Introduction
Cerebrovascular disease (CVD) is a leading cause of
morbidity and mortality worldwide (1) Hypertension, diabetes mellitus (DM),
dyslipidemia, and smoking are risk factors for ischemic CVD (ICVD)
(2–4). Atherosclerosis is the most common cause
of ICVD (5), but gene mutations are
also associated with this disease. CVD may result from genetic and
environmental factors (6,7). Sickle cell anemia, Fabry disease, and
cerebral autosomal dominant arteriopathy with subcortical infarcts
and leukoencephalopathy (CADASIL) are caused by single gene defects
associated with ICVD (6–8). In particular, CADASIL is a small-vessel
disease that can cause stroke. The four cardinal features of
CADASIL are aura, cerebrovascular ischemic events, mood
disturbances and dementia. Lacunar ICVD is frequently associated
with CADASIL and is found in ~85% of symptomatic CADASIL patients
(9).
Notch3 mutations have been shown to cause
CADASIL (10). Notch3 has 31
exons, although most of the mutations are located in exons 2–24,
which encode the extracellular domain (ECD) of the Notch3 receptor.
Mutations in the parts of exons 3 and 4 that code for the first
five epidermal growth factor-like repeats (EGFRs) were present in
70% of the subjects with CADASIL (11). These mutations result in a gain or
loss of cysteine residues in one of the 34 EGFRs in the ECD of the
Notch3 protein, which are very important in vascular system
development and maturity. Numerous polymorphisms have been
identified in the coding sequence of Notch3, some of which
have led to amino acid substitutions (10). However, whether these polymorphisms
affect the Notch3 signaling pathway or are involved in CVD is
unknown.
The aim of the present study was to examine the
association between single-nucleotide polymorphisms (SNPs) in
Notch3 exons 3–6 and lacunar ischemic stroke (LIS). Blood
samples were collected from the control subjects and those with LIS
to analyze Notch3 exons 3–6 to determine whether
Notch3 SNPs are associated with lacunar ICVD.
Patients and methods
Ethics approval
The study protocol was approved by the Ethics
Committee of Beijing Military General Hospital (Beijing, China).
All the participants included in the study provided written
informed consent.
Patients
A total of 140 Chinese Han patients, including 110
lacunar patients and 30 controls (>40 years old) admitted to the
Department of Neurology, Military General Hospital of Beijing
(Beijing, China) between June and December 2010 were included in
the study. Demographic factors (gender and age) were recorded.
Subjects with LIS were enrolled, as were subjects without
infarction (as controls). ICVD was confirmed by magnetic resonance
imaging (MRI) or computed tomography (CT) brain imaging.
Lacunar infarcts were defined as parenchymal defects
with a signal intensity corresponding to that of the cerebrospinal
fluid in all the sequences and were ≤15 mm in diameter, as
determined by brain imaging (MRI or CT). Leukoaraiosis was defined
as changes in white matter diffusion. These changes were observed
on CT scans as bilateral patchy or diffuse areas of hypodensity
with ill-defined margins or hyperintensities on T2-weighted MRI
involving the periventricular and centrum semiovale white
matter.
Clinical history
Clinical history of the participants included CVD,
myocardial infarction, atrial fibrillation, hypertension, DM, and
hyperlipidemia [hypercholesterolemia; high levels of triglycerides
(TGs) and low-density lipoprotein cholesterol (LDL-C)].
Hypertension was defined as a mean blood pressure of >140/90
mmHg or the use of an antihypertensive agent. DM was defined as a
fasting glucose level of ≥6.1 mmol/l, random non-fasting glucose
level of ≥11.1 mmol/l, or the use of anti-diabetic medication.
Hypercholesterolemia was defined as a total serum cholesterol level
of >5.6 mmol/l. High levels of TGs and LDL-C were defined as
total serum TG >1.7 mmol/l and total serum LDL-C >2.7 mmol/l,
respectively.
