Introduction
Subacute ruminal acidosis (SARA), which is a common
disease in high yielding dairy cows that receive highly digestible
diets, has a high economic impact (1). SARA increases the content of free
lipopolysaccharide (LPS) in the rumen by increasing the lysis of
gram-negative bacteria (2). LPS is
the primary component of the gram-negative bacterial outer
membrane, and is a key factor that induces the release of
proinflammatory cytokines, including tumor necrosis factor (TNF)-α,
interleukin (IL)-1β, and IL-6 (3),
which in turn activate hepatocytic receptors and initiate the
synthesis of acute phase proteins (4). In addition, LPS induces the activation
of nuclear factor (NF)-κB, which translocates into the nucleus and
regulates the expression of genes involved in cellular
differentiation, proliferation, inflammation and apoptosis (5,6).
Furthermore, LPS has been shown to regulate lactation and the
synthesis of milk fat (7); previous
studies associated rumen LPS-mediated inflammatory responses with
milk fat depression (MFD) syndrome in lactating dairy cows, which
is characterized by reduced milk fat synthesis and milk energy
efficiency (8,9). Previous in vitro experiments
have demonstrated that LPS is capable of inhibiting fatty acid
synthase (FAS), acetyl-coA carboxylase (ACC) and peroxisome
proliferator-activated receptor-γ (PPARG) gene expression levels,
thereby inhibiting the synthesis of fatty acids (10–12).
Therefore, inhibiting LPS-induced inflammatory cytokine production
is a formidable challenge that may improve milk fat content and
milk quality.
14-3-3γ is an influential member of the 14-3-3
family, which are localized to the cell nucleus (13) and have important roles in
coordinating the progression of cells (14,15).
Previous studies have reported that 14-3-3γ overexpression promotes
the viability of DCMECs (16) and
14-3-3γ may serve a crucial function in the regulation of
LPS-induced myocardial injury (17,18). Our
previous study (11) demonstrated
that 14-3-3γ was able to inhibit the production of LPS-induced
cytokines in dairy cow mammary epithelial cells (DCMECs) by
inhibiting the activation of NF-κB signaling pathways. However, to
the best of our knowledge, the mechanism underlying the role of
14-3-3γ in LPS-induced DCMEC injury, and the association between
14-3-3γ and milk fat synthesis in LPS-induced DCMECs, has yet to be
investigated.
The present study aimed to investigate the
protective effect of 14-3-3γ on LPS-induced DCMECs and the effects
of 14-3-3γ on milk fat synthesis. A grapevine chrome mosaic virus
(GCMV)/internal ribosome entry site (IRES)/enhanced green
fluorescent protein (EGFP)-14-3-3γ expression vector was
constructed and transfected into DCMECs in order to evaluate the
ability of 14-3-3γ to protect against LPS-induced cell damage, and
to determine its effect on the expression levels of milk fat
synthesis-associated genes.
Materials and methods
Ethics statement
All experiments in the present study were approved
by the Northeast Agricultural University Provincial Experimental
Animal Management Committee (Harbin, China) and were performed in
accordance with the guidelines of this committee.
Chemicals and reagents
LPS (Escherichia coli 0111:B4) and MTT were
obtained from Sigma-Aldrich (St. Louis, MO, USA). Fetal bovine
serum (FBS) and Dulbecco's modified Eagle's medium: F12 (DMEM/F12)
base were obtained from Thermo Fisher Scientific, Inc. (Gibco;
Waltham, MA, USA). Antibodies against PPARG, phosphorylated
(p)-PPARG, sterol regulatory element binding protein (SREBP1) and
β-actin were purchased from Santa Cruz Biotechnology, Inc. (Dallas,
TX, USA). Anti-p-SREBP1 was purchased from Cell Signaling
Technology Inc. (Danvers, MA, USA) and horseradish
peroxidase-conjugated goat anti-rabbit and anti-mouse secondary
antibodies were obtained from Beijing Biosynthesis Biotechnology
Co., Ltd. (Beijing, China). ACC (GMS50510.2 v.A) and FAS
(GMS50509.1 v.A) activity detection kits were obtained from
Shanghai GenePharma Co. Ltd. (Shanghai, China).
Culture of DCMECs
Purified DCMECs were obtained from the Key
Laboratory of Dairy Science of Education Ministry at Northeast
Agricultural University. The cells were incubated at 37°C in an
atmosphere containing 5% CO2 in basic culture medium
(DMEM/F12 base with 10% FBS, 100 U/ml penicillin (Harbin
Pharmaceutical Group Co., Ltd., Harbin, China) and 100 U/ml
streptomycin (Dalian Merro Pharmaceutical Factory, Dalian, China
(19,20).
Construction of
pGCMV/IRES/EGFP-14-3-3γ expression vectors
The full-length cDNA encoding bovine 14-3-3γ was
generated by polymerase chain reaction (PCR) using total RNA
extracted from DCMECs. The final reaction volume of 20 µl contained
10 µl SYBR® Premix Eq Taq™, 0.4 µl forward primer (10 µM),
0.4 µl reverse primer (10 µM), 0.4 µl Rox reference dye (50X), 2 µl
cDNA template and 6.8 µl diethylpyrocarbonate-treated water.
Reaction conditions were as follows: 94°C for 5 min, followed by 35
cycles of 94°C for 30 sec, 62°C for 30 sec and 72°C for 60 sec, and
finally 72°C for 10 min. Primers were purchased from the Beijing
Genomics Institute and the sequences were as follows: Forward,
5′-GATCATCCTCGTCCGG-3′ and reverse, 5′-CAGTCCACCTGGGGGC-3′. The PCR
products were digested with EcoRI and BamHI (both
Takara Biotechnology Co., Ltd., Dalian, China) and were
subsequently subcloned into the multiple cloning sites of the
pGCMV/IRES/EGFP expression vector (Ambion; Thermo Fisher
Scientific, Inc.), as previously described (16).
Transfection of
pGCMV/IRES/EGFP-14-3-3γ
DCMECs were cultured in 6-well plates
(3×105 cells/cm2) until 80–90% cell density
was reached. pGCMV/IRES/EGFP-14-3-3γ or pGCMV/IRES/EGFP expression
vectors were transfected into cells using Lipofectamine™ 2000
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Briefly, 4 µg pGCMV/IRES/EGFP-14-3-3γ plasmid and 10 µl
Lipofectamine™ 2000 were diluted using 250 µl Gibco Opti-MEM I
medium (Thermo Fisher Scientific, Inc.) and the complexes were
incubated at room temperature for 20 min, prior to addition to the
cell cultures. The cultures were then incubated with serum- and
antibiotic-free DMEM/F12 medium at 37°C for 4 h, after which the
medium was discarded and fresh culture medium was added. Following
transfection for 24 h, the mRNA expression levels of 14-3-3γ were
measured, as previously described (11).
