematic analysis of population health data.
Lancet. 367:1747–1757. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mathers CD and Loncar D: Projections of
global mortality and burden of disease from 2002 to 2030. PLoS Med.
3:e4422006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Libby P: Inflammation in atherosclerosis.
Nature. 420:868–874. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kroupis C, Theodorou M, Chaidaroglou A, et
al: The association between a common FCGR2A polymorphism and
C-reactive protein and coronary artery disease revisited. Genet
Test Mol Biomarkers. 14:839–846. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Samani NJ, Erdmann J, Hall AS, et al;
WTCCC and the Cardiogenics Consortium. Genomewide association
analysis of coronary artery disease. N Engl J Med. 357:443–453.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mangino M and Spector T: Understanding
coronary artery disease using twin studies. Heart. 99:373–375.
2013. View Article : Google Scholar
|
|
7
|
Dai J, Krasnow RE, Liu L, Sawada SG and
Reed T: The association between postload plasma glucose levels and
38-year mortality risk of coronary heart disease: the prospective
NHLBI Twin Study. PLoS One. 8:e693322013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vaccarino V, Goldberg J, Rooks C, et al:
Post-traumatic stress disorder and incidence of coronary heart
disease: a twin study. J Am Coll Cardiol. 62:970–978. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Georgopoulos K, Winandy S and Avitahl N:
The role of the Ikaros gene in lymphocyte development and
homeostasis. Annu Rev Immunol. 15:155–176. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kelley CM, Ikeda T, Koipally J, et al:
Helios, a novel dimerization partner of Ikaros expressed in the
earliest hematopoietic progenitors. Curr Biol: CB. 8:508–515. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chio KD, Vodyanik M and Slukvin II:
Hematopoietic differentiation and production of mature myeloid
cells from human pluripotent stem cells. Nat Protoc. 6:296–313.
2011. View Article : Google Scholar
|
|
12
|
Engelmann MG, Kolbe T, Faul C and
Steinbeck G: Hypereosinophilic syndrome associated with
heterozygous factor V gene mutation: an unusual combination
resulting in an acute coronary syndrome and recurrent cerebral
stroke - a case report. Angiology. 55:221–225. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Buysschaert ID, Grulois V, Eloy P, et al:
Genetic evidence for a role of IL33 in nasal polyposis. Allergy.
65:616–622. 2010. View Article : Google Scholar
|
|
14
|
Akhabir L and Sandford A: Genetics of
interleukin 1 receptor-like 1 in immune and inflammatory diseases.
Curr Genomics. 11:591–606. 2010. View Article : Google Scholar
|
|
15
|
Sekkach Y, Mekouar F, Jira M, et al:
Durable efficacity and remission after treatment with imatinib
mesylate for FIP1L1-PDGFRA transcript negative associated
eosinophilic cardiomyopathy. Ann Pharm Fr. 69:277–281. 2011.(In
French). View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Singh RK, Gupta S, Dastidar S and Ray A:
Cysteinyl leukotrienes and their receptors: molecular and
functional characteristics. Pharmacology. 85:336–349. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hogan SP, Rosenberg HF, Moqbel R, et al:
Eosinophils: biological properties and role in health and disease.
Clin Exp Allergy. 38:709–750. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Roivainen M, Viik-Kajander M, Palosuo T,
et al: Infections, inflammation, and the risk of coronary heart
disease. Circulation. 101:252–257. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sato Y, Fukunaga T, Hayashi T and Asada Y:
Hypereosinophilic syndrome associated with occlusive coronary
thrombosis and right ventricular thrombus. Pathol Int. 58:138–141.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sakai T, Inoue S, Matsuyama TA, et al:
Eosinophils may be involved in thrombus growth in acute coronary
syndrome. Int Heart J. 50:267–277. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gudbjartsson DF, Bjornsdottir US, Halapi
E, et al: Sequence variants affecting eosinophil numbers associate
with asthma and myocardial infarction. Nat Genet. 41:342–347. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lian J, Huang Y, Huang RS, et al:
Meta-analyses of four eosinophil related gene variants in coronary
heart disease. J Thromb Thrombolysis. 36:394–401. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
No authors listed. Nomenclature and
criteria for diagnosis of ischemic heart disease. Report of the
Joint International Society and Federation of Cardiology/World
Health Organization task force on standardization of clinical
nomenclature. Circulation. 59:607–609. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Chuang K, Ahluwalia M, et al:
High-throughput SNP genotyping by single-tube PCR with Tm-shift
primers. Biotechniques. 39:885–893. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuan F, Xu J, Ji LD, et al: Application of
Tm-shift genotyping method in genetic studies. Yi Chuan.
34:1484–1490. 2012.(In Chinese). View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Excoffier L and Lischer HE: Arlequin suite
ver 3.5: a new series of programs to perform population genetics
analyses under Linux and Windows. Mol Ecol Resour. 10:564–567.
2010. View Article : Google Scholar
|
|
27
|
Sham PC and Curtis D: Monte Carlo tests
for associations between disease and alleles at highly polymorphic
loci. Ann Hum Genet. 59:97–105. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dupont WD and Plummer WD Jr: Power and
sample size calculations. A review and computer program. Control
Clin Trials. 11:116–128. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu H, Huang XQ, Yang M, et al: MRAS
genetic variation is associated with atherothrombotic stroke in the
Han Chinese population. J Clin Neurol. 9:223–230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li X, He M, Zhu J, Yao P, Li X, Yuan J,
Min X, Lang M, Yang H, Hu FB, Wu T and Wei S: Higher carbohydrate
antigen 125 levels are associated with increased risk of coronary
heart disease in elderly chinese: a population-based case-control
study. PLoS One. 8:e813282013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiang D, Zheng D, Wang L, et al: Elevated
PLA2G7 gene promoter methylation as a gender-specific marker of
aging increases the risk of coronary heart disease in females. PLoS
One. 8:e597522013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
National Center for Biotechnology
Information. HapMap population allele frequency data. http://www.ncbi.nlm.nih.govuri.
Accessed February 7, 2014
|
|
33
|
Halsell SR and Kiehart DP: Second-site
noncomplementation identifies genomic regions required for
Drosophila nonmuscle myosin function during morphogenesis.
Genetics. 148:1845–1863. 1998.PubMed/NCBI
|
|
34
|
Jia LX, Qi GM, Liu O, Li TT, Yang M, Cui
W, Zhang WM, Qi YF and Du J: Inhibition of platelet activation by
clopidogrel prevents hypertension-induced cardiac inflammation and
fibrosis. Cardiovasc Drugs Ther. 27:521–530. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Paoletti R, Gotto AM Jr and Hajjar DP:
Inflammation in atherosclerosis and implications for therapy.
Circulation. 109(23 Suppl 1): III20–III26. 2004. View Article : Google Scholar : PubMed/NCBI
|