Introduction
Diabetes, including Type I and Type II, is the most
common non-communicable disease (NCD), with an overall prevalence
of 8.3% worldwide, and is the fourth or fifth leading cause of
mortality in developed countries (1,2). Type II
diabetes accounts for the majority (>85%) of the total diabetes
prevalence (3). It is estimated that
15–25% of patients with diabetes will develop a diabetic foot ulcer
(DFU) at some point during their lifetime (1), and this includes patients with Type I
or Type II diabetes (4). According
to the International Diabetic Federation Report of 2005 (5), 85% of diabetes-related lower extremity
amputations were preceded by DFUs.
Hyperglycemia causes microvascular complications,
including neuropathy, retinopathy and nephropathy (6). The primary function of normal, intact
skin is to control microbial populations that live on the skin
surface and to prevent underlying tissues from becoming colonized
and invaded by potential pathogens (7). In diabetes, a loss of sensation in the
lower extremities may occur, which is known as neuropathy (8). Neuropathic individuals are highly prone
to physical injuries in their lower extremities (9). Any such injury is a potential cause of
a DFU, since hyperglycemia reduces blood flow and the phagocytic
activity of neutrophils and macrophages (10). The most grave consequence of DFUs is
limb amputation, which occurs 10–30 times more frequently in
patients with diabetes than in the general population (46.1–9,600
individuals/105 vs. 5.8–31 individuals/105, respectively) (11–13). The
mortality rates following amputation are 13–40% in the first year,
35–65% in the first 3 years, and 39–80% in the first 5 years
(14).
In addition to the maintenance of glycemic control,
surgical debridement, wound care, pressure offloading and adequate
blood flow maintenance, it is also important to evaluate the type
of microorganisms in infected wounds (15). Infection can convert simple injuries
to gangrene and cause osteomyelitis, leading to lower extremity
amputation (16). The majority of
mild infections are monomicrobial and caused by aerobic
gram-positive cocci, such as Staphylococcus aureus (S.
aureus) and Streptococcus species (spp.). By contrast,
the most severe infections are polymicrobial and caused by aerobic
gram-positive cocci, gram-negative bacilli and anaerobes (17,18).
The emergence of antibiotic resistance in infecting
bacteria can complicate and prolong the treatment regime, and may
even cause chronic wounds to become gangrenous (19). It is noteworthy that
methicillin-resistant S. aureus (MRSA) infections are
associated with a higher mortality rate compared with
methicillin-susceptible S. aureus infections (20). The mecA gene, which is responsible
for methicillin resistance, encodes an altered penicillin-binding
protein (PBP2A) with a low affinity for β-lactam antibiotics
(21,22). Extended spectrum β-lactamases (ESBLs)
are a group of enzymes encoded by genes that are common among
Enterobacteriaceae (23). Most ESBLs
are mutants of Temoneira (TEM)-, sulfhydryl variable (SHV)- and
cefotaximase (CTX-M)-type lactamases, which hydrolyze cefotaxime
and ceftriaxone, and are weakly active against ceftazidime
(24,25). Metallo β-lactamases (MBLs), which
require divalent cations, usually zinc, as metal cofactors for
enzyme activity, are very broad spectrum β-lactamases with the
ability to hydrolyze virtually all classes of β-lactams, including
extended spectrum cephalosporins and carbapenems (26).
Genetic variations and the high incidence of
resistance genes in microbes hinder the control of infections in
DFUs and have an important role in the manifestation of the disease
(27). It is therefore important to
investigate the prevalence of these genes in patients with DFUs in
order to facilitate the prompt control of infection. The aim of the
present study was to investigate the phenotypic and genotypic
prevalence of mecA and ESBLs in bacteria isolated from DFU
samples.
Materials and methods
Sample size and inclusion and
exclusion criteria
In total, 50 diabetic patients with DFU were
included in the study between January 21, 2013 and July 20, 2013.
The study was approved by the Ethics Committee and Institutional
Review Board of the Atta-ur-Rahman School of Applied Biosciences
(Islamabad, Pakistan). Patients of all ages and genders with type 2
diabetes with a foot infection or DFU who visited the Diabetic Foot
Clinic and Out-Patient Department of Pakistan Institute of Medical
Sciences (Islamabad, Pakistan) were included. The exclusion
criteria were as follows: Non-diabetic patients with foot
infection, diabetic patients that had previously received
antibiotic treatment, patients with type 1 diabetes with foot
infection, and DFUs with a duration of >3 weeks. Information on
the duration of the diabetes and DFUs, glycemic control, and
history of previous hospitalization due to the DFU was obtained
from all patients. The patients were examined for the presence of
sensory neuropathy and peripheral vascular disease by measuring the
ankle brachial index. The foot ulcers were classified according to
the New University of Texas classification system (28).
Isolation, identification and
antimicrobial susceptibility testing of the microbes
Cultures of the specimens were obtained at the time
of admission, after the surface of the wound had been washed
vigorously with saline, followed by debridement of the superficial
tissue from the exudates to avoid the isolation of colonizing
flora. Bacteria were isolated through the inoculation of specimens
on a set of selective and non-selective media, such as blood agar
and MacConkey and chocolate agars (Difco; BD Biosciences, Franklin
Lakes, NJ, USA). All inoculated plates were incubated at 37°C for
24–48 h. The bacterial isolates were identified using conventional
biochemical tests.
The Kirby-Bauer disk diffusion method (29) was used for antibiotic susceptibility
testing. The Clinical and Laboratory Standard Institute (CLSI)
guidelines were followed for the selection of media, inoculum
turbidity and preparation of media plates along with the
application of disks and the interpretation of the zone of
inhibition. Suspension was inoculated on the media plate with the
assistance of a sterile glass spreader. Oxoid™ antibiotic disks
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) were applied
using sterile forceps. The zone of inhibition around the tested
antibiotics was measured, and interpretations were made using the
breakpoints elaborated in the CLSI guidelines (30).
