Introduction
Lung cancer is one of the most prevalent types of
malignant tumor in China and the overall five-year survival rate
remains unsatisfactory. Approximately 80–85% of all lung cancer
diagnoses are non-small cell lung cancer (NSCLC) (1). Adjuvant chemotherapy, for example
platinum agents combined with cytotoxic agents, including
vinorelbine, gemcitabine and taxane, has become a treatment
standard for NSCLC (2). However, the
resistance of tumor cells to these drugs leads to a poor prognosis
and survival rate. Therefore, the investigation of tumor biomarkers
associated with resistance to chemotherapeutic drugs is
required.
Excision repair cross-complementing 1 (ERCC1) is a
structure-specific DNA repair endonuclease responsible for the
5-incision during DNA excision repair. Clinical studies have
indicated that high ERCC1 expression is associated with
resistance to platinum-based chemotherapy (3). The ribonucleotide reductase subunit M1
(RRM1) gene, located on chromosome 11p15.5, encodes one of
two non-identical subunits of the ribonucleoside-diphosphate
reductase. The ribonucleoside-diphosphate reductase enzyme is
essential for the production of deoxyribonucleotides prior to DNA
synthesis. It has been reported that NSCLC patients with low
RRM1 expression have improved survival when treated with
gemicitabine-based therapy (4).
Thymidylate synthase (TYMS) is the enzyme used to generate
thymidine monophosphate (dTMP), which is subsequently
phosphorylated to thymidine triphosphate for use in DNA synthesis
and repair. TYMS inhibitors, including fluorinated pyrimidine
derivatives, are capable of inhibiting the activity of TYMS. Thus,
TYMS expression is associated with in vivo chemosensitivity
to TYMS inhibitors (5). Class III
β-tubulin (TUBB3) encodes a class III member of the β-tubulin
protein family. The overexpression of TUBB3 is associated
with resistance to docetaxel and paclitaxel (6,7).
Changes in the mRNA expression of the above tumor
biomarkers during tumor development may have an impact on the
prognosis of patients with NSCLC. In the present study, multiplex
branched-DNA liquidchip technology was used to analyze the mRNA
expression of the ERCC1, RRM1, TUBB3 and TYMS genes
in tumor tissue samples from patients with resected NSCLC prior to
chemotherapy. The multiplex branched-DNA technology is a sandwich
nucleic acid hybridization method in which targets are captured
through cooperative hybridization of multiple probes, then coupled
with a fluorescence signal amplification system (8,9). The
signals were detected using the Luminex® 200 system (Luminex,
Austin, TX, USA). It is a multiplex assay that enables the
measurement of the mRNA expression of >30 biomarkers in one test
well (8). The present study focused
on: i) the analysis of the correlation between gene expression and
different clinical characteristics; and ii) the analysis of the
correlation between gene expression and patient survival.
Materials and methods
Patient samples
A total of 72 patients with NSCLC at the General
Hospital of Peoples Liberation Army (Beijing, China) who underwent
curative surgery between March and October 2011 were enrolled in
the present study. Informed consent was obtained from all patients.
Patient information and clinical characteristics are shown in
Table I. A total of 72 tissue
samples were obtained following surgery and the tissues were fixed
in 10% neutral formalin for 24 h, desiccated and embedded in
paraffin. We confirmed the hematoxylin and eosin staining results
of the 72 formalin-fixed paraffin-embedded (FFPE) tissue samples
from the pathology department, which revealed that the tumor
sections contained large numbers of tumor cells (usually >60%).
For each FFPE tissue sample, the tumor tissue was cut using
microdissection techniques and sent to Guangzhou Surexam Medical
Test Center (SurExam Bio-Tech Co., Ltd., Guangzhou, China) for
ERCC1, RRM1, TUBB3 and TYMS mRNA
expression analysis. This study was approved by the ethics comittee
of the General Hospital of People's Liberation Army.
 | Table I.Patient information and clinical
characteristics. |
Table I.