Transfection of small interfering RNAs
(siRNAs)
14-3-3γ siRNAs and negative scrambled control siRNA
were purchased from Shanghai GenePharma Co. Ltd. The 14-3-3γ siRNA
had the following sequence: Sense, 5′-CCCUUAACUACUCCGUCUUTT-3′ and
antisense, 5′-AAGACGGAGUAGUUAAGGGTT-3′. The negative scrambled
control siRNA lacked significant sequence homology to any gene and
had the following sequence: Sense, 5′-UUCUUGAACGUGUCACGUTT-3′ and
antisense, 5′-ACGUGACACGUUCGGAGAATT-3′. DCMECs were cultured in
6-well plates until they reached a cell density of 80–90%.
siRNA-14-3-3γ or negative control were transfected into the cells
using Lipofectamine™2000, according to the manufacturer's protocol.
Briefly, 2 µl siRNA-14-3-3γ and 5 µl Lipofectamine™ 2000 were
diluted using 200 ml Opti-MEM I medium, after which the mixture was
incubated at room temperature for 20 min, prior to addition into
the wells. The cultures were then incubated with serum- and
antibiotic-free medium at 37°C for 4 h, after which the medium was
discarded and fresh culture medium with was added. Transfection
efficiency was observed under a fluorescence microscope (CKX41;
Olympus Corporation, Tokyo, Japan) according to the number of
fluorescent cells.
MTT assay
The viability of DCMECs was measured using the MTT
assay. Briefly, DCMECs were transfected with with
pGCMV/IRES/EGFP-14-3-3γ, pGCMV-IRES-EGFP, 14-3-3γ siRNA or negative
control siRNA for 24 h. Transfected cells were seeded into 96-well
plates at 1×105 cells/well, followed by 24 h culturing
in the presence or absence of 1 µg/ml LPS. Subsequently, the cells
were washed with D-Hanks, and MTT (5 mg/ml in PBS; all
Sigma-Aldrich) was added to each well and incubated for 4 h at
37°C. The reaction was stopped by the addition of 100 µl dimethyl
sulfoxide (Amresco, LLC, Solon, OH, USA). After the mixture was
oscillated for 15 min, absorbance was measured at 570 nm using a
spectrophotometer (DU800; Beckman Coulter, Inc., Brea, CA,
USA).
Lactate dehydrogenase (LDH) activity
assay
Following transfecion, DCMECs were incubated with
antibiotic-free DMEM/F12 medium for 24 h and subsequently treated
with 1 µg/ml LPS for 24 h, after which the cell-free supernatants
were collected for LDH viability assays using LDH Assay kits
(Nanjing Jiancheng Bioengineering Institute, Nanjing, China),
according to the manufacturer's protocol. The optical density (OD)
of the cell-free supernatants was measured using a Sunrise-Basic
microplate reader (Tecan Group, Ltd., Männedorf, Switzerland) at
440 nm.
Acridine orange (AO) double
staining
DCMECs were transfected for 24 h and subsequently
treated with 1 µg/ml LPS. After 24 h, the cells was collected and
washed three times with PBS and were adjusted to a density of
5×105 cells/ml. Subsequently, 4 µl AO dye liquor (100
mg/l in PBS; Amresco, LLC) was added to 96 µl cell suspension,
after which the specimens were incubated in the dark for 30 min at
room temperature. After rinsing three times with PBS, the cellular
morphology was visualized and photographed using a CKX41
fluorescence microscope (Olympus Corporation), as previously
described (6).
Reverse transcription-quantitative PCR
(RT-qPCR)
DCMECs were transfected for 24 h. Transfected cells
were treated with 1 µg/ml LPS for 24 h. Total RNA was isolated
using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the method described in a previous study (21). Total RNA was reverse transcribed into
cDNA using PrimeScript Reverse Transcriptase (Takara Biotechnology
Co., Ltd.), according to the manufacturer's protocol. The mRNA
expression levels of various genes were quantified using SYBR
premix Ex Taq™ (Takara Bio, Inc., Otsu, Japan), and the analysis
was performed by the ABI PRISM 7300 Real-Time PCR system (Applied
Biosystems, Thermo Fisher Scientific, Inc.), as previously
described (16). Primers used for
RT-qPCR analysis are presented in Table
I. The RT-qPCR conditions were as follows: 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec and 60°C for 31 sec,
according to the manufacturer's instructions. All target cDNA were
analyzed in triplicate. The relative mRNA expression levels were
quantified using the 2−ΔΔCq method.
 | Table I.Primer sequences for the reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primer sequences for the reverse
transcription-quantitative polymerase chain reaction.
| Gene name | Accession
number | Primer sequence
(5′-3′) | Product size
(bp) |
|---|
| 14–3–3γ | BC153255.1 | Forward:
GAATGAGCCACTGTCCAA | 171 |
|
|
| Reverse:
GCACATCCTGACATACGG |
| SREBP1 | NM_001113302.1 | Forward:
GTAGCAGCGGTGGAAGT | 67 |
|
|
| Reverse:
GCAGCGGCTCTGGATT |
|
| PPARG | NM_181024.2 | Forward:
GCAGCGGCTCTGGATT | 170 |
|
|
| Reverse:
ATAGTGGAACCCTGACG |
|
| CD36 | NM_001046239.1 | Forward:
GGAAAGGACGACATAAGCAAAG | 187 |
|
|
| Reverse:
TCAACAAAAGGTGGAAATGAGG |
|
| ACC | NM_174224.2 | Forward:
AGACAAACAGGGACCATT | 141 |
|
|
| Reverse:
AGGGACTGCCGAAACAT |
|
| FAS | NM_001012669.1 | Forward:
CCACGGCTGTCGGTAAT | 171 |
|
|
| Reverse:
CGCTCCCACTCATCCTG |
|
| FABP3 | NM_174313.2 | Forward:
GAACTCGACTCCCAGCTTGAA | 214 |
|
|
| Reverse:
AAGCCTACCACAATCATCGAAG |
|
| β-actin | NM_173979 | Forward:
CCGCAAGGACCTCTACGC | 206 |
|
|
| Reverse:
ATGCCAATCTCATCTCGTTTT |
|
Enzyme activity assay
The activities of FAS and ACC in the LPS-treated
transfected DCMECs were analyzed using the Enzyme Activity
Detection kit, according to the manufacturer's protocol. The OD of
the microplate was read at 340 nm using a Sunrise-Basic microplate
reader (Tecan Group, Ltd., Männedorf, Switzerland).