Brain imaging
MRI was conducted using a 3.0-T system (Discovery
MR750; GE Healthcare, Waukesha, WI, USA). The brain imaging
protocol (slice thickness, 5 mm; interslice thickness, 1.5 mm)
employed the following parameters: T1 fluid-attenuated inversion
recovery images (TR, 1,750 msec; TE, 23 msec; TI, 780 msec; FOV, 24
cm) and T2-weighted images (TR, 7,498 msec; TE, 105 msec; FOV, 24
cm). CT (GE LightSpeed VCT 64 system; GE Healthcare, Waukesha, WI,
USA) was conducted at a slice thickness of 9 mm.
Gene analyses
To identify Notch3 SNPs, genomic DNA was
obtained from blood using a Human Blood DNA kit (Qiagen, Hilden,
Germany). DNA was stored at −20°C prior to genotyping. The region
of interest was amplified by polymerase chain reaction (PCR) to
analyze the SNPs in Notch3 exons 3–6.
The primers for exons 3 and 4 were: forward:
5′-GTTTGCTGCTCTGTTTCCCTG-3′ and reverse:
5′-GGCACAGTCGTAAGTGAGGT-3′. PCR conditions consisted of one cycle
of 10 min at 95°C, 36 cycles of 30 sec at 94°C and 1 min at 65°C,
followed by 30 min at 72°C in a GeneAmp PCR system 2400
(PerkinElmer, Inc., Foster City, CA, USA). The PCR product was 656
base pairs. For the reverse reaction of exons 5 and 6, the primers
used were: forward: 5′-AGAAAACGGCCACTCACCAG-3′ and reverse
5′-ACACCGATGTCTCAATGGGG-3′, at an annealing temperature of 55°C.
The PCR product was 415 base pairs.
Direct sequencing of the PCR product was performed
by a genomic company in Chongqing, China (Genemine Biotechnology
Co., Ltd., Chongqing, China), and GeneTools software (Gene Tools,
LLC, Philomath, OR, USA) was used to identify the SNPs. The
position of the nucleotide sequence was based on the reference
sequence obtained from the National Center for Biotechnology
Information (NCBI) nucleotide database. The SNP database of the
NCBI database was used (www.ncbi.nlm.nih.gov/SNP/).
Statistical analysis
Data were presented as mean ± SD. The differences in
the genotype frequencies and other risk factors were analyzed by
the χ2 test. Mean ages in the two groups and allele
frequencies were compared using the independent samples t-test. The
χ2 tests were used to determine the relationship between
the SNPs and LIS. Statistical analyses were conducted using SPSS
v16.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate statistically significant results.
Results
Patient demographics and disease
characteristics
A total of 110 LIS (67 males) and 30 control (17
males) patients participated in the study. LIS patients were
sub-classified as ‘pure lacunar’ or ‘lacunar + leukoaraiosis’. The
age of the subjects in the lacunar + leukoaraiosis group
(73.24±9.43 years) was significantly greater than that of the
control (63.03±12.25 years) and pure lacunar (67.74±11.04 years)
groups (P<0.05). The characteristics of the subjects are
summarized in Table I.
 | Table I.Clinical characteristics of included
subjects. |
Table I.
Clinical characteristics of included
subjects.
| Characteristics | Control | Lacunar | Pure lacunar | Lacunar +
leukoaraiosis |
|---|
| Male:female | 17:13 | 67:43 | 53:27 | 14:16 |
| Age, years (mean ±
SD) | 63.03±12.25 | 69.05±10.89 | 67.74±11.04 |
73.24±9.43a |
| Hypertension, no.
(%) | 12 (40.0) | 62 (56.4) | 42 (52.5) | 20 (66.6) |
| DM, no (%) | 11 (36.7) | 35 (31.8) | 23 (28.8) | 12 (40.0) |
| Heart disease, no.
(%) | 10 (33.3) | 28 (25.4) | 20 (25.0) | 8
(26.6) |
| Hyperlipidemia, no.
