Introduction
Chronic hepatitis B (CHB) infection is a public
health issue that may develop into cirrhosis, hepatic
decompensation and hepatocellular carcinoma (HCC) (1,2). The
treatment of CHB has changed with the inception of nucleos(t)ide
analogues (NAs), including lamivudine (LAM), adefovir dipivoxil
(ADV), entecavir (ETV) and telbivudine, which target hepatitis B
virus (HBV) reverse transcriptase (RT) activity and inhibit viral
replication (3,4). These antiviral effects may improve the
virological, biochemical and histological status in the majority of
CHB patients. However, the effectiveness of NAs is limited by the
emergence of drug-resistant HBV strains, which may cause hepatitis
flare and hepatic failure (1,5,6).
Although LAM is not recommended as first-line
intervention by current guidelines due to the relatively low
genetic barrier to developing resistance, it was the first NA to be
marketed and has been widely used as the first-line monotherapy
drug for a decade in clinical practice (7–9). ETV, a
high genetic barrier antiviral agent, exhibits >1,500 times
greater potency compared with LAM in vitro (10,11). The
development of ETV resistance associated with virological
breakthrough in NA-naïve patients has been reported to be rare
during 5 years of monotherapy (12).
The development of resistance to ETV in HBV requires at least three
substitutions in the HBV RT gene, including the LAM-related
variants rtL180 and rtM204, in addition to at least one mutation at
rtT184, rtS202 or rtM250 (11,13–17).
Understanding the evolution of these drug-resistant
variants under different antiviral pressures may aid clinicians to
select the correct treatment strategies in a timely manner and to
prevent undesirable clinical outcomes. In a previous longitudinal
study (16), it was reported that
the selection of primary ETV resistance is a two-step process in an
NA-naïve patient and that the development of resistance is
primarily a result of poor compliance with treatment protocols.
Furthermore, a combined therapy of ADV plus ETV was considered to
be the optimal rescue strategy following previous ETV treatment
failure in numerous HBV-infected patients in China, where more
potent drugs, such as tenofovir, have not been approved or are not
affordable for the majority of the population (18). However, the evolution of ETV
resistance during the long-term rescue therapy of ETV plus ADV has
not yet been investigated.
To date, direct sequencing following polymerase
chain reaction (PCR) amplification is the most commonly used method
for detecting a drug-resistant variant; however, this technique is
unable to detect variants in <20% of the heterogeneous viral
population (19). Selecting an
alternative approach for clinically monitoring resistant variants
is a challenging topic in antiviral research. Pyrosequencing
facilitates the detection of small subpopulations of resistant
variants, provides quantitative sequence data and enables
clinicians to better monitor antiviral therapy (20).
On this basis, pyrosequencing was used in the
present study to characterize the evolution of ETV-resistant
variants in 2 patients with differing histories of LAM exposure,
who received ETV combined with ADV as a rescue therapy, with the
aim of improving CHB treatment.
Materials and methods
Patients
In total, 53 patients with CHB (21 NAs-naïve
patients, 32 LAM-treated patients; age, 16–60 years; 42 male, 12
female) were enrolled in this study between June 2007 and July 2008
in Beijing YouAn Hospital, Capital Medical University (Beijing,
China). Once enrolled the patients were treated with ETV once
daily. During the 60-month study period, 2 patients (one received
0.5 mg daily, the other received 1.0 mg daily) were identified as
ETV-resistant due to virological breakthrough, which was defined as
a confirmed increase in the HBV DNA level of >1 log10
copies/ml compared with the nadir HBV DNA level during therapy. The
2 patients received additional administration of ADV at a dose of
10 mg daily as a rescue therapy. Their baseline characteristics are
shown in Table I.
 | Table I.Baseline characteristics of patients A
and B. |
Table I.
Baseline characteristics of patients A
and B.