Patient information and clinical
characteristics.
| Variable | No. of patients | Percentage |
|---|
| Age |
|
|
| <60
years | 42 | 58.3 |
| ≥60
years | 30 | 41.7 |
| Gender |
|
|
| Male | 47 | 65.3 |
|
Female | 25 | 34.7 |
| Smoking |
|
|
| Yes | 42 | 58.3 |
| No | 30 | 41.7 |
| Histology |
|
|
|
Adenocarcinoma | 38 | 52.8 |
|
Non-adenocarcinoma | 34 | 47.2 |
|
Differentiation |
|
|
|
High-Mid | 40 | 58.3 |
|
Low | 32 | 41.7 |
| Staging |
|
|
|
I/II | 47 | 65.3 |
|
III | 20 | 27.8 |
| IV | 5 |
6.9 |
| Node
metastasis |
|
|
|
Yes | 25 | 34.7 |
| No | 47 | 65.3 |
mRNA expression analysis of ERCC1,
RRM1, TUBB3 and TYMS using multiplex branched-DNA liquidchip
technology
Multiplex branched-DNA liquidchip technology
measures mRNA expression without RNA extraction, reverse
transcription or polymerase chain reaction amplification. FFPE
tissue samples were homogenized at 65°C for 1 h, then the
supernatant was used for hybridization. Sandwich nucleic acid
hybridization was performed for 16 h using magnetic capture beads
containing probes for ERCC1, RRM1, TUBB3 and
TYMS, the homogenate and another set of capture probes for
ERCC1, RRM1, TUBB3 and TYMS. The
hybridization reaction was performed in a 96-well plate. The
unbound RNA was removed by washing three times with 250 µl wash
buffer (0.1X saline sodium citrate and 0.03% lithium lauryl
sulfate) under a vacuum system. Signal amplification steps were
performed through incubation with 100 µl pre-amplifier solution for
1 h at 50°C, followed by incubation with 100 µl amplifier solution
for 1 h at 50°C, then incubation with probes labeled with biotin
for 1 h at 50°C. Following washing, the samples were incubated with
100 µl streptavidin conjugated phycoerythrin solution at 50°C for
30 min, prior to analysis using the Luminex 200 system
(Luminex).
The fluorescence intensity value of ERCC1,
RRM1, TUBB3 and TYMS generated using the
Luminex 200 system was normalized to that of the house keeping
genes, including β 2-microglobulin, box binding protein and the
transferrin receptor. The normalized values of ERCC1,
RRM1, TUBB3 and TYMS were compared to the
cut-off value of each gene, which was provided by Guangzhou Surexam
Medical Test Center. ERCC1, RRM1, TUBB3 and
TYMS mRNA expression was considered to be positive (high
expression) if the normalized values were equal or higher than the
cut-off value; otherwise, the result was considered to be negative
(low expression).
Statistical analysis
Data were analyzed using the SPSS version 19.0
software package (SPSS, Inc., Chicago, IL, USA). The association
between gene expression and different clinical characteristics was
analyzed using the χ2 test. The correlation between mRNA
expression levels was analyzed using Spearman correlation
coefficients. The Kaplan-Meier method was used to analyze the
correlation between gene expression and patient survival. P<0.05
was considered to indicate a statistically significant
difference.
Results
Correlation between ERCC1, RRM1, TUBB3
or TYMS mRNA expression and clinical characteristics
Tumor specimens from 72 patients with NSCLC were
subjected to mRNA expression analysis. Among the 72 samples, the
incidence rate of a high mRNA expression level of the ERCC1
gene was 38.9% (28/72), RRM1 was 55.6% (40/72), TUBB3
was 47.2% (34/72) and TYMS was 62.5% (45/72; Table II). The correlation between the
expression of these four genes and different clinical
characteristics was assessed. The incidence rate of expression of
the ERCC1 gene in adenocarcinoma (34.2%) was found to be
significantly lower than that in non-adenocarcinoma (44.1%;
P<0.05). Furthermore, the incidence rates of TYMS and
TUBB3 expression in high-median differentiation tissue
samples were observed to be significantly lower than those in low
differentiation tissue samples (P<0.05). However, no correlation
was observed between ERCC1, RRM1, TUBB3 or
TYMS expression and age, gender, smoking status, TNM stage
or lymph node metastasis.
 | Table II.Correlation between mRNA expression
of ERCC1, RRM1, TUBB3 and TYMS and
clinical characteristics. |
Table II.