(%) | 6
(20.0) | 37 (33.6) | 28
(35.0)a | 9
(30.0) |
Approximately 25.4% (n=28), 56.4% (n=62), and 31.8%
(n=35) of subjects with LIS also had heart disease, hypertension,
and DM, respectively, whereas 33.3% (n=10), 40.0% (n=12), and 36.7%
(n=11) of the control group had these risk factors, respectively.
Significant differences were not observed in the two groups with
regard to clinical history. Hyperlipidemia was observed in 33.6%
(n=37) of subjects with lacunar ICVD and 20.0% (n=6) of individuals
in the control group. This difference was not significant, although
the rate of hyperlipidemia in subjects with LIS (35.0%, n=28) was
significantly higher than that in the control group (20.0%, n=6)
(P<0.05).
Notch3 SNPs and LIS
A total of 37 SNPs were identified in Notch3
exons 3–6. Of those SNPs, only rs146810942, rs135069047, rs3815188,
rs202157633, rs142778401, rs1043994, rs2285981 and rs149307620 were
present in the current study. Of these 8 SNPS, only rs3815188 and
rs1043994 were identified at higher frequencies. Thus, we only
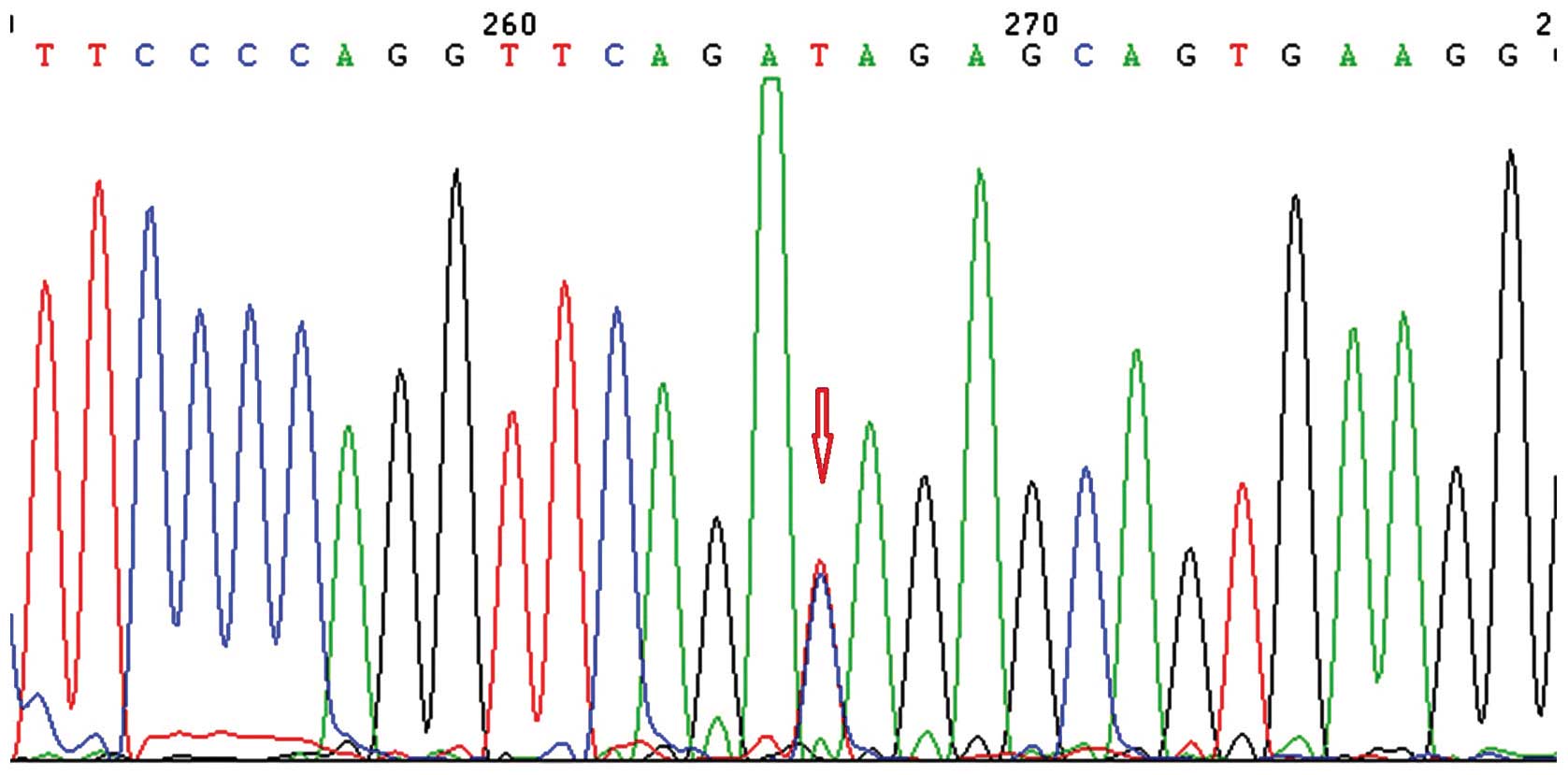