| Parameter | Patient A | Patient B | Normal range |
|---|
| Gender (M/F) | F | M | – |
| Age (years) | 43 | 49 | – |
| ALT (U/l) | 132.40 | 92.1 | 5–40 |
| AST (U/l) | 91.90 | 48.1 | 8–40 |
| TBil (µmol/l) | 20.70 | 25.5 | 5–20 |
| ALP (U/l) | 69.10 | 74.5 | 35–115 |
| BUN (mmol/l) | 3.17 | 4.97 | 2.29–7 |
| CREA (µmol/l) | 46.00 | 67.00 | 53–106 |
| ALB (g/l) | 41.80 | 45.80 | 36–55 |
| WBC
(109/l) | 4.46 | 5.60 | 4–10 |
| Hb (g/l) | 135 | 144 | 110–160 |
| PLT
(109/l) | 117 | 128 | 100–300 |
| Prothrombin
duration (sec) | 12.70 | 12.6 | 10.7–14.4 |
| INR (ratio) | 1.06 | 1.04 | – |
| CLIA |
|
|
|
|
HBsAg | >250
(positive) | >250
(positive) | <0.05 |
|
Anti-HBsAg | 2.06
(negative) | 0.00
(negative) | <10 |
|
HBeAg | 0.359
(negative) | 513.846
(positive) | <1 |
|
Anti-HBeAg | 0.01
(positive) | 15.81
(negative) | >1 |
|
Anti-HBcAb | 7.83
(positive) | 8.35
(positive) | <1 |
| LAM therapy
duration (months) | 6 | 22 | – |
| HBV DNA
(log10 copies/ml) | 7.80 | 5.31 | 2.46 |
| Genotype | C | C | – |
| Histology score
(inflammation/fibrosis)a | 15/5 | 13/4 | – |
During a health screening, patient A (female; age,
43 years) was diagnosed with asymptomatic CHB infection in the
immune-tolerant phase, at an age of 19 years. Between April 1999
and September 1999, at 35 years old, patient A was administered LAM
therapy in response to elevated alanine aminotransferase (ALT)
levels. Subsequently, partly due to poor medication compliance,
patient A ceased LAM therapy without consulting a doctor after the
elevated ALT level returned to the normal range. Between May 2004
and October 2004, patient A received interferon-α2a treatment due
to an increase in ALT levels, and subsequently received
interferon-α2b therapy between October 2004 and September 2006.
From July 2007, patient A was recruited in this observational study
and received a daily treatment of 0.5 mg ETV in response to
abnormal liver function.
During a health screening, patient B (male; age, 49
years) was diagnosed with asymptomatic CHB infection in the
immune-tolerant phase at an age of 40 years. In January 2002, at an
age of 44 years, patient B was treated with interferon-α1b and LAM
in response to elevated ALT levels. After 6 months of the
combination therapy, the interferon-α1b treatment was discontinued.
Due to poor medication compliance, patient B ceased LAM therapy
without consulting a doctor in January 2003. In March 2006, patient
B resumed LAM therapy due to liver enzyme fluctuations. After 1
year, a YMDD motif mutation was identified in the RT gene of
patient B. In March 2007, ADV was added to the therapy of the
patient. From July 2007, patient B was recruited into this
observational study and received a daily treatment of 1.0 mg ETV in
response to non-decreasing HBV DNA levels.
Patients A and B were diagnosed with CHB according
to the guidelines of the American Association for the Study of
Liver Diseases (7). Histology was
characterized according to the Ishak scoring system (21). Neither patient was co-infected with
hepatitis D virus, hepatitis C virus or human immunodeficiency
virus. The patients were consecutively monitored every 3 months
during the first year of therapy, and every 6 months thereafter,
throughout the treatment course. During each follow-up, the
patients visited their physicians at the hospital and serum
specimens were collected for liver function tests and HBV DNA