Correlation between mRNA expression
of ERCC1, RRM1, TUBB3 and TYMS and
clinical characteristics.
| Parameter | No. of
patients | ERCC1 high
expression subgroup, n, (%) | ERCC1 low
expression subgroup, n, (%) | RRM1 high
expression subgroup, n, (%) | RRM1 low
expression subgroup, n, (%) | TUBB3 high
expression subgroup, n, (%) | TUBB3 low
expression subgroup, n, (%) | TYMS high
expression subgroup, n, (%) | TYMS low
expression subgroup, n, (%) |
|---|
| Age |
|
|
|
|
|
|
|
|
|
| <60
years | 42 | 14 (33.3) | 28 (66.7) | 25 (59.5) | 17 (40.5) | 23 (54.8) | 19 (45.2) | 30 (71.4) | 12 (28.6) |
| ≥60
years | 30 | 14 (46.7) | 16 (53.3) | 15
(50) | 15
(50) | 11 (36.7) | 19 (63.3) | 15
(50) | 15
(50) |
| Gender |
|
|
|
|
|
|
|
|
|
|
Male | 47 | 19 (40.4) | 28 (59.6) | 22 (46.8) | 25 (53.2) | 21 (44.7) | 26 (55.3) | 30 (63.8) | 17 (36.2) |
|
Female | 25 | 9
(36.0) | 16 (64.0) | 15 (60.0) | 10 (40.0) | 13 (52.0) | 12 (48.0) | 15 (60.0) | 10 (40.0) |
| Smoking |
|
|
|
|
|
|
|
|
|
|
Yes | 42 | 15 (35.7) | 27 (64.3) | 17 (40.5) | 25 (59.5) | 16 (38.1) | 26 (61.9) | 24 (57.1) | 18 (42.9) |
| No | 30 | 13 (43.3) | 17 (56.7) | 18 (60.0) | 12 (40.0) | 18 (60.0) | 12 (40.0) | 21
(70) | 9
(30.0) |
| Histology |
|
|
|
|
|
|
|
|
|
|
Adenocarcinoma | 38 | 13 (34.2) | 25 (65.8) | 21 (55.3) | 17 (44.7) | 21 (55.3) | 17 (44.7) | 21 (55.3) | 17 (44.7) |
|
Non-adenocarcinoma | 34 | 15 (44.1) | 19 (55.9) | 19 (55.9) | 15 (44.1) | 13 (38.2) | 21 (61.8) | 24 (70.6) | 10 (29.4) |
|