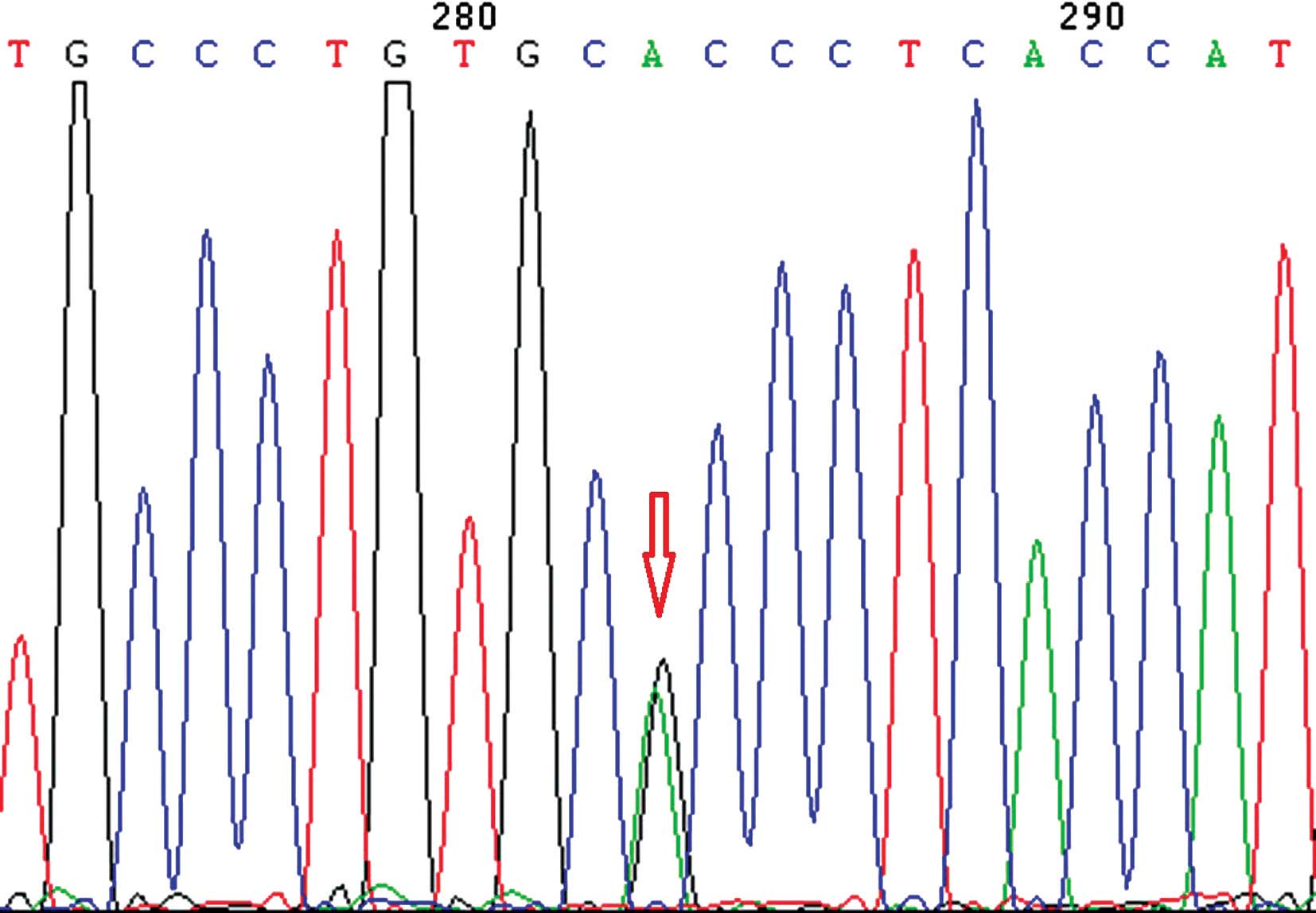
analyzed the association between LIS and the rs3815188 (Fig. 1) and rs1043994 (Fig. 2) SNPs. rs3815188 and rs1043994 were
identified in exons 3 and 4, respectively (Table II).
 | Table II.Genotype distributions. |
Table II.
Genotype distributions.
| Genotype | Control, no. (%) | Lacunar, no. (%) | Pure lacunar, no.
(%) | Lacunar +
leukoaraiosis, no. (%) |
|---|
| rs3815188 |
|
|
|
|
| CC | 15 (50) | 47
(42.7) | 34
(42.5) | 13 (43.3) |
| TT | 1
(3.3) | 22
(20.0) | 16
(20.0) | 6
(20.0) |
| CT | 14
(46.7) | 41
(37.3) | 30
(37.5) | 11 (36.7) |
|
rs1043994a |
|
|
|
|
| AA |
| 5
(4.5) | 5
(6.3) |
|
| GG | 21
(70.0) | 76
(69.1) | 62
(77.5) | 14 (46.7) |
| AG |
9 (30.0) | 29
(26.4) | 13
(16.3) | 16
(53.3)b |
The frequency of the TT genotype in rs3815188 was
higher in the LIS group than in the control group, albeit this
result was not statistically significant (P>0.05). However, we
did not observe any association between the allelic frequency of
rs3815188 and LIS. The frequency of the AG genotype in rs1043994