Phenotypic detection of ESBL
Isolates that showed intermediate resistance to
third-generation cephalosporin were screened to detect ESBL
production. A double disk synergy test (DDST) was performed for the
phenotypic detection of ESBL production (31). A third-generation cephalosporin disk
was placed on the plate with another disk containing amoxicillin +
clavulanic acid; other combination disks, such as ampicillin +
salbactam and piperacillin + tazobactam, were also tested. Plates
were incubated at 37°C for 18–24 h and the shape of the zone of
inhibition was noted. Isolates that exhibited a distinct
potentiation towards the amoxicillin + clavulanic acid disk were
considered potential ESBL producers. Escherichia coli (E.
coli) ATCC 25922 and Klebsiella pneumoniae (K.
pneumoniae) ATCC 700603 were used as negative and positive
controls for ESBL, respectively. Phenotypic detection of MBLs was
performed using an ethylenediaminetetraacetic acid (EDTA) disk
synergy test (32). For this, an
imipenem disk was placed on a plate inoculated with bacterial
suspension, and 5 µl 0.5 M EDTA was poured on another imipenem disk
and placed on the same plate; differences in the size of inhibition
zone were then observed.
Molecular detection of antibiotic
resistance genes
For the molecular detection of antibiotic resistance
genes, the phenol-chloroform method was used to extract DNA from
bacterial samples (33). Bacterial
cells were grown in Luria Bertani broth overnight at 37°C and
suspended in lysis buffer (0.2 mg/ml proteinase K in 1% sodium
dodecyl sulfate). The suspension was then incubated for 1 h at 55°C
and DNA was extracted twice using phenol-chloroform. Subsequently,
sodium acetate (0.3 M) and cold ethanol were added to precipitate
DNA. The precipitate was centrifuged (Sigma 1–14; Sigma
Laborzentrifugen GmbH, Osterode am Harz, Germany) at 14,462 × g for
2 min at room temperature, after which the DNA pellet was suspended
in 100 µl Tris-EDTA buffer. mecA, β-lactamase (bla)-SHV, bla-CTX-M,
bla-CTX-M-15, bla-TEM and bla-oxacillinase (OXA) genes were
detected using polymerase chain reaction (PCR). For PCR, a total
reaction mixture volume of 25 µl was prepared containing 2 mM
deoxyribonucleotide triphosphates, 10X PCR buffer, 50 mM MgCl2, 50
pM of each primer, 5 µl DNA sample, 1 unit of thermo stable Taq DNA
polymerase (Fermentas; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and nuclease-free water to adjust the final volume. The
reaction mixture was centrifuged (Sigma 1–14; Sigma
Laborzentrifugen GmbH) at 14,462 × g for 30 secs at room
temperature for thorough mixing. Subsequently, PCR was conducted
using the Swift™ MaxPro thermal cycler (Applied Biosystems; Thermo
Fisher Scientific, Inc., Foster City, USA). The primer sequences
are presented in Table I (34–37). The