quantification assays. The HBV DNA and ALT levels of the patients
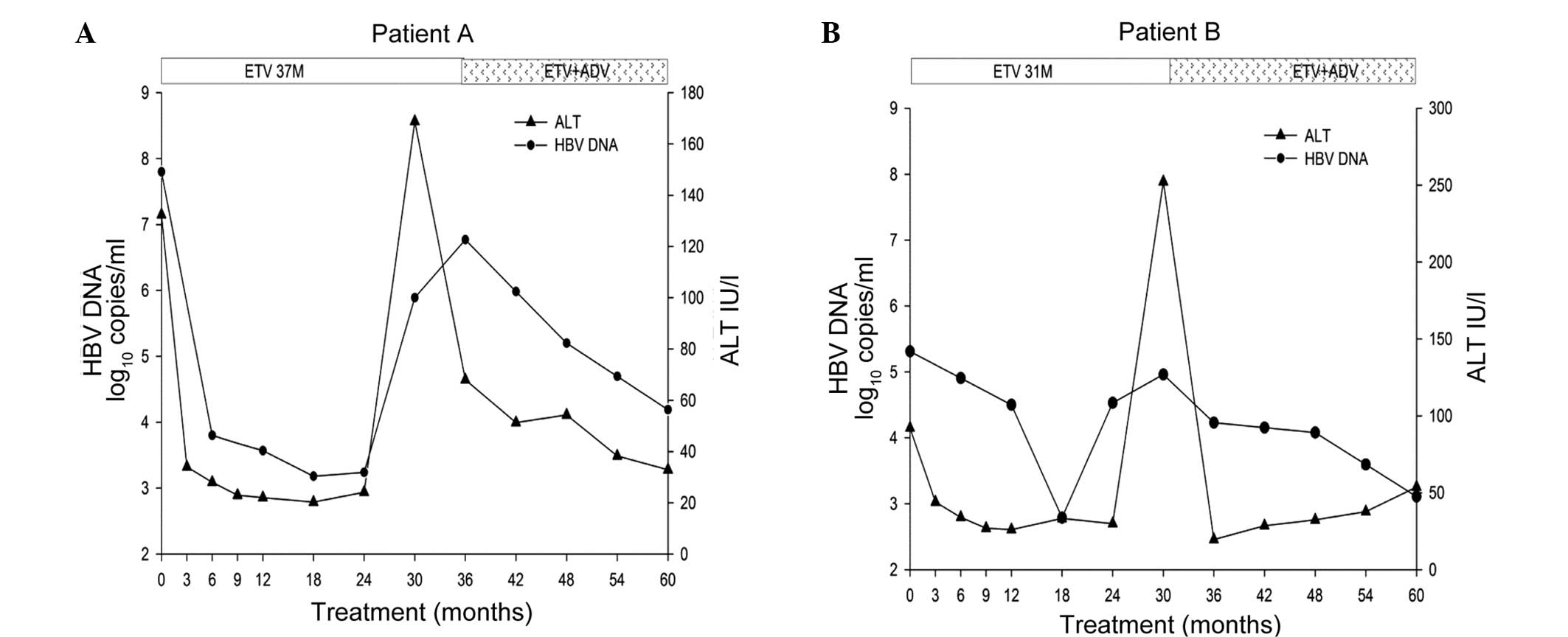
during the 60-month clinical course are shown in Fig. 1. There were no reported issues
concerning medication noncompliance. A total of 25 serum samples
were obtained from each patient, and any remaining serum samples
were stored at −80°C for subsequent research use.
This study was conducted in compliance with the
Declaration of Helsinki. The use of the collected serum samples was
approved by the Medical Ethics Review Committee of Beijing YouAn
Hospital (approval no. LL-2007-002S). Patients A and B provided
written informed consent authorizing access to their medical
records and to store the remaining serum specimens for research
use.
Measurement of liver function and HBV
DNA quantification
ALT and aspartate aminotransferase (AST) levels were
measured using kits purchased from Shanghai Kehua Bio-Engineering
Co., Ltd. (Shanghai, China) and an Olympus Automatic Biochemical
Analyzer (AU5400; Olympus Corporation, Tokyo, Japan) with a cut-off
value of 40 IU/L. The levels of viral markers, including hepatitis
B surface antigen (HBsAg), hepatitis B e-antigen (HBeAg) and
antibody against HBeAg (anti-HBe) were determined using commercial
chemiluminescent immunoassay kits (Beijing Wantai Biological
Pharmacy, Beijing, China) on an ARCHITECT i-20000SR automatic
chemiluminescence immunoassay analyzer purchased from Abbott
Laboratories (Chicago, IL, USA).
The serum HBV DNA level was determined using the
Cobas HBV Amplicor Monitor assay (Roche Molecular Diagnostics,
Pleasanton, CA, USA) at baseline, then every 6 months during the
first year of therapy and annually for the remaining of the
treatment. The lower limit of quantification was 50 IU/ml or 291
copies/ml. From the second year of treatment, the HBV DNA levels
were assessed using pyrosequencing (PyroMark Q24 Mdx system; Qiagen
GmbH, Hilden, Germany) at 18, 30, 42 and 54 months of
follow-up.
qPCR
HBV DNA was extracted from 200 µl serum samples
using QIAamp DNA Blood kit (Qiagen GmbH), according to the
manufacturer's instructions. Nested PCR was used to amplify the HBV
RT region. PCR was conducted using a ProFlex OCR Veriti 96 thermal
cycler purchased from Applied Biosystems (Thermo Fisher Scientific,
Inc., Waltham, MA, USA). All primers were synthesized by Sangon
Biotech Co., Ltd. (Shanghai, China). A total of 5 µl DNA extract
was added in the first 25 µl reaction using primers P5 [nt 63–84,
5′-GTGGCTCCAGTTCA(C)GGAACAGT-3′] and P2 (nt 1285–1264,
5′-CTAGGAGTTCCGCAGTATGGAT-3′). PCR conditions were as follows: 94°C
for 2 min; followed by 30 cycles at 94°C for 1 min, 63°C for 1 min
and 72°C for 1.25 min; and 72°C for 10 min. The second round of PCR
was performed in a 50 µl reaction containing 4 µl first-round PCR
product and primers CN1 (nt 301–319, 5′-TGGCCAAAATTCGCAGTCC-3′) and
CN2 (nt 1019-1000, 5′-GCAAAGCCCAAAAGACCCAC-3′). PCR conditions were
as follows: 94°C for 5 min; followed by 30 cycles at 94°C for 0.5
min, 61°C for 40 sec and 72°C for 1 min; and 72°C for 5 min. Both
PCR rounds shared the same final concentration of MgCl2
(1.5 mM), dNTP (200 µM), primers (0.8 uM each) and Taq Plus
DNA polymerase (50 U/ml) (Dongsheng Biotech Co., Ltd., Guangzhou,
China). A ~719 bp PCR fragment was purified and sequenced
commercially (Invitrogen Life Technologies, Beijing, China) using
primers CN1 and CN2.