was higher in the lacunar + leukoaraiosis group than in the control
group (P<0.05). The χ2 test revealed an association
between rs1043994 allelic frequency and LIS.
SNP characteristics and clinical history. The
association between rs3815188 and rs1043994 allelic frequencies and
hypertension, DM, hyperlipidemia, and heart disease were assessed,
albeit no significant difference was observed (Table III).
 | Table III.SNP characteristics and clinical
history. |
Table III.
SNP characteristics and clinical
history.
|
| rs3815388, no.
(%) | rs1043994, no.
(%) |
|---|
|
|
|
|
|---|
| Characteristics | CC | TT | CT | AA | GG | AG |
|---|
| Hypertension | 33 (44.6) | 13 (17.6) | 28 (37.8) | 1 (1.4) | 55 (74.3) | 18 (24.3) |
| DM | 18 (39.1) | 7
(15.2) | 21 (45.7) | 0 | 34 (73.9) | 12 (26.1) |
| Hyperlipidemia | 21 (48.8) | 10 (23.3) | 12 (27.9) | 2 (4.6) | 31 (72.1) | 10 (23.3) |
| Heart disease | 15 (39.5) | 4
(10.5) | 19 (50.0) | 2 (5.2) | 27 (71.1) | 9
(23.7) |
We also found a new SNP located at chromosome
position 15302941(C588T) that was not included in the NCBI SNP
database.