PCR products were separated by 1% agarose gel electrophoresis, in
which a 100 bp DNA ladder was used as a reference (Invitrogen;
Thermo Fisher Scientific, Inc.).
 | Table I.Primer sequences of mecA and extended
spectrum β-lactamase genes. |
Table I.
Primer sequences of mecA and extended
spectrum β-lactamase genes.
| Target gene | Primer | Sequence
(5′-3′) | Tm, °C | GC, % | Ref. |
|---|
| bla-SHV | F |
CTTTATCGGCCCTCACTCAA |
60.4 | 50 | (25) |
|
| R |
AGGTGCTCATCATGGGAAAG |
60.4 | 50 |
|
| bla-OXA | F |
GGCACCAGATTCAACTTTCAAG |
60.8 | 45 | (26) |
|
| R |
GACCCCAAGTTTCCTGTAAGTG |
62.7 | 50 |
|
| bla-CTX-M | F |
ATGTGCAGTACCAGTAAAGTGATGGC |
64.6 | 46 | (27) |
|
| R |
TGGGTAAAATAAGTCACCAGAATCAGCGG | 66 | 45 |
|
| bla-TEM | F |
CGCCGCATACACTATTCTCAGAATGA |
64.6 | 46 | (28) |
|
| R |
ACGCTCACCGGCTCCAGATTTAT |
64.6 | 52 |
|
| bla-CTX-M-15 | F |
AGGCAGACTGGGTGTGGCAT |
64.5 | 60 | – |
|
| R |
TTACCCAGCGTCAGATTCCG |
62.4 | 55 |
|
| mecA | F |
GTAGAAATGACTGAACGTCCGATAA |
54.4 | 40 | – |
|
| R |
ATTGGCCAATTCCACATTGTTTCG | 54 | 42 |
|
Results
Neuropathy and ulcer grade
A total of 50 patients fulfilled the inclusion
criteria and were included in the study. Out of those 50 subjects,
29 (58%) were men and 21 (42%) women (age range, 36–80 years; mean
age, 58 years). According to the University of Texas
Classification, 11 patients had stage-B grade-I ulcer, 16 had
stage-B grade-II ulcer, 7 had stage-B grade-III ulcer, 2 had
stage-C grade-I ulcer, 1 had stage-C grade-II, 1 had stage-D
grade-I ulcer, 5 had stage-D grade-II ulcer and 7 had stage-D
grade-III ulcer (Table II). The
specimens containing clinically significant pathogens included
wound swabs (37/50; 74%), tissue (7/50; 14%), pus (4/50; 8%) and
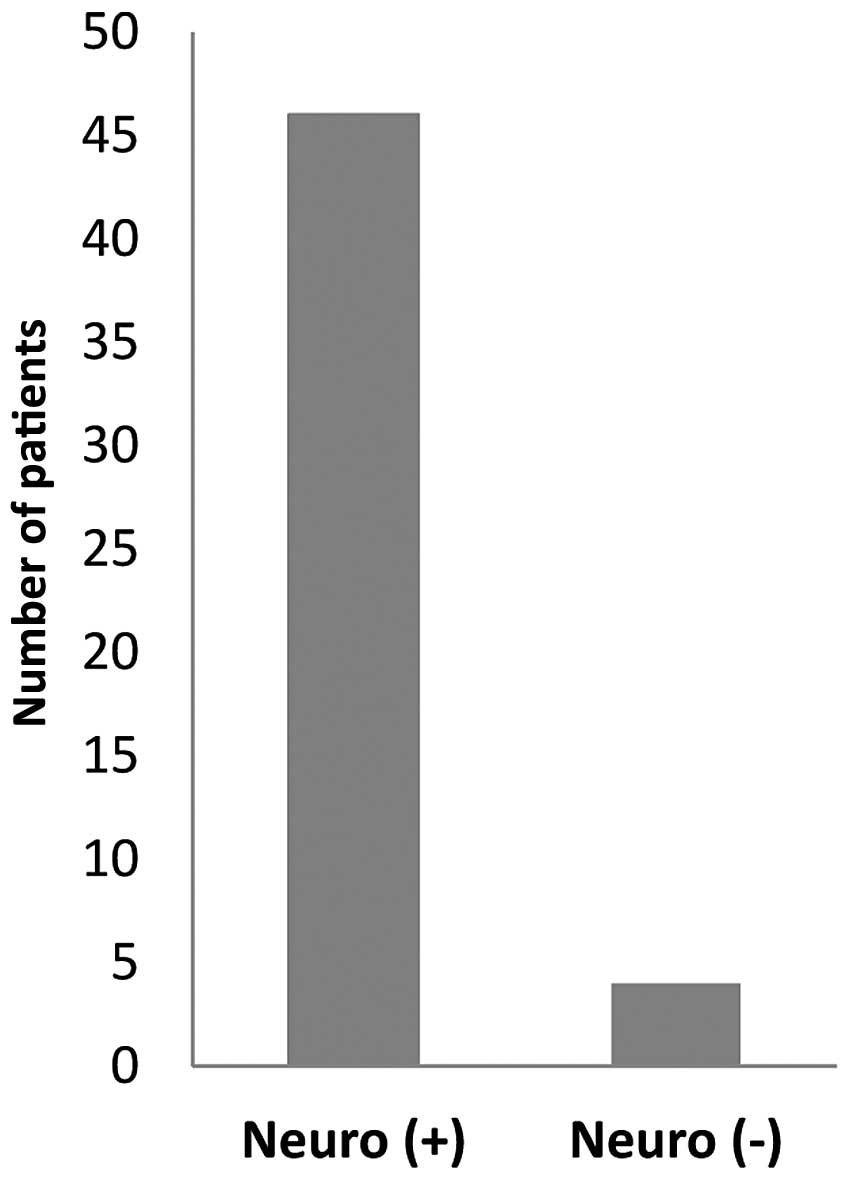
bone (2/50; 4%). Of the 50 patients, 46 patients (92%) had sensory
neuropathy while 4 patients (8%) did not (Fig. 1).
 | Table II.Stages and grades of DFU in the
patients according to the University of Texas Classification System
(19). |
Table II.
Stages and grades of DFU in the
patients according to the University of Texas Classification System
(19).
|
| DFU grade |
|
|---|
|
|
|
|
|---|
| Stage of ulcer | I | II | III | Total |
|---|
| A | 0 | 0 | 0 | 0 |
| B | 11 | 16 | 7 | 34 |
| C | 2 | 1 | 0 | 3 |
| D | 1 | 5 | 7 | 13 |
| Total | 14 | 22 | 14 | 50 |
Microbiology and antimicrobial
susceptibility testing of diabetic foot wounds sample
A total of 99 bacterial isolates were obtained from
the 50 patients with DFUs. In these patients, gram-negative bacilli
(59.59%) were isolated more frequently than gram-positive cocci
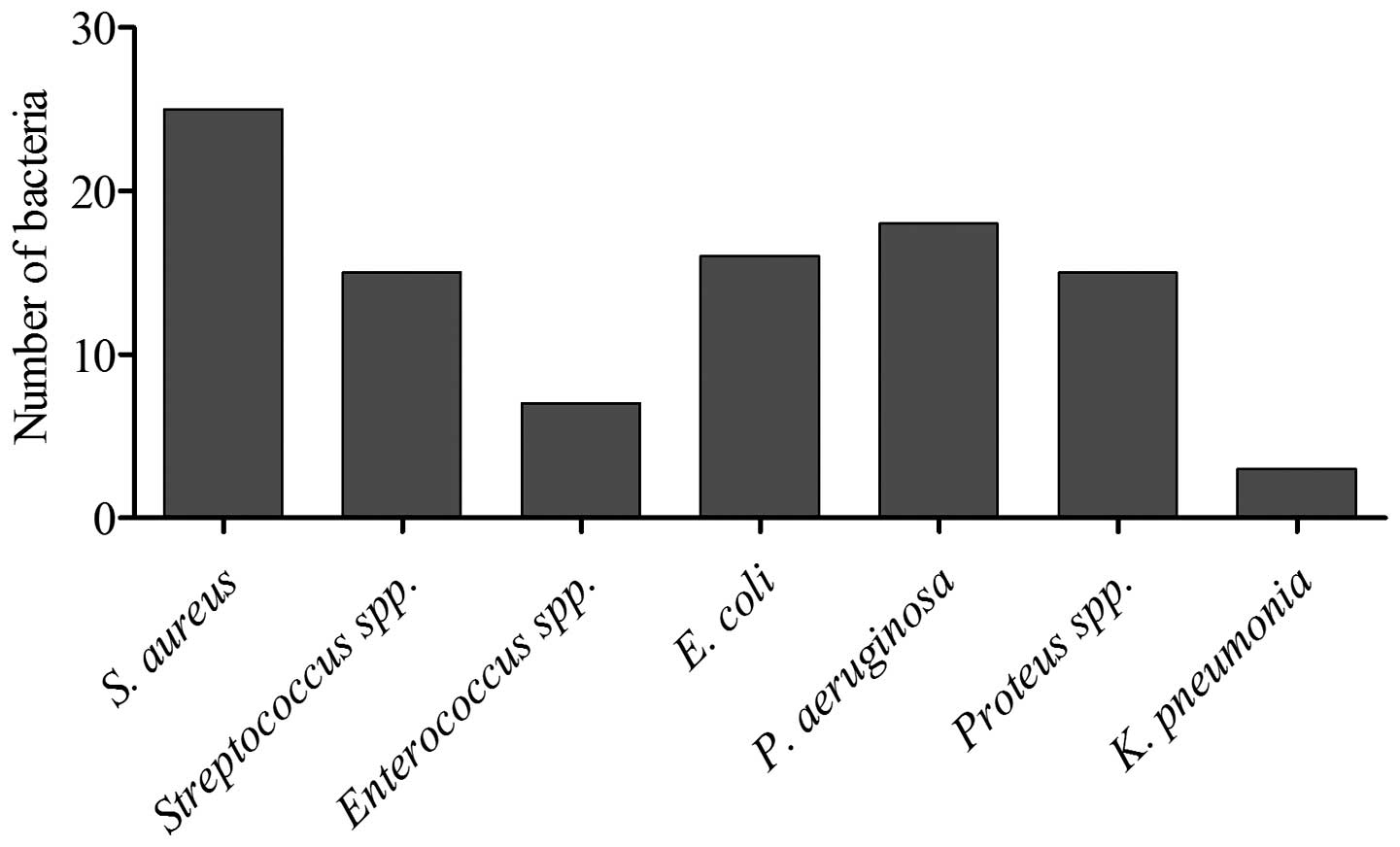
(40.4%). The most common isolate was S. aureus (25%),
followed by Pseudomonas aeruginosa (P. aeruginosa;
18.18%) and E. coli (16.16%). The other isolated organisms
were Streptococcus spp. (15.15%), Enterococcus spp.