Detection of antiviral-resistant
mutations
The pyrosequencing assay was performed according to
the standard protocol of the HBV Drug Resistance Test kit (Qiagen
Shenzhen Co., Ltd., Shenzhen, China) and the PyroMark Q24 MDx
system (Qiagen GmbH). In total, 10 mutation sites were
retrospectively analyzed, including rtL169, rtV173, rtL180, rtA181,
rtT184, rtA194, rtS202, rtM204, rtN236 and rtM250. These sites are
located on the RT domain of HBV DNA polymerase, and were previously
reported to be associated with HBV drug resistance (7,10,14,15,22,23).
For the clonal sequencing assay, the PCR products
were cloned into the pEASY-Blunt Clone vector (TransGen Biotech
Co., Ltd., Beijing, China) according to the manufacturer's
instructions. Following transformation into Escherichia coli
DH5α competent cells (TransGen Biotech Co., Ltd.), 27–30 colonies
per serum sample were selected and the plasmid DNA inserts were
sequenced.
Results
Evolution of ETV-resistant variants
associated with the response in patient A
Patient A initially received ETV therapy when the
serum HBV DNA and ALT levels were 7.80 log10 copies/ml
and 132.40 IU/l, respectively. The viral load was rapidly reduced
to 3.80 log10 copies/ml after 6 months of treatment,
while the ALT level returned to the normal range by month 3. The
HBV DNA level decreased to the lowest level (3.24 log10
copies/ml) at month 24, and then increased to 5.89 log10
copies/ml at month 30, indicating virological breakthrough.
Simultaneously, the ALT level peaked at 168.8 IU/l by month 30,
indicating a biochemical breakthrough. The increase in HBV DNA was
confirmed at the next re-examination 2 months apart using qPCR
(6.63 log10 copies/ml). The increase in HBV DNA was
confirmed at month 36 using a Cobas HBV Amplicor Monitor assay
(Roche Molecular Diagnostics). Therefore, patient A began a
combination therapy of 0.5 mg ETV and 10 mg ADV daily at month 37.
After 23 months of combination therapy, the HBV DNA and ALT levels
decreased to 4.19 log10 copies/ml and 32.90 IU/l,
respectively (Fig. 1). Compared to
the baseline score of 15/5 (inflammation grade/fibrosis stage), the
inflammation and fibrosis scores of percutaneous liver biopsy were
6 and 5 after the 60 months of therapy, according to the Ishak
classification (21).
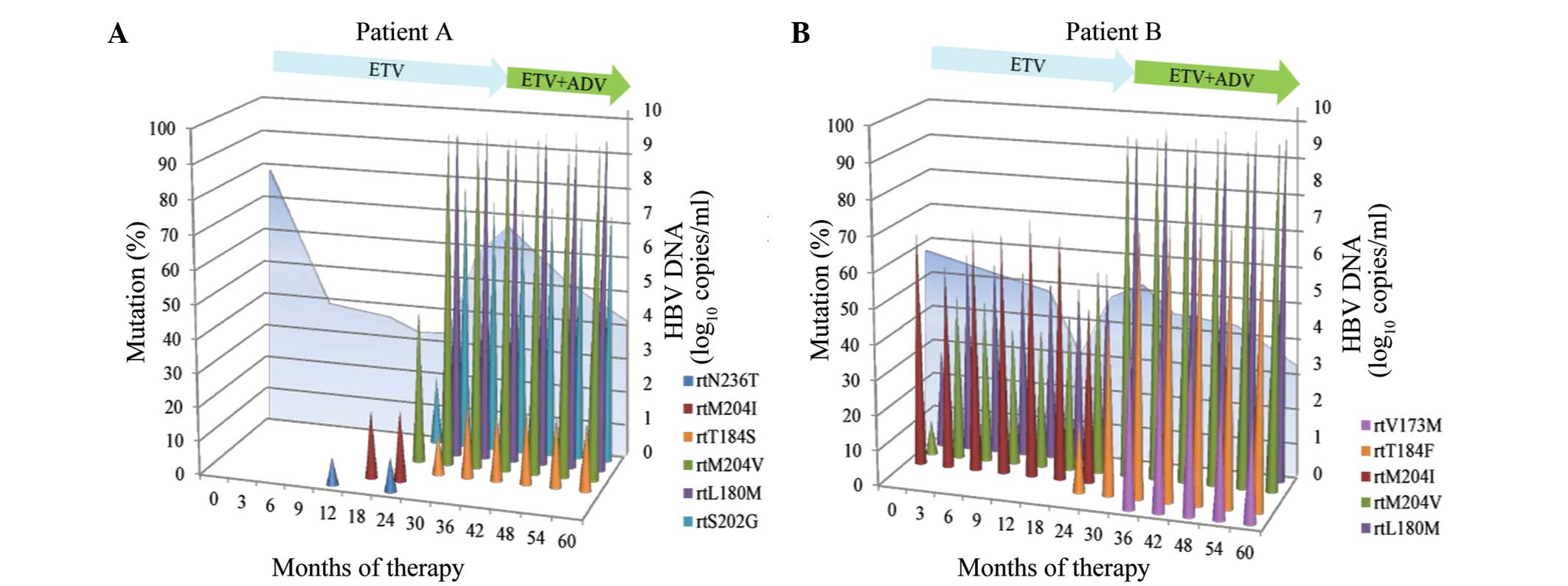
The pyrosequencing analysis shown in Fig. 2A is summarized as follows: i) No
substitution was detected in the baseline sample; ii) the rtN236T
substitution was present in ~10% of the viral population, whereas
the wild-type virus was predominantly repressed at month 12. The
rtN236T reappeared at month 24 with the viral load fluctuation and
was undetectable when outgrowth of the ETV-resistant variants was
observed; iii) among the LAM-resistant variants, the rtM204I
mutation emerged earlier than rtM204V, at month 18. rtM204I was
outcompeted by other mutants, and became undetectable at month 30,