Discussion
CADASIL is a hereditary micro-angiopathic condition
that can cause stroke in young adults. It is a non-atherosclerotic,
non-amyloid angiopathy that affects the small penetrating arteries
in the white matter (12). The mean
age at onset is approximately 45 years (range, 30–70 years), and
most ischemic stroke patients with CADASIL present with lacunar
syndromes (pure motor stroke, pure sensory stroke, sensorimotor
paralysis, ataxic hemiparesis, or dysarthria) (13). Notch3 mutations are the main
cause of CADASIL. Notch3 is located on chromosome
19q13.1–13.2, and the Notch3 receptor protein is a single
transmembrane protein (14) with
intracellular and intercellular domains. Most mutations result in a
gain or loss of cysteine residues in one of the 34 EGFRs in the ECD
of the Notch3 protein. These changes subsequently affect small
vessel structural integrity (15).
A number of SNPs were identified in the coding
sequence of Notch3, some of which led to amino acid
substitutions. In Notch3 exons 3–6, there are 37 SNPs, the
majority of which do not lead to amino acid substitutions. However,
whether these SNPs affect the vessel wall remains to be determined.
We found that 8 SNPs were present in our patients, and for most of
these, the frequency was low. Of these, only rs3815188 and
rs1043994 had higher frequencies, although they did not cause amino
acid substitution. In lacunar patients, rs3815188TT frequency was
higher than that in the pure lacunar and lacunar + leukoaraiosis
groups. Additionally, there was no significant difference in the
rs3815188 frequency when compared to the control, as determined by
the χ2 test. There were significant differences between
the control and lacunar patients with respect to the rs1043994
frequency. However, in the lacunar + leukroarasis patients,
rs1043994AG had a frequency higher than that in the control
subjects. The χ2 test revealed that rs1043994 was
significantly associated with LIS.
Previous findings (16,17)
showed that some Notch3 SNPs had no association with CVD.
However, these studies only analyzed 1 SNP, C381T, and the type of
stroke was not identified. However, in another study (18) all of the common Notch3 SNPs
(n=888), and 4 of these (rs1043994, rs10404382, rs10423702 and
rs1043997) were screened and found to be significantly associated
with the presence and progression of white matter lesions. Our
results are consistent with that study, showing that Notch3
mutations and SNPs are important for the integrity and function of
small vessels. Additionally, findings of a large case-control study
of Chinese patients showed that individuals with a combination of
MTHFR 677TT, ALOX5AP 2354AA, and NOTCH3
381TT/TC had a significantly higher risk of thrombotic stroke
(19) (OR=3.165, 95% confidence
interval: 1.461–6.858; P=0.003).
Moreover, in a study on CADASIL from Chinese
patients it was identified that the mutational spectrum and primary
clinical characteristics of patients with CADASIL were similar to
those in Caucasians (20,21). However, migraine with aura and
abnormal white matter in the temporal pole were less common than in
Caucasians, while brainstem involvement was more common. That study
revealed that Notch3 mutations in Chinese patients had different
effects on vessels.
Our findings have shown that, although the 2 SNPs on
Notch3 exons 3–6 did not result in amino acid substitutions,
the effect of these 2 SNPs on vascular development remains to be
determined, although rs1043994 was associated with LIS. The Notch3
receptor participates in vessel development and maturation
(22). Notch receptor signaling is
involved in the control of proliferation of smooth muscle cells
(SMCs) and maintenance of undifferentiated SMCs. In arteries,
Notch3 protein is the predominant receptor in vascular SMCs, and
signaling is initiated following binding to its ligand, Jagged1
(23). Notch3 gene mutation
results in arteriopathy and leukoencephalopathy (24). However, how ligand-presenting
strategies affect Notch signaling and subsequent upregulation of
SMC differentiation remain to be elucidated. If Notch3
abnormalities are detected earlier, it may be possible to prevent
CVD.