(9%), Proteus spp. (15.15%) and K. pneumoniae (3%)
(Fig. 2). Single organisms were
isolated from 14 samples (28%) and mixed bacterial growths were
identified in 36 samples (72%). The details of the organisms
isolated from the infected foot lesions are presented in Fig. 2. The mean number of isolates per
culture-positive sample was 1.9.
All S. aureus isolates were resistant to
penicillin, out of which 90% exhibited resistance against
oxacillin, cefoxitin and ceftazidime. Vancomycin showed inhibitory
effects for only 20% of the S. aureus isolates. By contrast,
100% of the Streptococcus spp. isolates exhibited resistance
to oxicillin, cefotaxime, ceftriaxone, cefepime, cefoxitin,
ceftazidime and penicillin, while 40% showed susceptibility to
vancomycin. According to the sensitivity data, 70% of the S.
aureus and 80% of Streptococcus spp. isolates were
multidrug-resistant (Table III).
All P. aeruginosa and K. pneumoniae isolates, as well
as 83.33% of the Proteus spp. and 75% of the E. coli
isolates were found to be resistant to ceftazidime. All
gram-negative isolates exhibited high susceptibility to
chloramphenicol and meropenem. K. pneumoniae and P.
aeruginosa were found to be the most resistant, with >60% of
strains exhibiting antibiotic resistance, whereas for
Proteus spp. and E. coli, <55% of strains were
resistant (Table IV).
 | Table III.Antibiotic resistance (%) pattern of
gram-positive bacteria. |
Table III.
Antibiotic resistance (%) pattern of
gram-positive bacteria.
| Antibiotics | S.
aureus |
Streptococcus spp. |
|---|
| Oxicillin | 90 | 100 |
| Cefotaxime | 80 | 100 |
| Ceftriaxone | 80 | 100 |
| Cefepime | 70 | 100 |
| Cefoxitin | 90 | 100 |
| Ceftazidime | 90 | 100 |
| Aztreonam | 30 | 80 |
| Imepenem | 50 | 80 |
| Gentamicin | 30 |
0 |
| Cefoperazone | 50 | 60 |
| Amoxicillin +
clavulanic acid | 40 | 60 |
| Ampicillin +
sulbactam | 30 | 60 |
| Tazobactam +
piperacillin |
0 | 40 |
|
Chloramphenicol |
0 | 20 |
| Penicillin | 100 | 100 |
| Vancomycin | 80 | 60 |
 | Table IV.Antibiotic resistance (%) pattern of
gram-negative bacteria. |
Table IV.
Antibiotic resistance (%) pattern of
gram-negative bacteria.
| Antibiotics | K.
pneumoniae | P.
aeruginosa | Proteus
spp. | E. coli |
|---|
| Oxicillin | 100 |
66.7 |
83.3 | 50 |
| Cefotaxime | 80 | 100 |
83.3 | 75 |
| Ceftriaxone | 100 | 100 | 100 | 50 |
| Cefepime | 100 | 100 |
83.3 | 50 |
| Cefoxitin | 80 | 100 | 100 | 75 |
| Ceftazidime | 100 | 100 |
83.3 | 75 |
| Aztreonam | 60 |
33.3 | 50 | 50 |
| Imepenem | 60 |
66.7 |
33.3 | 50 |
| Cefoperazone | 60 | 100 |
66.7 | 50 |
| Amoxicillin +
clavulanic acid | 80 | 100 |
16.7 | 50 |
| Ampicillin +
sulbactam | 60 |
66.7 |
33.3 | 50 |
| Tazobactam +
piperacillin | 80 |
66.7 |
33.3 | 75 |
| Amikacin | 60 |
33.3 |
33.3 | 25 |
| Meropenim | 40 | 0 |
16.7 | 25 |
|
Chloramphenicol | 20 | 0 | 0 | 25 |
Phenotypic and genotypic detection of
MRSA and ESBL
The phenotypic prevalence of the mecA gene was found
to be 88%. No inhibition zone was observed around the 30-µg
cefoxitin disk. Out of 25 S. aureus strains, 21 were found
to be positive for the mecA gene PCR, with an overall prevalence of
84%. DDST results showed that 66.66, 33.33, 66.7 and 50% of the
Proteus spp., P. aeruginosa, K. pneumoniae and
E. coli populations, respectively, were ESBL producers. In
the case of the cefotaxime + clavulanic acid/ceftazidime +
clavulanic acid combination disk test for ESBL detection, a
>5-mm increase was observed in the size of the zone of
inhibition around the cefotaxime or ceftazidime disk containing the
ESBL inhibitor clavulanic acid, as compared with the simple
cefotaxime or ceftazidime disk. According to the cefotaxime +
clavulanic acid/ceftazidime + clavulanic acid combination disk
test, K. pneumoniae and E. coli had the highest
prevalence (100%) of ESBL producers, while the EDTA disk synergy
test confirmed P. aeruginosa and K. pneumoniae as
having the highest prevalence of MBL producers; E. coli was
found to have the lowest prevalence, with only 37% of strains
producing MBL (Table V).
 | Table V.Phenotypic detection of methicillin
resistance, extended spectrum β-lactamase and metallo β-lactamase
production (%). |
Table V.
Phenotypic detection of methicillin
resistance, extended spectrum β-lactamase and metallo β-lactamase
production (%).
| Bacterial
isolates | Double disk synergy
test | CTX + CL/CAZ + CL
combination disk test | EDTA combinationt
disk test |
|---|
| Proteus
spp. |
66.7 | 73 | 80 |
| P.
aeruginosa |
33.3 | 33 | 100 |
| K.
pneumoniae |
66.7 | 100 | 100 |
| E. coli | 50 | 100 | 37 |
PCR showed that all K. pneumoniae and E.
coli isolates were positive for bla-CTX-M, bla-CTX-M-15,
bla-TEM, bla-OXA and bla-SHV resistance genes (Fig. 3). In addition, 72.2% of the P.
aeruginosa strains were positive for bla-SHV and 66.7% for
bla-CTX-M, bla-CTX-M-15 and bla-TEM. However, only 33.3% of the
P. aeruginosa strains were positive for the bla-OXA gene. In
Proteus spp., the resistance gene with the highest
prevalence (80%) was bla-SHV (Table
VI).