Western blot analysis
DCMECs were transfected for 24 h, after which the
cells were stimulated with 1 µg/ml LPS for 24 h. Subsequently, the
cells were washed with cold PBS and ice-cold lysis buffer (Cell
Signaling Technology Inc.), which contained 20 mM Tris-HCl (pH7.5),
150 mM NaCl, 1mM Na2 EDTA, 1mM EGTA, 1% NP-40, 1% sodium
decxycholate, 2.5 mM β-glycerophosphate, 1 mM
Na3VO4 and 1 µg/ml leupeptin. The cells were
scraped and collected in a microfuge tube. Total protein was
isolated from DCMECs using a method described in a previous study
(22). Briefly, the protein
concentration was measured using the bicinchoninic acid assay
(Beyotime Institute of Biotechnology, Haimen, China), after which
30 µg protein was separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and transferred onto
nitrocellulose membranes using glycine transfer buffer (192 mM
glycine, 25 mM Tris-HCl (pH 8.8), 20% methanol) (both Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The membranes were blocked
using blocking buffer (5% nonfat dry milk) for 1.5 h at room
temperature, and incubated with the following specific primary
polyclonal antibodies at 4°C overnight at a dilution of 1:200:
rabbit PPARG (sc-7196) and p-PPARG (sc-28001-R), and mouse SREBP1
(sc-365513), p-SREBP1 (9874) and β-actin (sc-8432). Following
washing with TBST three times, the membranes were subsequently
incubated with goat anti-mouse (bs-0296G) or anti-rabbit (bs-0295G)
horseradish peroxidase-conjugated secondary antibodies at 37°C for
1.5 h at a dilution of 1:1,000. Antibody complexes were detected by
enhanced chemiluminescence using the Super ECL Plus (Applygen
Technologies, Inc., Beijing, China). Subsequently, the western
blotting results were analyzed using a Tanon 1600R Gel Imaging
System (Tanon Science and Technology Co., Ltd., Shanghai,
China).
Statistical analysis
Quantitative data from the experiments are presented
as the mean ± standard deviation of triplicate experiments.
Differences between mean values of normally distributed data were
analyzed using one-way analysis of variance. Statistical analyses
were conducted using the SPSS software, version 17.0 (SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
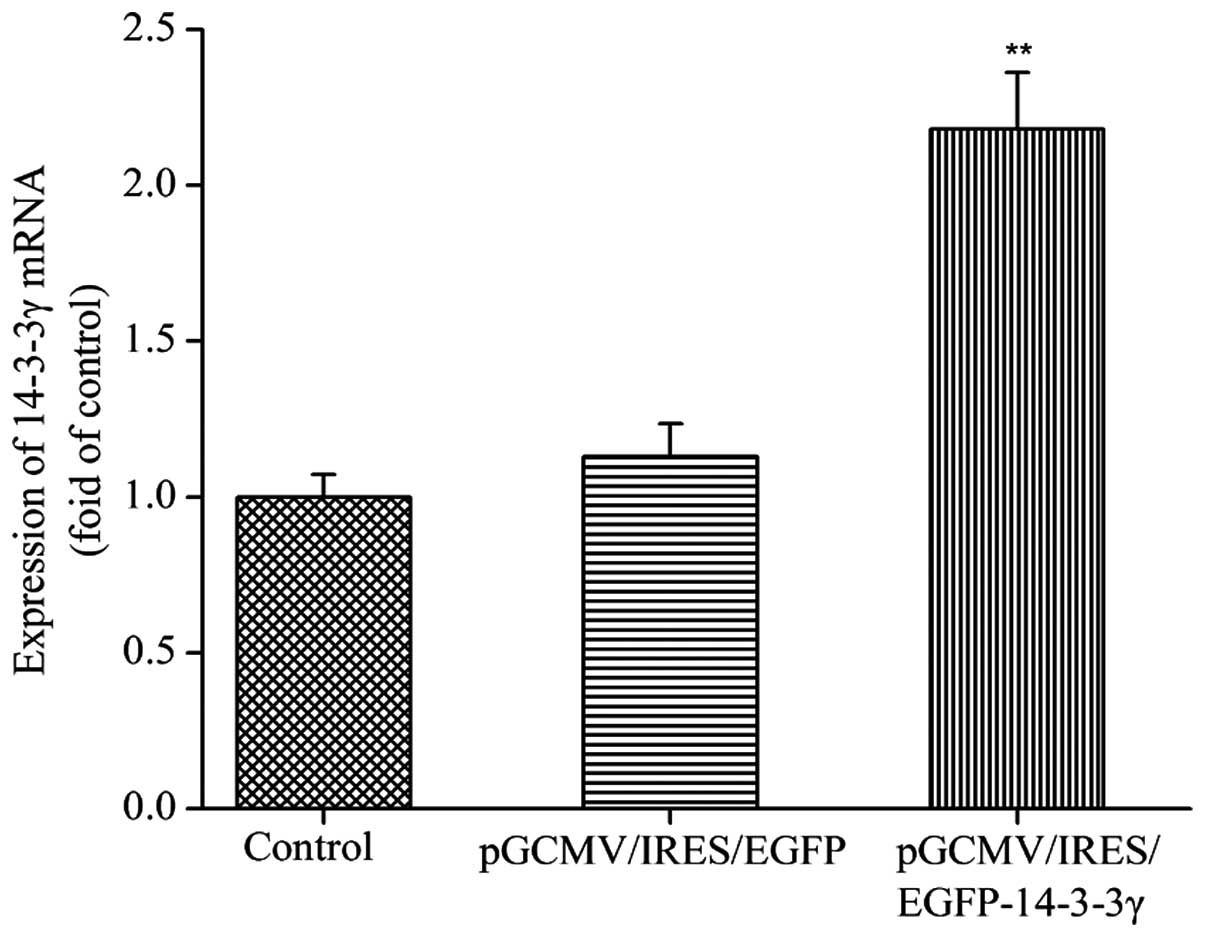
mRNA expression levels of 14-3-3γ in
transfected DCMECs
In order to determine whether the recombinant
pGCMV/IRES/EGFP-14-3-3γ expression vector had been successfully
constructed, the mRNA expression levels of 14-3-3γ were detected.
DCMECs were transfected with pGCMV/IRES/EGFP-14-3-3γ or
pGCMV/IRES/EGFP for 24 h, after which total RNA was extracted and
analyzed by RT-qPCR The relative mRNA expression levels of 14-3-3γ
in the DCMECs transfected with pGCMV/IRES/EGFP-14-3-3γ were
significantly increased, as compared with the control group and
pGCMV/IRES/EGFP group (P<0.01; Fig.
1). These results suggested that the pGCMV/IRES/EGFP-14-3-3γ
expression vector was effectively expressed in cultured DCMECs.