whereas rtM204V and other mutant strains became dominant in the
viral population; iv) at month 30 the rtM204V, rtL180M, rtS202G and
rtT184S variants were present in ~100, 100, 80 and 15% of the viral
population, respectively, which was accompanied by a virological
breakthrough. Prior to the virological breakthrough, the
ETV-associated variant, rtS202G, had been detected at month 24 and
was present in ~20% of the viral population; v) after ADV therapy
was initiated at month 37, the viral load gradually declined; the
rtL180M, rtT184S, rtS202G and rtM204V variants were persistently
dominant in the viral population during the combination
therapy.
A clonal analysis of the samples at month 30, the
point of virological breakthrough, revealed that rtL180M and rtM204
were always co-localized in the same viral strain. At this time,
the viral strains of rtL180M + rtM204V + rtS202G, rtL180M + rtM204V
+ rtT184S and rtL180M + rtM204V + rtS202G + rtT184S were
co-existent and represented 83% (25/30), 10% (3/30) and 7% (2/30)
of the viral population, respectively.
Evolution of ETV-resistant variants
associated with the response in patient B
The ETV treatment reduced the HBV DNA load from 5.31
log10 copies/ml at baseline to a nadir of 2.79
log10 copies/ml at month 18. The ALT levels returned to
the normal range at month 6. Subsequently, the HBV DNA level
increased to 4.53 log10 copies/ml at month 24,
indicating virological breakthrough, which was supported by the
results at month 30. In addition, the ALT levels increased from
29.90 IU/l at month 24 to 252.3 IU/l at month 30, indicating
biochemical breakthrough. From month 31, the patient received a
combination therapy consisting of 1.0 mg ETV and 10 mg ADV daily.
Subsequently, the ALT levels were normalized at month 36, and the
HBV DNA level decreased with the extended therapy (Fig. 1). Compared with the baseline score of
13/4 (inflammation grade/fibrosis stage), the inflammation and
fibrosis scores of percutaneous liver biopsy after a 60-month
therapy were 4 and 3, respectively, according to the Ishak
classification.
The pyrosequencing analysis shown in Fig. 2B may be summarized as follows: i) At
baseline, rtL180M, rtM204V and rtM204I were detected in ~30, 10 and
70% of the viral population, respectively; ii) at month 24, rtT184F
was present in ~20% of the viral population, within the background
of rtL180M and rtM204I/V, which was accompanied by a virological
breakthrough; iii) with the outgrowth of rtL180M, rtM204V and
rtT184F, rtM204I was gradually outcompeted by other viral mutants
and was undetectable in the viral population; iv) following the
initiation of ADV treatment at month 31, the quantity of HBV
started to decline gradually. rtV173M was detectable in ~50% of the
viral population at month 36, while rtL180M, rtM204V and rtT184F
were persistently dominant in the viral population.