No differences were identified with regard to
Notch3 SNPs between the pure lacunar and lacunar +
leukoaraiosis groups. The prevalence of hypertension was greater in
the lacunar + leukoaraiosis group than that in the control and pure
lacunar groups, suggesting that hypertension is an important factor
in small-vessel disease. The mean age of the lacunar +
leukoaraiosis group was higher than that of the pure lacunar and
control groups (P<0.05). Thus, age may be another risk factor
for leukoaraiosis. rs1043994AG had a higher frequency in the
lacunar + leukoaraiosis group than that in the control and pure
lacunar groups, suggesting that this gene type is important for
small vessels.
The primary limitation of this study was the low
number of participants (especially in the control group).
Additional studies conducted in larger populations are required to
confirm our findings. Additionally, we did not study SNPs from
whole Notch3 sequences, which is imperative to elucidate the
association between the Notch3 SNPs and lacunar infarctions.
The mechanism by which Notch3 SNPs affects small vessels
should also be investigated.
In conclusion, the results of the present study
suggest that Notch3 SNPs are likely to be associated with
lacunar infarction. In particular, we found that rs1043994 was
associated with lacunar infarctions.
References
|
1
|
Wang YJ, Zhang SM, Zhang L, Wang CX, Dong
Q, Gao S, Huang RX, Huang YN, Lv CZ, Liu M, et al: Chinese
guidelines for the secondary prevention of ischemic stroke and
transient ischemic attack 2010. CNS Neurosci Ther. 18:93–101. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartsch JA, Teare GF, Neufeld A, Hudema N
and Muhajarine N: Secondary prevention of stroke in Saskatchewan,
Canada: ηypertension control. Int J Stroke. 8:32–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hankey GJ, Wong KS, Chankrachang S, Chen
C, Crimmins D, Frayne J, Kim JS, Li Y, Liou CW, Merican JS, et al:
Working Group on Stroke and Lipids Management in Asia Consensus
Panel: Management of cholesterol to reduce the burden of stroke in
Asia: consensus statement. Int J Stroke. 5:209–216. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Allen CL and Bayraktutan U: Risk factors
for ischaemic stroke. Int J Stroke. 3:105–116. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim JS, Nah HW, Park SM, Kim SK, Cho KH,
Lee J, Lee YS, Kim J, Ha SW, Kim EG, et al: Risk factors and stroke
mechanisms in atherosclerotic stroke: Intracranial compared with
extracranial and anterior compared with posterior circulation
disease. Stroke. 43:3313–3318. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Meschia JF, Worrall BB and Rich SS:
Genetic susceptibility to ischemic stroke. Nat Rev Neurol.
7:369–378. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu Y, Wang J, Wu Y, Wang G and Hu B: Two
novel mutations in NOTCH3 gene causes cerebral autosomal dominant
arteriopathy with subcritical infarct and leucoencephalopathy in
two Chinese families. Int J Clin Exp Pathol. 8:1321–1327.
2015.PubMed/NCBI
|
|
8
|
Bevan S and Markus HS: Genetics of common
polygenic ischaemic stroke: current understanding and future
challenges. Stroke Res Treat. 2011:1790612011.PubMed/NCBI
|
|
9
|
Viswanathan A, Gschwendtner A, Guichard
JP, Buffon F, Cumurciuc R, O'Sullivan M, Holtmannspötter M, Pachai
C, Bousser MG, Dichgans M, et al: Lacunar lesions are independently
associated with disability and cognitive impairment in CADASIL.
Neurology. 69:172–179. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pescini F, Nannucci S, Bertaccini B, et
al: The cerebral autosomal-dominant arteriopathy with subcortical
infarcts and leukoencephalopathy (CADASIL) scale. Stroke.