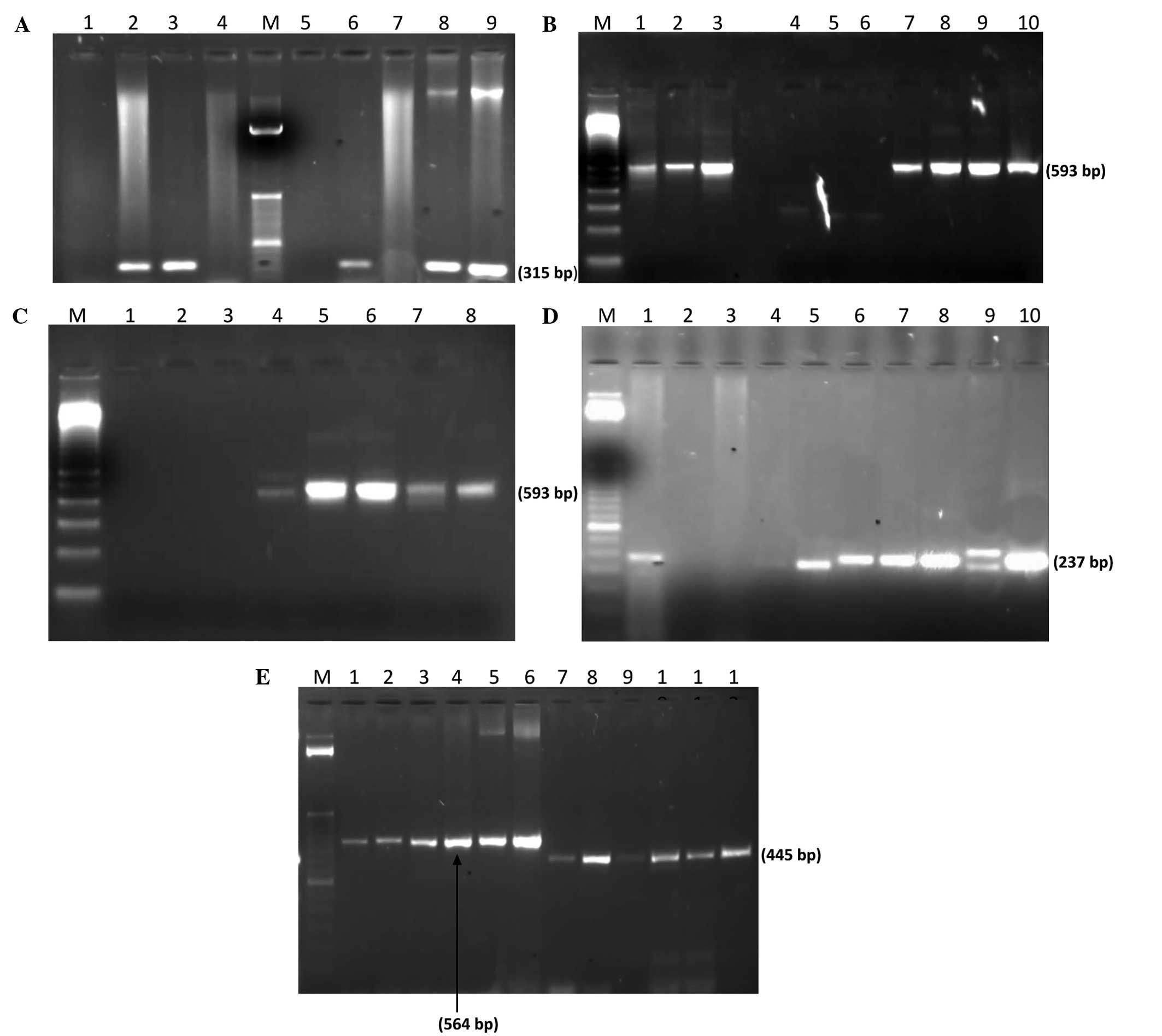 | Figure 3.Molecular detection of antibiotic
resistance genes. (A) PCR amplification of mecA gene: Lane M; 100
bp ladder (Invitrogen); lane 1, negative control (K.
pneumoniae ATCC BAA-1144); lane 2, positive control (S.
aureus ATCC BAA-1026); lanes 3, 6, 8 and 9, mecA gene
expression in S. aureus. (B) PCR amplification of bla-CTXM
gene: Lane M, 100 bp Ladder; lane 1, Proteus spp.; lanes 2
and 3, E. coli; lane 7, P. aeruginosa; lanes 8–11,
K. pneumoniae. (C) PCR amplification of bla-CTXM-15 gene:
Lane M, 100 bp ladder; lane 1, negative control (K.
pneumoniae ATCC BAA-1144); lane 4, P. aeruginosa; lanes
5 and 6, E. coli; lane 7, K. pneumoniae; lane 8,
Proteus spp. (D) PCR amplification of bla-SHV gene: Lane M,
50 bp ladder (Invitrogen); lane 10, positive control (K.
pneumoniae ATCC 700603); lane 2, negative control (E.
coli ATCC 25922); lane 5, P. aeruginosa; lanes 6–8,
E. coli; lanes 9 and 10, K. pneumoniae. (E) PCR
amplification of bla-TEM and bla-OXA genes: Lane M; 50 bp ladder;
lane 1, Proteus spp.; lane 2, P. aeruginosa; lanes 3
and 4, K. pneumoniae; lanes 5 and 6, E. coli; lane 7,
Proteus spp.; lane 8, P. aeruginosa; lanes 9 and 10,
K. pneumoniae; lane 11 and 12, E. coli. PCR,
polymerase chain reaction; bla, β-lactamase; CTX-M, cefotaximase;
SHV, sulfhydryl variable; TEM, Temoneira; OXA, oxacillinase; ATCC,
American Type Culture Collection; K. pneumoniae, Klebsiella
pneumoniae, S. aureus, Staphylococcus aureus; P. aeruginosa,
Pseudomonas aeruginosa; E. coli, Escherichia coli, spp.,
species. |
 | Table VI.Molecular detection of extended
spectrum β-lactamase production (%). |
Table VI.