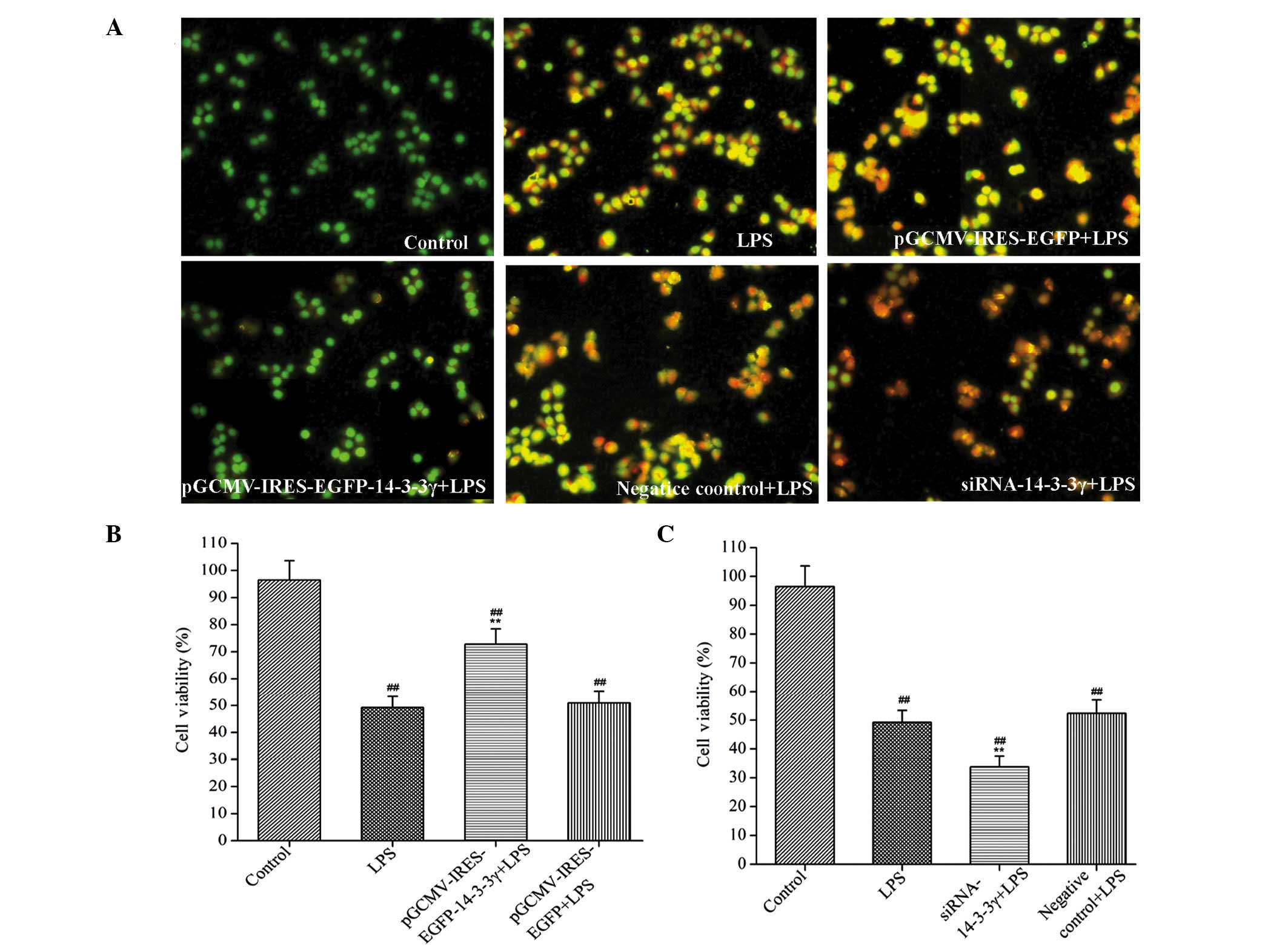
14-3-3γ regulates the viability of
LPS-induced DCMECs
In order to investigate whether 14-3-3γ was able to
confer protection against LPS-induced DCMEC damage, pre-transfected
cells were treated with 1 µg/ml LPS for 24 h and MTT and LDH
activity assays were conducted to evaluate cell damage. Cell
viability was significantly reduced, and LDH activity was
significantly increased, in the LPS-treated DCMECs, as compared
with the control group (P<0.01). However, 14-3-3γ overexpression
significantly increased cell viability and decreased LDH activity
in the LPS-induced DCMECs by 34.29 and 16.30% respectively, as
compared with the LPS-treated group (P<0.01 and P<0.05),
respectively (Table II).
Conversely, treatment with 14-3-3γ siRNA significantly reduced the
cell viability and increased the LDH activity in LPS-induced DCMECs
by 15.74 and 18.10% respectively, as compared with the LPS-treated
group (P<0.05), respectively (Table
III). These findings suggested that 14-3-3γ was capable of
increasing cell viability in LPS-induced DCMECs.
 | Table II.Effects of 14–3–3γ overexpression on
cell viability and LDH activity in LPS-induced dairy cow mammary
epithelial cells. |
Table II.
Effects of 14–3–3γ overexpression on
cell viability and LDH activity in LPS-induced dairy cow mammary
epithelial cells.
| Groups | Viability (%) | LDH Viability
(U/l) |
|---|
| Control | 95.53±0.96 | 52.05±4.31 |
| LPS |
56.21±0.73a |
125.7±12.13a |
|
pGCMV/IRES/EGFP-14–3–3γ + LPS |
70.49±0.81b |
105.2±5.63c |
| pGCMV/IRES/EGFP +
LPS |
59.89±0.81a |
129.12±10.38a |
 | Table III.Effects of 14–3–3γ siRNA on cell
viability and LDH activity in LPS-induced dairy cow mammary
epithelial cells. |
Table III.
Effects of 14–3–3γ siRNA on cell
viability and LDH activity in LPS-induced dairy cow mammary
epithelial cells.
| Groups | Viability (%) | LDH Viability
(U/l) |
|---|
| Control | 93.73±0.83 | 60.05±5.81 |
| LPS |
60.01±0.63a |
131.76±14.15a |
|
siRNA-14–3–3γ+LPS |
50.56±0.59b |
155.62±8.64c |
| Negative
control+LPS |
58.79±0.91a |
138.14±11.18a |
14-3-3γ regulates LPS-induced
apoptosis in DCMECs
In order to evaluate the effect of 14-3-3γ on
LPS-induced DCMEC apoptosis, the extent of apoptosis was analyzed
morphologically by staining the cells with AO and performing
fluorescence microscopy. Normal viable cells were dyed with AO and
emitted uniform green fluorescence. Conversely, cells undergoing
apoptosis emitted a yellow-green fluorescence and exhibited
condensed or fragmented chromatin. In addition, 200 stained cells
were randomly counted and viability was calculated as the
percentage of the number of living cells, as compared with the
total number of cells. LPS treatment significantly reduced the
viability and increased the extent of apoptosis of DCMECs, as
compared with the control group (P<0.01). Conversely, 14-3-3γ
overexpression significantly increased the viability of the cells
and decreased the extent of cell apoptosis, as compared with the
LPS-treated group (P<0.01; Fig. 2A
and B). Following treatment with 14-3-3γ siRNA, opposite
results were observed, as compared with those for the 14-3-3γ
recombinant plasmid (Fig. 2A and
C).