Differentiation |
|
|
|
|
|
|
|
|
|
|
High-median | 40 | 15 (37.5) | 25 (62.5) | 20
(50) | 20
(50) | 11 (27.5) | 29 (72.5) | 19 (47.5) | 21 (52.5) |
|
Low | 32 | 13 (40.6) | 19 (59.4) | 20 (62.5) | 12 (37.5) | 23 (71.9) | 9
(28.1) | 26 (81.3) | 6
(18.7) |
| Staging |
|
|
|
|
|
|
|
|
|
|
I/II | 47 | 16 (34.0) | 31 (66.0) | 23 (48.9) | 24 (51.1) | 22 (46.8) | 25 (53.2) | 30 (63.8) | 17 (36.2) |
|
III | 20 | 10 (50.0) | 10 (50) | 14 (70.0) | 6
(30.0) | 11 (55.0) | 9
(45.0) | 12 (60.0) | 8
(40.0) |
| IV | 5 | 2
(40.0) | 3
(60) | 3
(60.0) | 2
(40.0) | 1
(20.0) | 4
(80.0) | 3
(60.0) | 2
(40.0) |
| Node
metastasis |
|
|
|
|
|
|
|
|
|
|
Yes | 25 | 12 (48.0) | 13 (52.0) | 17 (68.0) | 8
(32.0) | 12
(48) | 13 (52.0) | 15 (60.0) | 10 (40.0) |
| No | 47 | 16 (34.0) | 31 (66.0) | 23 (48.9) | 24 (51.1) | 22 (46.8) | 25 (53.2) | 30 (63.8) | 17 (36.2) |
Correlation between ERCC1, RRM1, TUBB3
or TYMS mRNA expression and patient survival
Following surgery, all 72 patients with NSCLC were
treated with platinum-based doublet chemotherapy. The median number
of chemotherapy cycles was 4.5. The follow-up duration was 379
days. There were eight cases of mortality. The overall survival
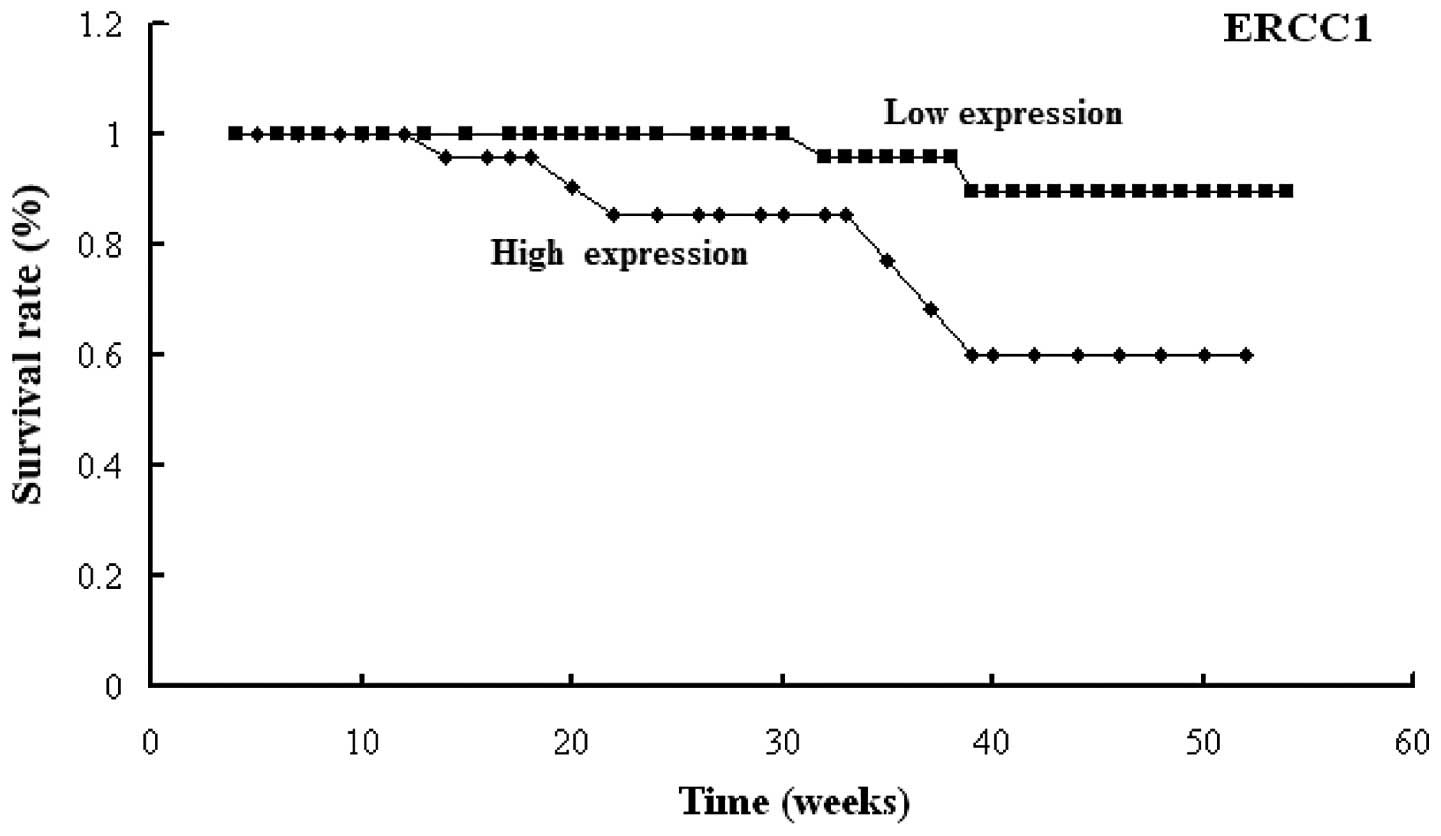
rate was found to be 88.9%. The results indicated that high
expression of ERCC1 was associated with poor prognosis
(P<0.001) and the one-year survival rates of the patients with
high ERCC1 expression and low ERCC1 expression were
78.6 and 95.4%, respectively (Fig.