A clonal analysis of the samples at month 24 (a
virological breakthrough) and at month 36 (ETV + ADV combination
therapy) revealed that rtL180 M and rtM204V were co-localized in
the same viral strain. At month 24, the rtL180M + rtM204V, rtM204I
and wild-type viral strains co-existed and were present in 17%
(5/30), 47% (14/30) and 37% (11/30) of the viral population,
respectively. At month 36, rtT184F emerged and was co-localized
with L180M + M204V in the same virus-isolate clone. The rtL180M +
rtT184F + rtM204V and rtV173M + rtL180M + rtT184F + rtM204V viral
strains represented 33% (9/27) and 67% (18/27) of the viral
population, respectively.
Discussion
In the present longitudinal study, pyrosequencing
was used to characterize the evolution of ETV-resistant variants in
2 patients that were previously treated with LAM. The addition of
ADV to ongoing ETV treatment for ETV resistance did not appear to
suppress the ETV-resistant variants in LAM-treated patients. A
clonal analysis of the virological breakthrough samples further
revealed that rtT184F or rtS202G were linked with rtL180M and
rtM204V and co-localized in the same viral strain. Therefore, the
present results suggest that LAM therapy should be cautiously
prescribed for NA-naïve patients in the clinical setting.
There is a high genetic barrier to ETV resistance in
NA-naïve patients, and only 0.8% of ETV resistance associated with
virological breakthrough was observed during the 5 years of therapy
(12). In the present observational
cohort study, 2/53 patients presented with ETV resistance
associated with virological breakthrough during 5 years of
follow-up. Previous LAM exposure may contribute to the higher
incidence of ETV resistance. Serum ALT levels may remain normal for
a number of weeks following virological breakthrough (10); however, virological and biochemical
breakthrough were simultaneously detected at month 30 in patient A.
This result does not demonstrate that virological and biochemical
breakthrough occurred simultaneously, but indicates that a 6-month
interval of follow-up may not be sufficient for certain patients in
clinical practice.
Pyrosequencing for resistant variants suggested a
two-step process of ETV resistance in LAM-treated patients. In
patient A, although previously exposed to LAM for 6 months, no
LAM-resistant mutants were detected in the baseline sample,
indicated that LAM-resistant mutants were replaced by the wild-type
virus after cessation of LAM therapy or that resistant variants had
not been selected (24). However,
the LAM-resistant variant, rtM204I, was detected after an 18-month
period of ETV therapy, which is consistent with previous studies
that observed that ETV-resistant variants preceded by LAM-resistant
variants (25,26). At month 24, the resistant variants
rtM204V, rtL180M and rtS202G co-existed in the viral population and
were co-localized in the same viral strain, while virological
breakthrough occurred 6 months later, at month 30 of the therapy.
The present results were inconsistent with a previous report, which
indicated that a new resistant variant, rtS202G, emerged within the
backgrounds of rtM204V and rtL180M, and was accompanied by
virological breakthrough (16). This
discrepancy may be attributed to the difference of ETV-resistant
variants that were proportional in the viral populations.
A previous study reported that ETV resistance
occurred more frequently in LAM-treated patients with LAM-resistant
variant compared with patients without detectable LAM-resistant
variants during ETV monotherapy (27). Prolonged ETV treatment has been
associated with an increased risk of acquiring ETV resistance
(28,29). Compared with the wild-type virus, the
LAM-resistant variant is 8- to 10-fold less sensitive to ETV
(13,30,31). In
patient B, the baseline sample contained the rtL180M and rtM204I/V
variants. At month 24, a new resistant strain that carried all
three mutations (rtM204V, rtL180M and rtT184F) emerged, which was
accompanied by a virological breakthrough. The results of patient B
support the perspective that the LAM-resistant variants were
persistently presented during ETV monotherapy (13,14,32,33).
This result does not support a previous study, which reported that
LAM-resistant variants revert to wild-type HBV during ETV
monotherapy (25). The reversion of
resistant variants to wild-type HBV was considered to be a good
response to rescue therapy. However, a number of studies have
proposed a different explanation for this phenomenon, suggesting
that it is only an intermediate step in the selection of novel
drug-resistant variants (25,34,35).
There are limited studies concerning the treatment
of patients with ETV resistance (13). By using pyrosequencing to detect and
quantify the ETV-resistant variants, the present study revealed
that the ETV + ADV combination therapy may not suppress the
replication of the ETV-resistant strain. This conclusion is
suggested by the gradual decline in the serum HBV DNA levels and
persistent dominance of ETV-resistant variants in the viral
population during the ETV + ADV combination therapy. The
histological benefits may be offset by the emergence of resistance
of ETV during NA antiviral therapy (36). In the present study, patients
underwent liver biopsy at baseline and at month 60 of therapy, and
exhibited improved necroinflammation scores; however, neither
patient demonstrated a significantly improved fibrosis score.