14-3-3γ regulates the expression of
genes associated with milk fat synthesis in LPS-induced DCMECs at
the transcriptional level
In order to investigate the effects of 14-3-3γ on
the mRNA expression levels of genes associated with milk fat
synthesis in LPS-induced DCMECs, RT-qPCR was conducted. LPS
significantly decreased the mRNA expression levels of SREBP1,
PPARG, cluster of differentiation 36 (CD36), ACC, FAS and fatty
acid binding protein (FABP), as compared with the control group
(P<0.05). Conversely, the mRNA expression levels of these genes
were significantly increased in LPS-induced DCMECs overexpressing
14-3-3γ, as compared with the LPS group (P<0.01; Fig. 3A), whereas treatment with
siRNA-14-3-3γ significantly reduced the mRNA expression levels of
these genes (P<0.05; Fig.
3B).
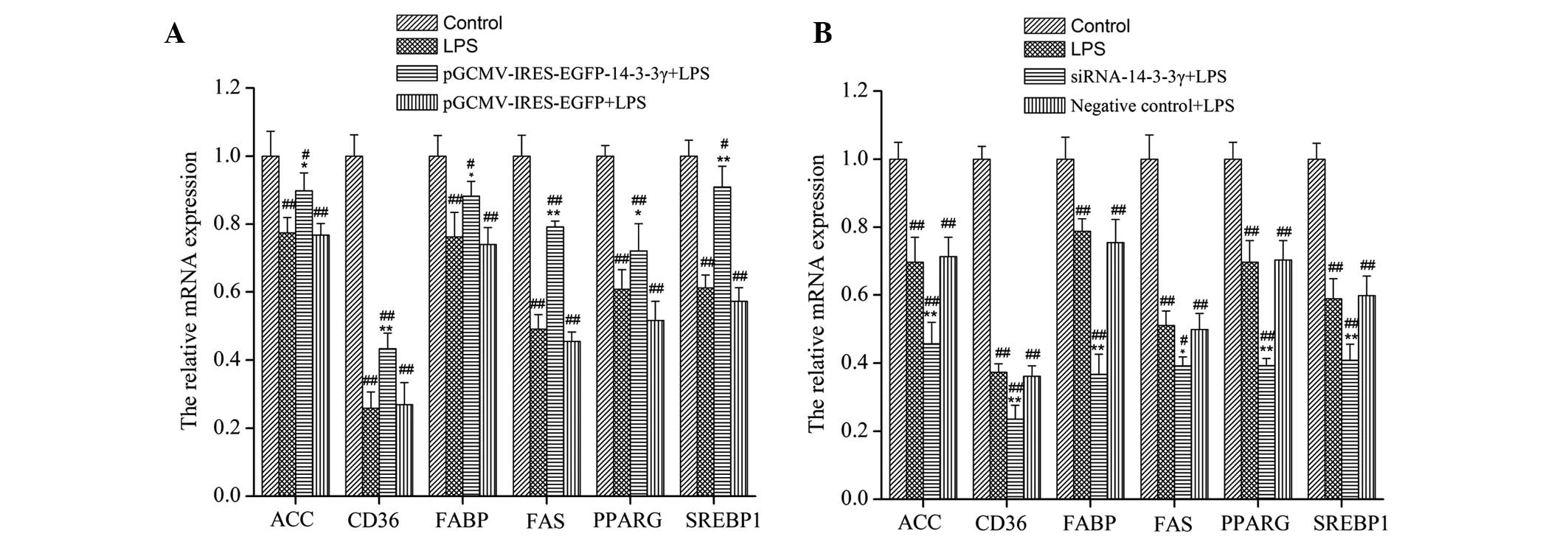 | Figure 3.Effects of 14-3-3γ on the mRNA
expression levels of genes associated with milk fat synthesis in
LPS-induced dairy cow mammary epithelial cells (DCMECs). (A) DCMECs
were transfected with pGCMV/IRES/EGFP-14-3-3γ or (B) siRNA-14-3-3γ
for 24 h, after which the cells were stimulated with LPS (1 µg/ml)
for 24 h. The mRNA expression levels of SREBP1, PPARG, CD36, ACC,
FAS and FABP were measured using reverse transcription-quantitative
polymerase chain reaction, and were normalized to the β-actin gene.
Data are presented as the mean ± standard deviation of three
independent experiments. *P<0.05 and **P<0.01 vs. the
LPS-treated group; #P<0.05 and ##P<0.01
vs. the control group. LPS, lipopolysaccharide; SREBP1, sterol
regulatory element binding protein-1; PPARG, peroxisome
proliferator-activated receptor-γ; CD36, cluster of differentiation
36; ACC, acetyl-CoA carboxylase; FAS, fatty acid synthase; FABP,
fatty acid binding protein; GCMV, grapevine chrome mosaic virus;
IRES, internal ribosome entry site; EGFP, enhanced green
fluorescent protein. |
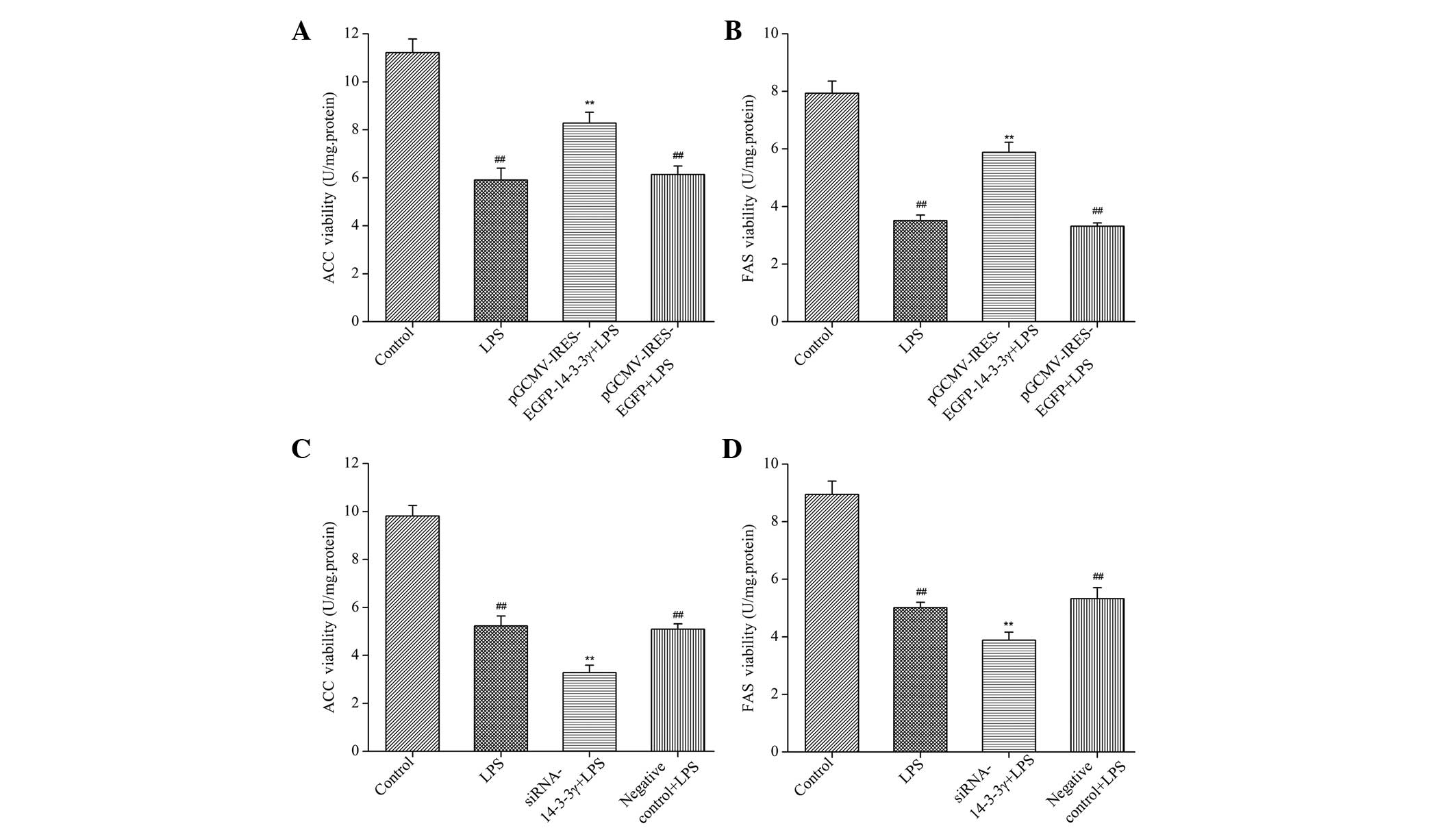
14-3-3γ regulates the activities of
FAS and ACC in LPS-induced DCMECs