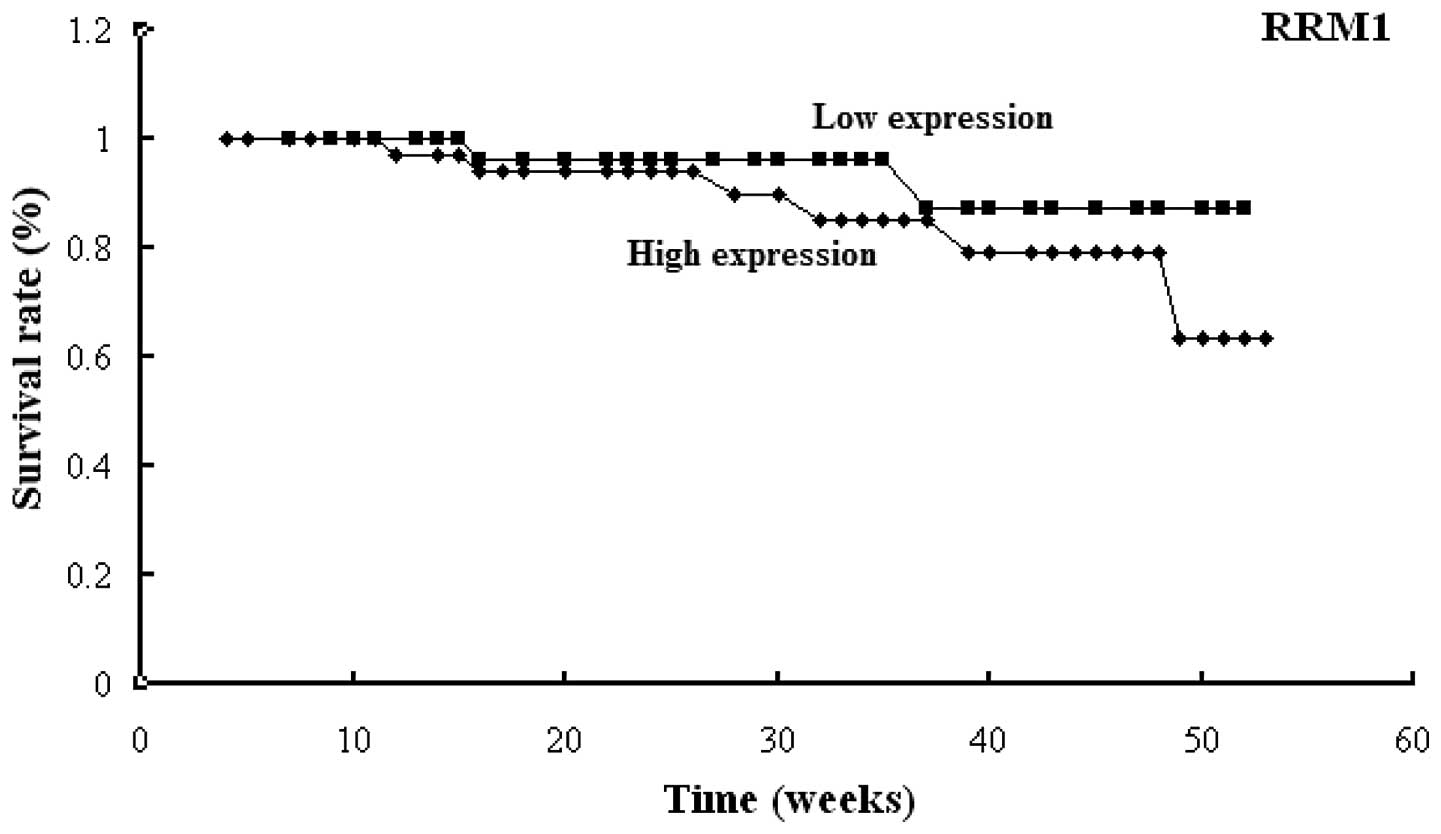
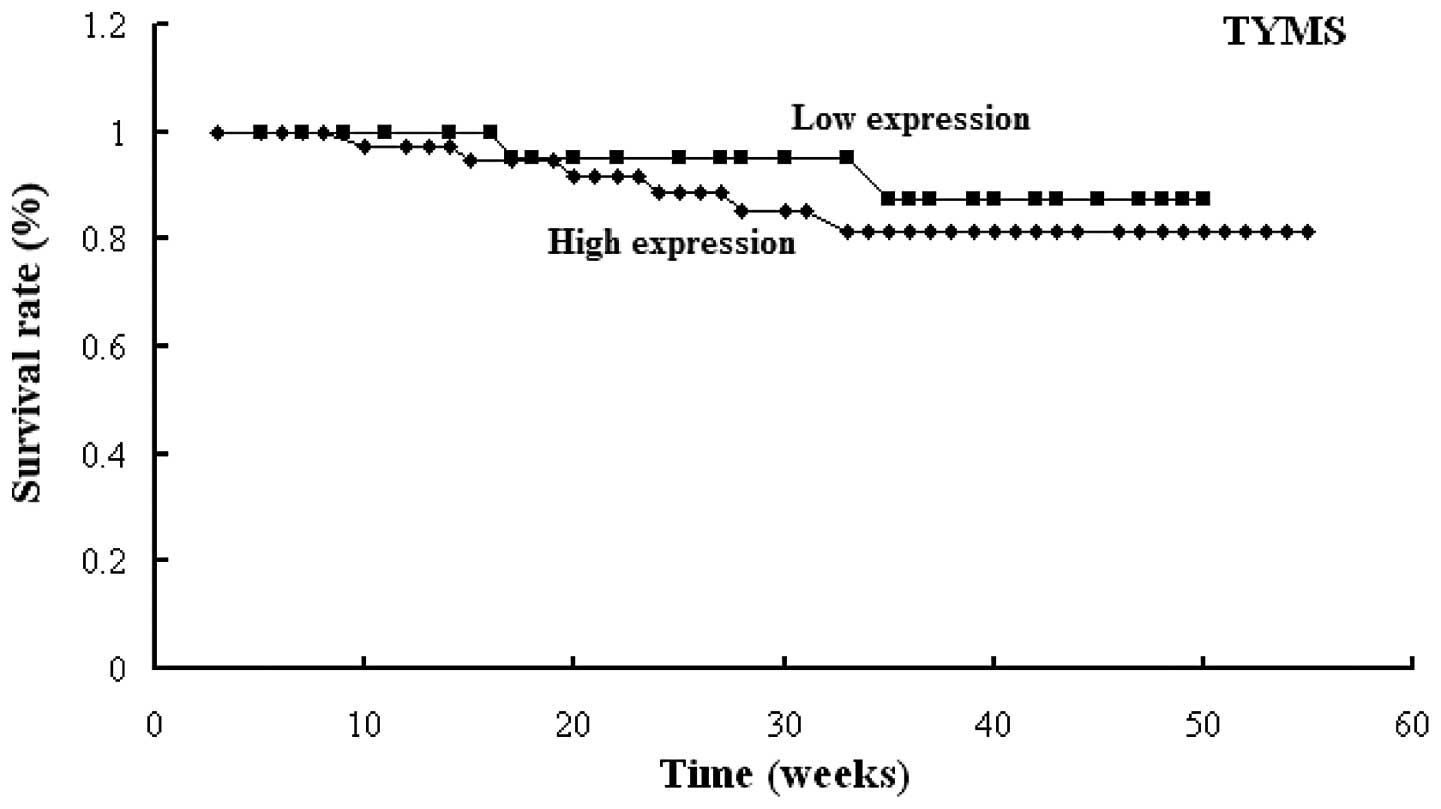
1). Similar results were found for the RRM1,
TUBB3 and TYMS genes (Figs. 2–4).
High expression levels of RRM1, TUBB3 or TYMS
were observed to be significantly associated with poor prognosis
(P=0.001, P=0.001 and P=0.001, respectively). The one-year survival
rates of the patients were: 85 and 93.7% for those with high and
low RRM1 expression, respectively; 85.3 and 94.7% for those
with high and low TUBB3 expression, respectively; and 86.7
and 92.6% in those with high and low TYMS expression,
respectively.