43:2871–2876. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dichgans M, Ludwig H, Müller-Höcker J,
Messerschmidt A and Gasser T: Small in-frame deletions and missense
mutations in CADASIL: 3D models predict misfolding of Notch3
EGF-like repeat domains. Eur J Hum Genet. 8:280–285. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Okeda R, Arima K and Kawai M: Arterial
changes in cerebral autosomal dominan arteriopathy with subcortical
infarcts and leukoencephalopathy (CADASIL) in relation to
pathogenesis of diffuse myelin loss of cerebral white matter.
Stroke. 33:2565–2569. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang SC, Jeng JS, Lee MJ and Yip PK: Notch
signaling and CADASIL. Acta Neurol Taiwan. 18:81–90.
2009.PubMed/NCBI
|
|
14
|
Joutel A, Monet M, Domenga V, Riant F and
Tournier-Lasserve E: Pathogenic mutations associated with cerebral
autosomal dominant arteriopathy with subcortical infarcts and
leukoencephalopathy differently affect Jagged1 binding and Notch3
activity via the RBP/JK signaling Pathway. Am J Hum Genet.
74:338–347. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Domenga V, Fardoux P, Lacombe P, Monet M,
Maciazek J, Krebs LT, Klonjkowski B, Berrou E, Mericskay M, Li Z,
et al: Notch3 is required for arterial identity and maturation of
vascular smooth muscle cells. Genes Dev. 18:2730–2735. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ito D, Tanahashi N, Murata M, Sato H,
Saito I, Watanabe K and Fukuuchi Y: Notch3 gene polymorphism and
ischaemic cerebrovascular disease. J Neurol Neurosurg Psychiatry.
72:382–384. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mizuno T, Makino M, Fujiwara Y and
Nakajima K: Lack of association between NOTCH3 gene polymorphism
and cerebrovascular disease in Japanese patients. Ann N Y Acad Sci.
977:252–257. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schmidt H, Zeginigg M, Wiltgen M,
Freudenberger P, Petrovic K, Cavalieri M, Gider P, Enzinger C,
Fornage M, Debette S, et al: CHARGE consortium Neurology working
group: Genetic variants of the NOTCH3 gene in the elderly and
magnetic resonance imaging correlates of age-related cerebral small
vessel disease. Brain. 134:3384–3397. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu J, Sun K, Bai Y, Zhang W, Wang X, Wang
Y, Wang H, Chen J, Song X, Xin Y, et al: Association of three-gene
interaction among MTHFR, ALOX5AP and NOTCH3 with thrombotic stroke:
a multicenter case-control study. Hum Genet. 125:649–656. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Z, Yuan Y, Zhang W, Lv H, Hong D,
Chen B, Liu Y, Luan X, Xie S and Wu S: NOTCH3 mutations and
clinical features in 33 mainland Chinese families with CADASIL. J
Neurol Neurosurg Psychiatry. 82:534–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liao YC, Hsiao CT, Fuh JL, et al:
Characteriazation of CADASIL among the Han Chinese in Taiwan:
Distinct Genotypic and Phenotypic Profiles. PLoS One.
10:e01365012015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Boulos N, Helle F, Dussaule JC, Placier S,
Milliez P, Djudjaj S, Guerrot D, Joutel A, Ronco P, Boffa JJ, et
al: Notch3 is essential for regulation of the renal vascular tone.
Hypertension. 57:1176–1182. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Caolo V, Schulten HM, Zhuang ZW, Murakami
M, Wagenaar A, Verbruggen S, Molin DG and Post MJ: Soluble Jagged-1
inhibits neointima formation by attenuating Notch-Herp2 signaling.
Arterioscler Thromb Vasc Biol. 31:1059–1065. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pippucci T, Maresca A, Magini P, Cenacchi
G, Donadio V, Palombo F, Papa V, Incensi A, Gasparre G, Valentino
ML, et al: Homozygous NOTCH3 null mutation and impaired NOTCH3
signaling in recessive early-onset arteriopathy and cavitating
leukoencephalopathy. EMBO Mol Med. 7:848–858. 2015. View Article : Google Scholar : PubMed/NCBI
|