Molecular detection of extended
spectrum β-lactamase production (%).
| Bacterial
isolates | bla-CTX-M | bla-CTX-M15 | bla-TEM | bla-OXA | bla-SHV |
|---|
| Proteus
spp. | 60 | 60 |
53.3 |
33.3 | 80 |
| P.
aeruginosa |
66.7 |
66.7 |
66.7 |
33.3 |
72.2 |
| K.
pneumoniae | 100 | 100 | 100 | 100 | 100 |
| E. coli | 100 | 100 | 100 | 100 | 100 |
| Total |
76.9 |
76.9 | 75 |
57.7 |
84.6 |
Discussion
The high occurrence of DFUs and amputation within
the diabetic population has become an increasingly alarming public
health concern in the developed and developing worlds (38). These complications begin with
neuropathy, which occurs as a result of hyperglycemia and involves
a loss of sensation in the lower extremities (8). Neuropathic individuals are highly prone
to physical injuries in their lower extremities (9), which lead to diabetic foot infections
(DFIs), which in turn may result in the amputation of the lower
extremities and subsequent mortality (11–13).
DFUs impose a tremendous medical and financial burden on the health
care system in the USA, with a cost as high as $45,000 per patient;
in addition, the impaired mobility and substantial loss of
productivity associated with DFUs affects the quality of life of
the patient (39).
Diabetic neuropathy, along with poor blood
circulation in the lower extremities, nerve damage and foot wounds,
constitutes one of the leading causes of DFUs (40), which is consistent with previous
studies in which up to 92% of patients with DFUs also had
neuropathy (41,42). The treatment prognosis is exacerbated
when an ulcer is infected with multiple microbes (43), since little is known about
multi-species interactions or the ideal antibiotic regimen for the
treatment of multi-species infections. In the present study
gram-positive bacteria, including S. aureus, were observed
to be dominant in these infections; which is consistent with
previous reports (44,45). The assays performed in the present
study also identified high percentages of multidrug-resistant P.
aeruginosa, which is troubling as P. aeruginosa is an
aggressive gram-negative bacillus (46).
High rates of antibiotic resistance have previously
been reported in patients with diabetes (47). Richard et al (48) found that the most common causative
agent of DFIs, S. aureus, represented 36.5% of isolates from
DFUs and, notably, 37.4% of these were MRSA. In the present study,
however, it was found that 84% of S. aureus isolates were
MRSA, while 20% were vancomycin resistant. S. aureus
isolates have been associated with prolonged bacteremia, greater
rates of infection-associated complications and vancomycin
treatment failure (49). Among the
K. pneumoniae isolates of the present study, a very high
incidence of ESBL in the bacteria was detected by phenotypic
testing (100% by the cefotaxime + clavulanic acid/ceftazidime +
clavulanic acid combination disk test), which is comparable with
previous studies, where up to 97% prevalence has been reported
(50,51). The highest prevalence of MBL
producers was found in P. aeruginosa and K.
pneumoniae (100% in the EDTA combination disk test). In a study
from India, 74.5% of the ceftazidime-resistant P. aeruginosa
isolates were found to be MBL producers (52). In another study from Iran, 53% of the
94 ceftazidime resistant P. aeruginosa isolates were found
to be MBL producers (53).
The PCR analysis conducted in the present study
revealed that 21 out of 25 S. aureus isolates (84%) harbored
the mecA gene, a considerably high proportion compared with that
observed in the study by Bukhari et al (54), which found an MRSA prevalence of
41.9% in clinical isolates in Lahore, Pakistan. In addition, in the
present study all K. pneumoniae and E. coli isolates
were 100% positive for all ESBL genes. The high prevalence of the
CTX-M gene in the present study was in concordance with the results
of the study of Šeputienė et al (55), who reported CTX-M encoding genes in
the majority of E. coli (96%) and K. pneumoniae (71%)
isolates showing the ESBL phenotype. ESBL-positive E. coli
isolates investigated in Sweden encoded mainly CTX-Ms (92%),
followed by TEM-type (63%), OXA-type (59%) and SHV-type (6%)
β-lactamases (34). According to a
study by Bali et al (56),
TEM-type ESBLs were found in 72.72% of E. coli and 75% of
K. pneumoniae. A study conducted by Umadevi et al
(57) observed that ESBL production
in P. aeruginosa is less prevalent than that in
Enterobacteriaceae, which is consistent with the results of the
present study. This finding limits treatment options considerably,
causing great concern regarding the lack of adequate treatment and
the spread of mecA- and ESBL-carrying isolates in DFIs.
In conclusion, infections caused by
multidrug-resistant bacteria that produce mecA and ESBL enzymes
have been reported with an increasing frequency in DFIs and are
associated with amputation. Epidemiological information helps in
the design of better programs for infection control. Due to the
resistance of DFIs to numerous antimicrobial agents, treatment can
be challenging. Hence, it is recommended that active surveillance
for ESBL-producing pathogens in populations at high-risk for DFUs
is performed using appropriate antimicrobial techniques.
Acknowledgements
This study was supported by the Higher Education
Commission and Ministry of Science and Technology of Pakistan. The
authors would like to thank Mr. Muhammad Shafique from the
Microbiology Laboratory of the Pakistan Institute of Medical
Sciences, for generously helping with the isolation and
characterization of bacterial strains from DFU samples.
References
|
1
|
Singh N, Armstrong DG and Lipsky BA:
Preventing foot ulcers in patients with diabetes. JAMA.
293:217–228. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mutlu F, Bener A, Eliyan A, Delghan H,
Nofal E, Shalabi L and Wadi N: Projection of Diabetes Burden
through 2025 and Contributing Risk Factors of Changing Disease
Prevalence: An Emerging Public Health Problem. J Diabetes Metab.
5:3412014.
|
|
3
|
Forouhi NG and Wareham NJ: Epidemiology of
diabetes. Medicine. 38:602–606. 2010. View Article : Google Scholar
|
|
4
|
Cusick M, Meleth AD, Agrón E, Fisher MR,
Reed GF, Knatterud GL, Barton FB, Davis MD, Ferris FL and Chew EY:
Early Treatment Diabetic Retinopathy Study Research Group:
Associations of mortality and diabetes complications in patients
with type 1 and type 2 diabetes: Early treatment diabetic
retinopathy study report no. 27. Diabetes Care. 28:617–625. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Clayton W and Elasy TA: A Review of the
Pathophysiology, Classification, and Treatment of Foot Ulcers in
Diabetic Patients. Clin Diabetes. 27:52–58. 2009. View Article : Google Scholar
|
|
6
|
Fowler MJ: Microvascular and macrovascular
complications of diabetes. Clinical diabetes. 26:77–82. 2008.