Discussion
The combination of two cytotoxic drugs, for example
platinum agents combined with non-platinum agents, including
gemcitabine, has become the standard first-line treatment for
patients with NSCLC (10). However,
drug resistance is a major problem in NSCLC. Not all patients
benefit from chemotherapy; thus, it is important to identify
predictive markers for response to chemotherapy.
In the present study, the ERCC1, RRM1, TUBB3
and TYMS genes were investigated. ERCC1 is a key factor
involved in nuclear excision repair for platinum-induced adducts.
There is increasing evidence that reduced DNA repair capacity is
correlated with enhanced survival with platinum-based chemotherapy
(11,12). However, certain studies have reported
that the overexpression of ERCC1 is associated with improved
prognosis (13,14). These contradictory reports may be due
to regional disparity or differences in experimental procedures.
The present study found that patients with NSCLC with high
(positive) ERCC1 expression had a poor prognosis and that
the incidence rate of ERCC1 expression in non-adenocarcinoma
was higher than that in adenocarcinoma (P<0.05). Like ERCC1,
RRM1 also has a role in DNA repair processes. RRM1 is the
predominant cellular determinant of the efficacy of the nucleoside
analog gemcitabine, and high expression of RRM1 is
associated with chemoresistance to gemcitabine-based therapy
(15). Clinical studies have shown
that RRM1 expression is correlated with the expression of
ERCC1 (11,16). The present study indicated that
RRM1 expression may be correlated with ERCC1
expression, but not TUBB3 and TYMS expression;
however, no statistical significance was found (r=0.193; P>0.05;
data not shown). Statistical significance may increase with
expansion of the sample size. These results indicate that the
patients with NSCLC with high RRM1 expression had a poor
prognosis. TUBB3 is a structural protein and it has been reported
that high expression of TUBB3 is correlated with low
response rates in patients with NSCLC treated with chemotherapy
regimens using an anti-tubulin drug (17). However, studies have shown
conflicting results, possibly due to variation in trial design or
tumor stage analysis (18). The
present study suggested that overexpression of TUBB3 was
associated with poor prognosis in patients with NSCLC. Furthermore,
the incidence rate of TUBB3 expression in high-median
differentiation tissue samples was found to be significantly lower
than that in the low differentiation tissue samples (P<0.05).
TYMS is important for maintaining the dTMP pool for DNA synthesis
and repair. Various studies have reported that high expression of
TYMS is an unfavorable prognostic factor (19–21);
however, it has also been reported to be a favorable prognostic
factor (22). The findings of the
present study indicate that patients with NSCLC with high
TYMS expression had a poor prognosis and that the incidence
rate of TYMS expression in high-median differentiation
tissue samples was significantly lower than that in the low
differentiation tissue samples (P<0.05).
In conclusion, although the sample size used in the
present study was small, the findings indicate that ERCC1, RRM1,
TUBB3 and TYMS are prognostic factors for survival and may be
predictive biomarkers for platinum-based chemotherapy in patients
with NSCLC. Detecting the mRNA expression of these four genes may
be useful for predicting the efficacy of chemotherapy and may
enhance survival in NSCLC.
References
|
1
|
Chen W, Zhang S and Zou X: Evaluation on
the incidence, mortality and tendency of lung cancer in China.
Thoracic Cancer. 1:35–40. 2010. View Article : Google Scholar
|
|
2
|
Jazieh AR, Bamefleh H, Demirkazik A, et
al: MENA Lung Cancer Regional Guidelines Committee: Modification
and implementation of NCCN guidelines on non-small cell lung cancer
in the Middle East and North Africa region. J Natl Compr Canc Netw.