In order to determine whether 14-3-3γ affects milk
fat synthesis-associated enzymes in LPS-induced DCMECs, the
activities of FAS and ACC were analyzed using enzyme activity
assays. The activities of FAS and ACC were significantly increased
in LPS-induced DCMECs overexpressing 14-3-3γ, as compared with the
LPS group (P<0.01). Conversely, 14-3-3γ siRNA significantly
decreased the activities of FAS and ACC, as compared with the LPS
group (P<0.01; Fig. 4). These
findings suggested that 14-3-3γ was capable of increasing the
activities of milk fat synthesis-associated enzymes in LPS-induced
DCMECs.
14-3-3γ regulates the expression of
proteins associated with milk fat synthesis in LPS-induced
DCMECs
In order to investigate whether proteins involved in
milk fat synthesis signaling pathways are regulated by 14-3-3γ in
LPS-induced DCMECs, the protein expression levels of SREBP1,
p-SREBP1, PPARG and p-PPARG were analyzed by western blotting. The
protein expression levels of SREBP1 and PPARG in LPS-treated DCMECs
were significantly decreased, as compared with the control group
(P<0.05). Conversely, the protein expression levels of SREBP1
and PPARG were significantly increased in LPS-induced DCMECs
overexpressing 14-3-3γ, as compared with the LPS group (P<0.05;
Fig. 5A), whereas 14-3-3γ siRNA
significantly inhibited the expression of SREBP1 and PPARG proteins
in LPS-induced DCMECs, as compared with the LPS group (P<0.01;
Fig. 5B). These findings suggested
that 14-3-3γ was capable of increasing the expression of proteins
associated with milk fat synthesis in LPS-induced DCMECs.
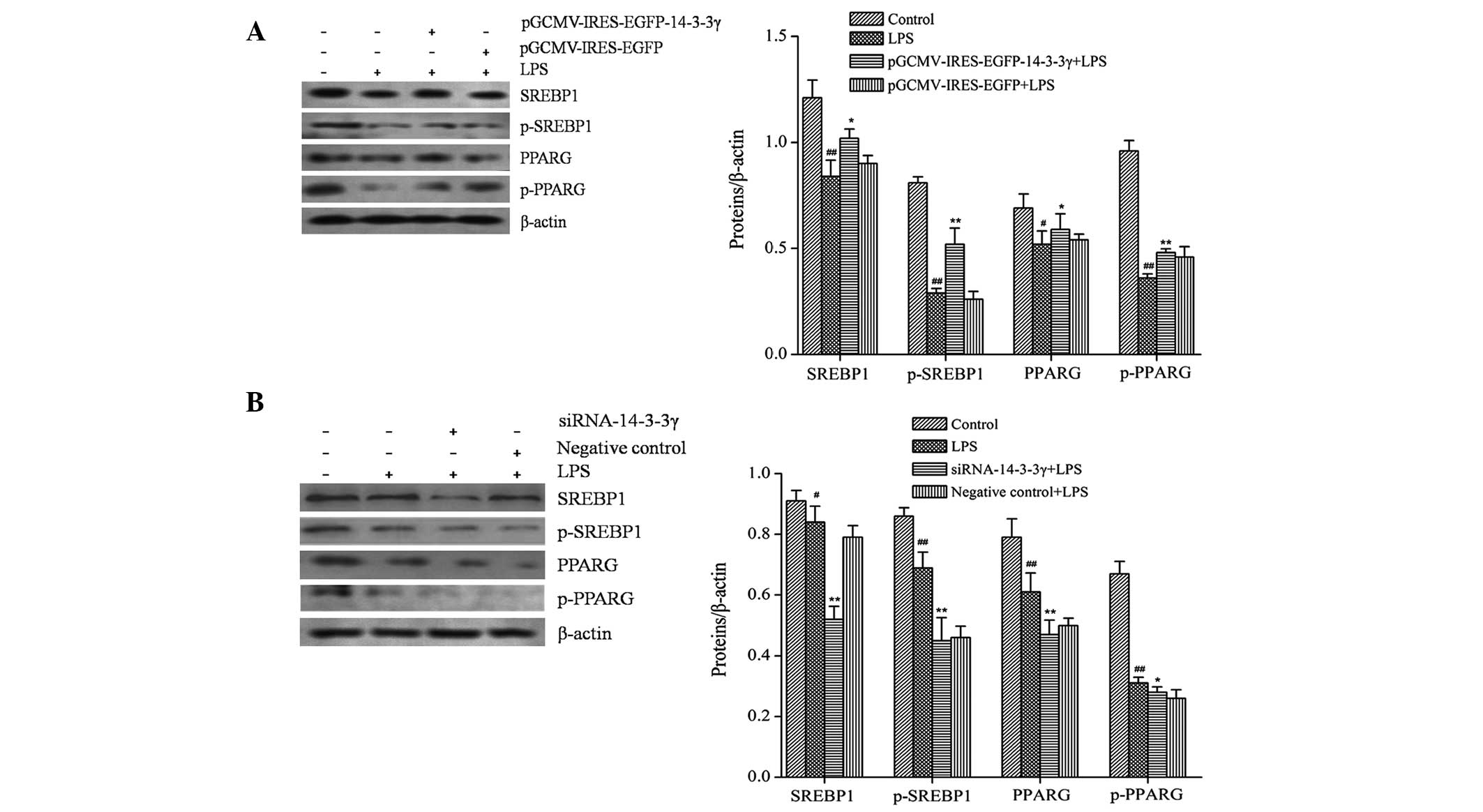 | Figure 5.Effects of 14-3-3γ on the expression
levels of milk fat synthesis-associated proteins in LPS-induced
dairy cow mammary epithelial cells (DCMECs). DCMECs were
transfected with (A) the pGCMV/IRES/EGFP-14-3-3γ or pGCMV/IRES/EGFP
expression vectors or (B) siRNA-14-3-3γ or a negative control for
24 h, after which the cells were treated with 1 µg/ml LPS for 24 h.