View Article : Google Scholar
|
|
7
|
Bowler PG, Duerden BI and Armstrong DG:
Wound microbiology and associated approaches to wound management.
Clin Microbiol Rev. 14:244–269. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Argoff CE, Cole BE, Fishbain DA and Irving
GA: Diabetic peripheral neuropathic pain: Clinical and
quality-of-life issues. Mayo Clin Proc. 81(Suppl 4): S3–S11. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Podwall D and Gooch C: Diabetic
neuropathy: Clinical features, etiology, and therapy. Curr Neurol
Neurosci Rep. 4:55–61. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Delamaire M, Maugendre D, Moreno M, Le
Goff MC, Allannic H and Genetet B: Impaired leucocyte functions in
diabetic patients. Diabet Med. 14:29–34. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Siitonen OI, Niskanen LK, Laakso M,
Siitonen JT and Pyörälä K: Lower-extremity amputations in diabetic
and nondiabetic patients: A population-based study in eastern
Finland. Diabetes care. 16:16–20. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Trautner C, Haastert B, Giani G and Berger
M: Incidence of lower limb amputations and diabetes. Diabetes care.
19:1006–1009. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paneni F, Beckman JA, Creager MA and
Cosentino F: Diabetes and vascular disease: Pathophysiology,
clinical consequences, and medical therapy. Eur Heart J.
34:2436–2443. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Reiber GE: Epidemiology of foot ulcers and
amputations in the diabetic foot. The diabetic foot. 6:13–32.
2001.
|
|
15
|
Tiwari S, Pratyush DD, Dwivedi A, Gupta
SK, Rai M and Singh SK: Microbiological and clinical
characteristics of diabetic foot infections in northern India. J
Infec Dev Ctries. 6:329–332. 2012.
|
|
16
|
Lipsky B, Pecoraro R and Wheat L: The
diabetic foot. Soft tissue and bone infection. Infect Dis Clin
North Am. 4:409–432. 1990.PubMed/NCBI
|
|
17
|
Frykberg RG: An evidence-based approach to
diabetic foot infections. Am J Surg. 186(Suppl): S44–S54. 2003.
View Article : Google Scholar
|
|
18
|
Benwan K, Al Mulla A and Rotimi VO: A
study of the microbiology of diabetic foot infections in a teaching
hospital in Kuwait. J Infect Public Health. 5:1–8. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roberts RR, Hota B, Ahmad I, Scott RD 2nd,
Foster SD, Abbasi F, Schabowski S, Kampe LM, Ciavarella GG, Supino
M, et al: Hospital and societal costs of antimicrobial-resistant
infections in a Chicago teaching hospital: Implications for
antibiotic stewardship. Clin Infect Dis. 49:1175–1184. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cosgrove SE, Sakoulas G, Perencevich EN,
Schwaber MJ, Karchmer AW and Carmeli Y: Comparison of mortality
associated with methicillin-resistant and methicillin-susceptible
Staphylococcus aureus bacteremia: A meta-analysis. Clin Infect Dis.
36:53–59. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grundmann H, Aires-de-Sousa M, Boyce J and
Tiemersma E: Emergence and resurgence of meticillin-resistant
Staphylococcus aureus as a public-health threat. Lancet.
368:874–885. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Askari E, Soleymani F, Arianpoor A,
Tabatabai SM, Amini A and Nasab Naderi M: Epidemiology of
mecA-methicillin resistant Staphylococcus aureus (MRSA) in Iran: A
systematic review and meta-analysis. Iran J Basic Med Sci.
15:1010–1019. 2012.PubMed/NCBI
|
|
23
|
Poole K: Resistance to beta-lactam
antibiotics. Cell Mol Life Sci. 61:2200–2223. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bonnet R: Growing group of
extended-spectrum beta-lactamases: The CTX-M enzymes. Antimicrob
Agents Chemother. 48:1–14. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Perez F, Endimiani A, Hujer KM and Bonomo
RA: The continuing challenge of ESBLs. Curr Opin Pharmacol.
7:459–469. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Walsh TR, Toleman MA, Poirel L and
Nordmann P: Metallo-beta-lactamases: The quiet before the storm?
Clin Microbiol Rev. 18:306–325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shahi SK, Singh VK and Kumar A: Detection
of Escherichia coli and associated β-lactamases genes from diabetic
foot ulcers by multiplex PCR and molecular modeling and docking of
SHV-1, TEM-1, and OXA-1 β-lactamases with clindamycin and
piperacillin-tazobactam. PLoS One. 8:e682342013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Oyibo SO, Jude EB, Tarawneh I, Nguyen HC,
Harkless LB and Boulton AJ: A comparison of two diabetic foot ulcer
classification systems: The Wagner and the University of Texas
wound classification systems. Diabetes care. 24:84–88. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bauer AW, Perry DM and Kirby WM:
Single-disk antibiotic-sensitivity testing of Staphylococci: An
analysis of technique and results. AMA Arch Intern Med.
104:208–216. 1959. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Clinical and Laboratory Standards
Institute (CLSI). Performance Standards for Antimicrobial
Susceptibility Testing: Twenty-first Informational Supplement
(M100-S21) (Wayne, PA). CLSI. 2011.
|
|
31
|
Singhal S, Mathur T, Khan S, Upadhyay DJ,
Chugh S, Gaind R and Rattan A: Evaluation of methods for AmpC
beta-lactamase in gram negative clinical isolates from tertiary
care hospitals. Indian J Med Microbiol. 23:120–124. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yong D, Lee K, Yum JH, Shin HB, Rossolini
GM and Chong Y: Imipenem-EDTA disk method for differentiation of
metallo-beta-lactamase-producing clinical isolates of Pseudomonas
spp. and Acinetobacter spp. J Clin Microbiol. 40:3798–3801. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shahcheraghi F, Nikbin VS and Feizabadi
MM: Prevalence of ESBLs genes among multidrug-resistant isolates of
Pseudomonas aeruginosa isolated from patients in Tehran. Microb
Drug Resist. 15:37–39. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fang H, Ataker F, Hedin G and Dornbusch K:
Molecular epidemiology of extended-spectrum beta-lactamases among
Escherichia coli isolates collected in a Swedish hospital and its
associated health care facilities from 2001 to 2006. J Clin
Microbiol. 46:707–712. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dallenne C, Da Costa A, Decré D, Favier C
and Arlet G: Development of a set of multiplex PCR assays for the
detection of genes encoding important beta-lactamases in
Enterobacteriaceae. J Antimicrob Chemother. 65:490–495. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Boyd DA, Tyler S, Christianson S, McGeer
A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P and Mulvey
MR: Complete nucleotide sequence of a 92-kilobase plasmid harboring
the CTX-M-15 extended-spectrum beta-lactamase involved in an
outbreak in long-term-care facilities in Toronto, Canada.