Furthermore, the results indicated that the ETV + ADV combination
rescue therapy partially restored the antiviral efficacy of ETV and
thus may contribute to the improvement of histology.
Although sensitive pyrosequencing methods were used
to analyze consecutive time-point serum samples of up to 60 months
of treatment, the present study has some limitations: The small
number of patients with ETV resistance, the relatively short
duration of ETV + ADV combination therapy, and the testing of only
knwon point mutations. It remains a possibility that novel
mutations associated with antiviral drug resistance influenced the
evolution of resistant variants in the present study. Furthermore,
with prolonged treatment, whether the ADV-resistant variant
(rtA181V/T or rtN236T) will be selected and linked with the
ETV-resistant strain warrants additional investigation.
In conclusion, the additional administration of ADV
in combination with ongoing ETV treatment for ETV resistance may
not suppress the ETV-resistant variants in patients previously
treated with LAM. Although ETV + ADV combination therapy partially
restored the antiviral efficacy of ETV, the ETV-resistant variants
remained the predominant strains during the 60-month therapy
period. The present results suggest that LAM therapy should be
cautiously prescribed for NA-naïve patients in clinical practice.
The additional benefit of quantifying variants using pyrosequencing
may serve as an useful monitoring technique for use in antiviral
therapy.
Acknowledgements
This study was supported by grants from the National
Science and Technology Key Project on ‘Major Infectious Diseases
such as HIV/AIDS, Viral Hepatitis Prevention and Treatment’ (nos.
2012ZX10002004-006, 2012ZX10004904-003-001, 2013ZX10002002-006-001
and 2012ZX10002005), High Technical Personnel Training Item in
Beijing Health System (no. 2011-3-083), Beijing Municipal Science
& Technology Commission (no. Z131107002213019), Special
Scientific Research Fund for Capital Health Development (no.
2011-2018-04) and the Beijing Nova Program (no. Z121107002512056).
The authors would like to thank the subjects who participated in
this study. The authors are also grateful to Lirong He and Yuping
Tang of the R&D Center Asia Pacific [Qiagen (Shenzhen) Co.,
Ltd.] for their assistance with the laboratory work and data
processing.
References
|
1
|
Peng CY, Chien RN and Liaw YF: Hepatitis B
virus-related decompensated liver cirrhosis: benefits of antiviral
therapy. J Hepatol. 57:442–450. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liaw YF and Chu CM: Hepatitis B virus
infection. Lancet. 373:582–592. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Buti M: HBeAg-positive chronic hepatitis
B: Why do I treat my patients with Nucleos(t)ide analogs? Liver
Int. 34(Suppl 1): 108–111. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Viganò M, Mangia G and Lampertico P:
HBeAg-negative chronic hepatitis B: Why do I treat my my patients
with nucleos(t)ide analogs? Liver Int. 34(Suppl 1): 120–126. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dienstag JL, Goldin RD, Heathcote EJ, et
al: Histological outcome during long-term lamivudine therapy.
Gastroenterology. 124:105–117. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zoulim F and Locarnini S: Hepatitis B
virus resistance to nucleos(t)ide analogues. Gastroenterology.
137:1593–1608, e1-e2. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lok AS and McMahon BJ: Chronic hepatitis
B: update 2009. Hepatology. 50:661–662. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
European Association For The Study of The
Liver: EASL clinical practice guidelines: Management of chronic
hepatitis B virus infection. J Hepatol. 57:167–185. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liaw YF, Kao JH, Piratvisuth, et al:
Asian-Pacific consensus statement on the management of chronic
hepatitis B: A 2012 update. Hepatol Int. 6:531–561. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lok AS, Zoulim F, Locarnini S, et al:
Antiviral drug-resistant HBV: standardization of nomenclature and
assays and recommendations for management. Hepatology. 46:254–265.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kobashi H, Fujioka S, Kawaguchi M, et al:
Two cases of development of entecavir resistance during entecavir
treatment for nucleoside-naive chronic hepatitis B. Hepatol Int.
3:403–410. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tenney DJ, Rose RE, Baldick CJ, et al:
Long-term monitoring shows hepatitis B virus resistance to
entecavir in nucleoside-naive patients is rare through 5 years of
therapy. Hepatology. 49:1503–1514. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tenney DJ, Levine SM, Rose RE, et al:
Clinical emergence of entecavir-resistant hepatitis B virus
requires additional substitutions in virus already resistant to
Lamivudine. Antimicrob Agents Chemother. 48:3498–3507. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Villet S, Ollivet A, Pichoud C, et al:
Stepwise process for the development of entecavir resistance in a
chronic hepatitis B virus infected patient. J Hepatol. 46:531–538.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee HW, Kim HJ, Hong SP, et al:
Simultaneous emergence of entecavir resistance mutations in a
nucleoside-naive chronic hepatitis B patient. Intervirology.