The protein expression levels of SREBP1, p-SREBP1, PPARG and
p-PPARG were analyzed by western blotting. β-actin was used as an
internal control. Data are presented as the mean ± standard
deviation of three independent experiments. *P<0.05 and
**P<0.01 vs. the LPS-treated group; #P<0.05 and
##P<0.01 vs. the control group. LPS,
lipopolysaccharide; SREBP1, sterol regulatory element binding
protein-1; PPARG, peroxisome proliferator-activated receptor-γ;
p-SREBP1, phosphorylated-SREBP1; p-PPARG, phosphorylated-PPARG;
siRNA, small interfering RNA; GCMV, grapevine chrome mosaic virus;
IRES, internal ribosome entry site; EGFP, enhanced green
fluorescent protein. |
Discussion
The present study investigated the protective
effects of 14-3-3γ overexpression against LPS-induced damage in
DCMECs, and demonstrated that a high level of 14-3-3γ protein was
able to attenuate LPS-induced apoptosis or death of DCMECs, and
promote milk fat synthesis. Conversely, treatment of DCMECs with
14-3-3γ siRNA revealed opposed results; thus suggesting that
14-3-3γ may have a potential role in preventing SARA-induced
MFD.
Bacterial LPS concentrations are markedly increased
during SARA (23). LPS, which is a
major component of the outer membrane of gram-negative bacteria, is
a highly efficient pro-inflammatory response factor that triggers a
series of immune responses and results in the production of
cytokines, including TNF-α, IL-6 and IL-1β (24). In addition, LPS has been shown to
significantly increase the mRNA expression levels of NF-κB (25.26).
It has been reported that LPS is among the most potent microbial
inducer of inflammation, which is a cascade of intracellular events
that may initiate cell death (27).
Furthermore, LPS has been shown to induce the activation of
executioner caspases and other signaling cascades that ultimately
lead to apoptosis and destruction of cells (28). Previous studies have implicated
14-3-3 proteins, a set of highly conserved scaffolding proteins, in
the regulation of numerous important cellular processes, including
the cell cycle, apoptosis and mitogenic signaling (29,30). In
addition, 14-3-3 proteins have been shown to have critical roles in
various vital physiological and pathological processes by
controlling the activity of their target proteins (31).
Our previous study (11) demonstrated that 14-3-3γ may exert
promising anti-inflammatory activity by downregulating the NF-κB
and mitogen-activated protein kinase signaling pathways. In the
present study, a pGCMV/IRES/EGFP-14-3-3γ expression vector was
successfully constructed and transfected into DCMECs. After 24 h,
the cells were treated with 1 µg/ml LPS for 24 h, after which the
effects of LPS on DCMECs were analyzed using the MTT, LDH activity
and AO double staining assays. It has previously been reported that
the ability to produce milk is determined by the number and
activity of secreting cells in ruminants (32). In the present study, LPS reduced the
viability of DCMECs, increased the activity of LDH, and promoted
the apoptosis of DCMECs; thus indicating that LPS induced DCMECs
injury. However, in the DCMECs transfected with
pGCMV/IRES/EGFP-14-3-3γ, 14-3-3γ overexpression significantly
increased the viability of the LPS-induced cells and decreased the
extent of apoptosis. Conversely, the inhibition of 14-3-3γ using
14-3-3γ siRNA significantly decreased the viability of LPS-induced
DCMECs; thus suggesting that 14-3-3γ may regulate LPS-induced cell
viability. The results of the present study suggested that 14-3-3γ
overexpression may have protective effects against LPS-induced
DCMECs injury.
SARA has been shown to affect both the viability of
DCMECs, as well as their ability to lactate; in particular it has
been shown to inhibit milk fat synthesis (10). It has been suggested that SARA may
initiate the activation of systemic inflammatory responses and other
essential metabolic disturbances of the host, leading to MFD. MFD
is a syndrome characterized by a reduction in milk fat content
(33–36). In a previous study, LPS stimulated
lipolysis in adipose cells via the Toll-like receptor 4 and
MEK1/2-ERK1/2 signaling pathway in the process of inhibiting milk
fat synthesis (37). Previous
studies have reported that LPS may affect milk fat synthesis, and
indirectly downregulate the expression levels of the fatty acid
transporter, via the release of proinflammatory cytokines (38–40). Our
previous study demonstrated that 14-3-3γ overexpression was able to
suppress the production of TNF-α and IL-6 in cell culture
supernatants. Therefore, since 14-3-3γ overexpression has been
shown to suppress inflammation and increase the activity of cells,
the present study hypothesized it may also improve the milk fat
synthesis. In the present study, LPS significantly decreased the
activities of key enzymes associated with de novo fatty acid
synthesis, including FAS and ACC, in LPS-induced DCMECs, and this
is consistent with a previous study (8). However, when the 14-3-3γ protein was
overexpressed in DCMECs, the activities of FAS and ACC were
significantly increased, whereas, the opposite results were
observed when 14-3-3γ was inhibited using siRNA. These results
suggested that 14-3-3γ overexpressed was able to promote de
novo fatty acid synthesis.
SREBP1 and PPARG have important roles in gene
networks associated with milk fat synthesis, and have been shown to
regulate the expression of numerous genes associated with milk fat
synthesis (41). PPARG, which is a
member of the nuclear receptor superfamily of transcription factors
that have been shown to be responsible for the transcriptional
regulation of fatty acid metabolism (42), is activated by lipophilic ligands and
is important for maintaining the quality of milk by inhibiting the
production of inflammatory lipids in lactating mammary glands
(43). SREBP1 is a member of the
basic helix-loop-helix transcription factor family, which are the
major nuclear transcription factors that contribute to the
regulation of milk fat synthesis (44). In the present study, LPS
significantly decreased the expression levels of SREBP1 and PPARG
in DCMECs, and this affect was attenuated by 14-3-3γ overexpression
in these cells. Conversely, the expression levels of SREBP1 and
PPARG were significantly decreased in LPS-induced DCMECs
transfected with pGCMV/IRES/EGFP-14-3-3γ following the inhibition
of 14-3-3γ expression using siRNA. These results suggested that
14-3-3γ was able to regulate genes associated with fatty acid
metabolism in order to improve milk fat synthesis by increasing the
expression levels of SREBP1 and PPARG in LPS-induced DCMECs.