Antimicrob Agents Chemother. 48:3758–3764. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Monstein HJ, Ostholm-Balkhed A, Nilsson M,
Nilsson M, Dornbusch K and Nilsson L: Multiplex PCR amplification
assay for the detection of blaSHV, blaTEM and blaCTX-M genes in
Enterobacteriaceae. APMIS. 115:1400–1408. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rodrigues BT, Gilhotra RA, Vangaveti VN
and Malabu UH: Prevalence and risk factors of lower limb amputation
amongst diabetic food ulcer patients at the Townsville Hospital.
Annals of the ACTM. 15:572014.
|
|
39
|
Stockl K, Vanderplas A, Tafesse E and
Chang E: Costs of lower-extremity ulcers among patients with
diabetes. Diabetes Care. 27:2129–2134. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gardner SE, Hillis SL, Heilmann K, Segre
JA and Grice EA: The neuropathic diabetic foot ulcer microbiome is
associated with clinical factors. Diabetes. 62:923–930. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lipsky BA, Berendt AR, Deery HG, Embil JM,
Joseph WS, Karchmer AW, LeFrock JL, Lew DP, Mader JT, Norden C, et
al: Diagnosis and treatment of diabetic foot infections. Clin
Infect Dis. 39:885–910. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
42
|
Monteiro-Soares M, Boyko EJ, Ribeiro J,
Ribeiro I and Dinis-Ribeiro M: Risk stratification systems for
diabetic foot ulcers: A systematic review. Diabetologia.
54:1190–1199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
James GA, Swogger E, Wolcott R, Pulcini
ED, Secor P, Sestrich J, Costerton JW and Stewart PS: Biofilms in
chronic wounds. Wound Repair Regen. 16:37–44. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mantey I, Hill R, Foster A, Wilson S, Wade
JJ and Edmonds ME: Infection of foot ulcers with Staphylococcus
aureus associated with increased mortality in diabetic patients.
Commun Dis Public Health. 3:288–290. 2000.PubMed/NCBI
|
|
45
|
Dang CN, Prasad YD, Boulton AJ and Jude
EB: Methicillin-resistant Staphylococcus aureus in the diabetic
foot clinic: A worsening problem. Diabet Med. 20:159–161. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gadepalli R, Dhawan B, Sreenivas V, Kapil
A, Ammini AC and Chaudhry R: A clinico-microbiological study of
diabetic foot ulcers in an Indian tertiary care hospital. Diabetes
care. 29:1727–1732. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Trivedi U, Parameswaran S, Armstrong A,
Burgueno-Vega D, Griswold J, Dissanaike S and Rumbaugh KP:
Prevalence of Multiple Antibiotic Resistant Infections in Diabetic
versus Nondiabetic Wounds. J Pathog. 2014:1730532014.PubMed/NCBI
|
|
48
|
Richard JL, Sotto A and Lavigne JP: New
insights in diabetic foot infection. World J Diabetes. 2:24–32.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Satola SW, Lessa FC, Ray SM, Bulens SN,
Lynfield R, Schaffner W, Dumyati G, Nadle J and Patel JB: Active
Bacterial Core surveillance (ABCs) MRSA Investigators: Clinical and
laboratory characteristics of invasive infections due to
methicillin-resistant Staphylococcus aureus isolates demonstrating
a vancomycin MIC of 2 micrograms per milliliter: Lack of effect of
heteroresistant vancomycin-intermediate S. aureus phenotype. J Clin
Microbiol. 49:1583–1587. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Harish BN, Menezes GA, Shekatkar S and
Parija SC: Extended-spectrum beta-lactamase-producing Klebsiella
pneumoniae from blood culture. J Med Microbiol. 56:999–1000. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Parveen RM, Khan MA, Menezes GA, Harish
BN, Parija SC and Hays JP: Extended-spectrum β-lactamase producing
Klebsiella pneumoniae from blood cultures in Puducherry, India.
Indian J Med Res. 134:392–395. 2011.PubMed/NCBI
|
|
52
|
Umadevi S, Joseph NM, Kumari K, Easow JM,
Kumar S, Stephen S, Srirangaraj S and Raj S: Detection of extended
spectrum beta lactamases, AmpC beta lactamases and
metallobetalactamases in clinical isolates of ceftazidime resistant
Pseudomonas aeruginosa. Braz J Microbiol. 42:1284–1288.
2011.PubMed/NCBI
|
|
53
|
Saderi H, Karimi Z, Owlia P, Bahar MA and
Rad SMBA: phenotypic detection of metallo-beta-lactamase producing
Pseudomonas aeruginosa strains isolated from burned patients.
Iranian Journal of Pathology. 3:20–24. 2008.
|
|
54
|
Bukhari SZ, Ahmed S and Zia N:
Antimicrobial susceptibility pattern of Staphylococcus aureus on
clinical isolates and efficacy of laboratory tests to diagnose
MRSA: MRSA: A multi-centre study. J Ayub Med Coll Abbottabad.
23:139–142. 2011.PubMed/NCBI
|
|
55
|
Šeputienė V, Linkevičius M, Bogdaitė A,
Povilonis J, Plančiūnienė R, Giedraitiene A, Pavilonis A and
Sužiedėlienė E: Molecular characterization of extended-spectrum
β-lactamase-producing Escherichia coli and Klebsiella pneumoniae
isolates from hospitals in Lithuania. J Med Microbiol.
59:1263–1265. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bali EB, Accedil L and Sultan N:
Phenotypic and molecular characterization of SHV, TEM, CTX-M and
extended-spectrum-lactamase produced by Escherichia coli,
Acinobacter baumannii and Klebsiella isolates in a Turkish
hospital. African Journal of Microbiology Research. 4:650–654.
2010.
|
|
57
|
Umadevi S, Kandhakumari G, Joseph NM,
Kumar S, Easow JM, Stephen S and Singh UK: Prevalence and
antimicrobial susceptibility pattern of ESBL producing gram
negative bacilli. J Clin Diagn Res. 5:236–239. 2011.
|