8(Suppl 3): S16–S21. 2010.PubMed/NCBI
|
|
3
|
Li J, Li ZN, Yu LC, et al: Association of
expression of MRP1, BCRP, LRP and ERCC1 with outcome of patients
with locally advanced non-small cell lung cancer who received
neoadjuvant chemotherapy. Lung Cancer. 69:116–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rosell R, Danenberg K, Alberola V, et al:
Spanish Lung Cancer Group: Ribonucleotide reductase messenger RNA
expression and survival in gemcitabine/cisplatin-treated advanced
non-small cell lung cancer patients. Clin Cancer Res. 10:1318–1325.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peters GJ, Backus HH, Freemantle S, et al:
Induction of thymidylate synthase as a 5-fluorouracil resistance
mechanism. Biochim biophys Acta. 1587:194–205. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Azuma K, Sasada T, Kawahara A, et al:
Expression of ERCC1 and class III beta-tubulin in non-small cell
lung cancer patients treated with carboplatin and paclitaxel. Lung
Cancer. 64:326–333. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Burkhart CA, Kavallaris M and Band Horwitz
S: The role of beta-tubulin isotypes in resistance to antimitotic
drugs. Biochim Biophys Acta. 1471:O1–O9. 2001.PubMed/NCBI
|
|
8
|
Flagella M, Bui S, Zheng Z, et al: A
multiplex branched DNA assay for parallel quantitative gene
expression profiling. Anal Biochem. 352:50–60. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren GJ, Zhao YY, Zhu YJ, et al: Tumor gene
mutations and messenger RNA expression: correlation with clinical
response to icotinib hydrochloride in non-small cell lung cancer.
Chin Med J (Engl). 124:19–25. 2011.PubMed/NCBI
|
|
10
|
Crinò L, Weder W, van Meerbeeck J and
Felip E: ESMO Guidelines Working Group: Early stage and locally
advanced (non-metastatic) non-small-cell lung cancer: ESMO Clinical
Practice Guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 21(Suppl 5): 103–115. 2010. View Article : Google Scholar
|
|
11
|
Zheng Z, Chen T, Li X, et al: DNA
synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J
Med. 356:800–808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee KH, Min HS, Han SW, et al: ERCC1
expression by immunohistochemistry and EGFR mutations in resected
non-small cell lung cancer. Lung Cancer. 60:401–407. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gurubhagavatula S, Liu G, Park S, et al:
XPD and XRCC1 genetic polymorphisms are prognostic factors in
advanced non-small-cell lung cancer patients treated with platinum
chemotherapy. J Clin Oncol. 22:2594–2601. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou W, Gurubhagavatula S, Liu G, et al:
Excision repair cross-complementation group 1 polymorphism predicts
overall survival in advanced non-small cell lung cancer patients
treated with platinum-based chemotherapy. Clin Cancer Res.
10:4939–4943. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Davidson JD, Ma L, Flagella M, et al: An
increase in the expression of ribonucleotide reductase large
subunit 1 is associated with gemcitabine resistance in non-small
cell lung cancer cell lines. Cancer Res. 64:3761–3766. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ceppi P, Volante M, Novello S, et al:
ERCC1 and RRM1 gene expressions but not EGFR are predictive of
shorter survival in advanced non-small-cell lung cancer treated
with cisplatin and gemcitabine. Ann Oncol. 17:1818–25. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sève P, Reiman T, Lai R, et al: Class III
beta-tubulin is a marker of paclitaxel resistance in carcinomas of
unknown primary site. Cancer Chemother Pharmacol. 60:27–34. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sève P and Dumontet C: Is class III
beta-tubulin a predictive factor in patients receiving
tubulin-binding agents? Lancet Oncol. 9:168–75. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ceppi P, Rapa I, Lo Iacono M, et al:
Expression and pharmacological inhibition of thymidylate synthase
and Src kinase in nonsmall cell lung cancer. Int J Cancer.
130:1777–1786. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Grimminger PP, Schneider PM, Metzger R, et
al: Low thymidylate synthase, thymidine phosphorylase, and
dihydropyrimidine dehydrogenase mRNA expression correlate with
prolonged survival in resected non-small-cell lung cancer. Clin
Lung Cancer. 11:328–34. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakagawa T, Otake Y, Yanagihara K, et al:
Expression of thymidylate synthase is correlated with proliferative
activity in non-small cell lung cancer (NSCLC). Lung Cancer.
43:145–149. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng Z, Li X, Schell MJ, et al:
Thymidylate synthase in situ protein expression and survival in
stage I nonsmall-cell lung cancer. Cancer. 112:2765–73. 2008.
View Article : Google Scholar : PubMed/NCBI
|