55:380–384. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee GH, Inoue M, Toh JK, et al: Two-step
evolution of the hepatitis B drug-resistant mutations in a patient
who developed primary entecavir resistance. Liver Int. 33:642–646.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tenney DJ, Rose RE, Baldick CJ, et al:
Two-year assessment of entecavir resistance in
Lamivudine-refractory hepatitis B virus patients reveals different
clinical outcomes depending on the resistance substitutions
present. Antimicrob Agents Chemother. 51:902–911. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ko SY, Kim BK, Kwon SY, et al: Clonal
evolution of hepatitis B virus polymerase gene mutations during
lamivudine-adefovir combination treatment. World J Gastroenterol.
18:6437–6446. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Degertekin B and Lok AS: Monitoring
antiviral resistance in patients receiving nucleos(t)ide analog
therapies for hepatitis B: which method should be used? J Hepatol.
48:892–894. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lindström A, Odeberg J and Albert J:
Pyrosequencing for detection of lamivudine-resistant hepatitis B
virus. J Clin Microbiol. 42:4788–4795. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Goodman ZD: Grading and staging systems
for inflammation and fibrosis in chronic liver diseases. J Hepatol.
47:598–607. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pallier C, Castéra L, Soulier A, et al:
Dynamics of hepatitis B virus resistance to lamivudine. J Virol.
80:643–653. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pallier C, Rodriguez C, Brillet R,
Nordmann P, Hézode C and Pawlotsky JM: Complex dynamics of
hepatitis B virus resistance to adefovir. Hepatology. 49:50–59.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chayama K, Suzuki Y, Kobayashi M, et al:
Emergence and takeover of YMDD motif mutant hepatitis B virus
during long-term lamivudine therapy and re-takeover by wild type
after cessation of therapy. Hepatology. 27:1711–1716. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng XL, Li QL and Guo JJ: Dynamics of
lamivudine-resistant hepatitis B virus strains in patients with
entecavir rescue therapy. Virus Genes. 47:1–9. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo JJ, Li QL, Shi XF, et al: Dynamics of
hepatitis B virus resistance to entecavir in a
nucleoside/nucleotide-naive patient. Antiviral Res. 81:180–183.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee JH, Cho Y, Lee DH, et al: Prior
exposure to lamivudine increases entecavir resistance risk in
chronic hepatitis B Patients without detectable lamivudine
resistance. Antimicrob Agents Chemother. 58:1730–1737. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ono A, Suzuki F, Kawamura Y, et al:
Long-term continuous entecavir therapy in nucleos(t)ide-naïve
chronic hepatitis B patients. J Hepatol. 57:508–514. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao P, Wang C, Huang L, Xu D and Li T:
Comparison of rescue strategies in lamivudine-resistant patients
with chronic hepatitis B. Antiviral Res. 96:100–104. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sherman M, Yurdaydin C, Simsek H, et al:
Entecavir therapy for lamivudine-refractory chronic hepatitis B:
Improved virologic, biochemical and serology outcomes through 96
weeks. Hepatology. 48:99–108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Colonno RJ, Rose R, Baldick CJ, et al:
Entecavir resistance is rare in nucleoside naive patients with
hepatitis B. Hepatology. 44:1656–1665. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yim HJ, Hussain M, Liu Y, Wong SN, Fung SK
and Lok AS: Evolution of multi-drug resistant hepatitis B virus
during sequential therapy. Hepatology. 44:703–712. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zoulim F: Hepatitis B virus resistance to
entecavir in nucleoside naive patients: Does it exist? Hepatology.
44:1404–1407. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ijaz S, Arnold C, Dervisevic S, et al:
Dynamics of lamivudine-resistant hepatitis B virus during adefovir
monotherapy versus lamivudine plus adefovir combination therapy. J
Med Virol. 80:1160–1170. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Villeneuve JP, Durantel D, Durantel S, et
al: Selection of a hepatitis B virus strain resistant to adefovir
in a liver transplantation patient. J Hepatol. 39:1085–1089. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chang TT, Liaw YF, Wu SS, et al: Long-term
entecavir therapy results in the reversal of fibrosis/cirrhosis and
continued histological improvement in patients with chronic
hepatitis B. Hepatology (Baltimore, Md.). 52:886–893. 2010.
View Article : Google Scholar : PubMed/NCBI
|