In conclusion, the present study demonstrated that a
high level of 14-3-3γ protein exerted protective effects against
LPS-induced DCMEC injury, by increasing cell viability and
promoting milk fat synthesis through the upregulation of SREBP1,
PPARG, FAS, ACC, CD36 and FABP genes. It has previously been
demonstrated that the expression levels of 14-3-3γ were affected by
estrogen and prolactin (16); thus
suggesting that the addition of nutrients to the dairy cow diet
during lactation may allow the expression levels of 14-3-3γ to be
regulated, in order to prevent SARA-induced MFD.
Acknowledgements
The present study was supported by the Major State
Basic Research Development Program of China (program 973; grant no.
2011CB100804) and the National High Technology Research and
Development Program of China (program 863; grant no.
2006AA10Z1A4).
Glossary
Abbreviations
Abbreviations:
|
LPS
|
lipopolysaccharide
|
|
DCMEC
|
dairy cow mammary epithelial cell
|
|
SARA
|
subacute ruminal acidosis
|
|
MFD
|
milk fat depression
|
References
|
1
|
Plaizier JC, Krause DO, Gozho GN and
McBride BW: Subacute ruminal acidosis in dairy cows: The
physiological causes, incidence and consequences. Vet J. 176:21–31.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gozho GN, Krause DO and Plaizier JC:
Ruminal lipopolysaccharide concentration and inflammatory response
during grain-induced subacute ruminal acidosis in dairy cows. J
Dairy Sci. 90:856–866. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Waldron MR, Nishida T, Nonnecke BJ and
Overton TR: Effect of lipopolysaccharide on indices of peripheral
and hepatic metabolism in lactating cows. J Dairy Sci.
86:3447–3459. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sweet MJ and Hume DA: Endotoxin signal
transduction in macrophages. J Leukoc Biol. 60:8–26.
1996.PubMed/NCBI
|
|
5
|
Zandi E, Rothwarf DM, Delhase M, Hayakawa
M and Karin M: The IkappaB kinase complex (IKK) contains two kinase
subunits, IKKalpha and IKKbeta, necessary for IkappaB
phosphorylation and NF-kappaB activation. Cell. 91:243–252. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ansari N, Khodagholi F, Amini M and
Shaerzadeh F: Attenuation of LPS-induced apoptosis in
NGF-differentiated PC12 cells via NF-kB pathway and regulation of
cellular redox status by an oxazine derivative. Biochimie.
93:899–908. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ling B and Alcorn J: LPS-induced
inflammation downregulates mammary gland glucose, fatty acid, and
L-carnitine transporter expression at different lactation stages.
Res Vet Sci. 89:200–202. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zebeli Q and Ametaj BN: Relationships
between rumen lipopolysaccharide and mediators of inflammatory
response with milk fat production and efficiency in dairy cows. J
Dairy Sci. 92:3800–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Waggoner JW, Löest CA, Turner JL, Mathis
CP and Hallford DM: Effects of dietary protein and bacterial
lipopolysaccharide infusion on nitrogen metabolism and hormonal
responses of growing beef steers. J Anim Sci. 87:3656–3668. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang YD, Wang JQ and Hu T: Infusion of
lipopolysaccharide into external pudendal artery of lactating dairy
cows: Effects on milk composition and milk fat constituents. Dong
Wu Ying Yang Xue Bao. 23:1317–1323. 2011.
|
|
11
|
Liu L, Lin Y, Liu L, Bian Y, Zhang L, Gao
X and Li Q: 14-3-3γ regulates lipopolysaccharide-induced
inflammatory responses and lactation in dairy cow mammary
epithelial cells by inhibiting NF-κB and MAPKs and up-regulating
mTOR signaling. Int J Mol. 16:16622–16641. 2015. View Article : Google Scholar
|
|
12
|
López-Soriano FJ and Williamson DH: Acute
effects of endotoxin (lipopolysaccharide) on tissue lipid
metabolism in the lactating rat. The role of delivery of intestinal
glucose. Mol Cell Biochem. 141:113–120. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hosing AS, Kundu ST and Dalal SN: 14-3-3
Gamma is required to enforce both the incomplete S phase and G2 DNA
damage checkpoints. Cell Cycle. 7:3171–3179. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pozuelo-Rubio M: Proteomic and biochemical
analysis of 14-3-3-binding proteins during C2-ceramide-induced
apoptosis. FEBS J. 277:3321–3342. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van Hemert MJ, Steensma HY and van Heusden
GP: 14-3-3 proteins: Key regulators of cell division, signalling
and apoptosis. Bioessays. 23:936–946. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang JG: Proteomic analysis of the
lactation regulation in DCMECs treated with different hormones
(unpublished PhD thesis). Harbin, China: Northeast Agricultural
University. 2012.
|
|
17
|
Liu D, Yin D, Yan GH, Sun D, Xu M and He
M: Protective role of 14-3-3γ in burn and LPS-induced rat
myocardial injury. Zhong Guo Bing Li Sheng Li Za Zhi. 28:1160–1165.
2012.(In Chinese).
|
|
18
|
Liu D, Yin D, Sun D, Xu M and He M: The
Role of Bax in the 14-3-3γ protection against cardiomyocyte damage
induced by LPS. Tian Jin Yi Yao. 40:1222–1225. 2012.(In
Chinese).
|
|
19
|
Zhao K, Liu HY, Zhou MM and Liu JX:
Establishment and characterization of a lactating bovine mammary
epithelial cell model for the study of milk synthesis. Cell Biol
Int. 34:717–721. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li HM, Wang CM, Li QZ and Gao XJ: MiR-15a
decreases bovine mammary epithelial cell viability and lactation
and regulates growth hormone receptor expression. Molecules.
17:12037–12048. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu LM, Li QZ, Huang JG and Gao XJ:
Proteomic and functional analyses reveal MAPK1 regulates milk
protein synthesis. Molecules. 18:263–275. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Q, Zhao XH and Wang ZJ: Cytotoxicity
of flavones and flavonols to a human esophageal squamous cell
carcinoma cell line (KYSE-510) by induction of G2/M arrest and
apoptosis. Toxicol In Vitro. 23:797–807. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Khafipour E, Krause DO and Plaizier JC: A
grain-based subacute ruminal acidosis challenge causes
translocation of lipopolysaccharide and triggers inflammation. J
Dairy Sci. 92:1060–1070. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bruckmaier RM: Gene expression of factors
related to the immune reaction in response to intramammary
Escherichia coli lipopolysaccharide challenge. J Dairy Res.
72:120–124. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheng G, Zhao Y, Li H, Wu Y, Li X, Han Q,
Dai C and Li Y: Forsythiaside attenuates lipopolysaccharide-induced
inflammatory responses in the bursa of Fabricius of chickens by
downregulating the NF-kB signaling pathway. Exp Ther Med.
7:179–184. 2014.PubMed/NCBI
|
|
26
|
Reddy DB and Reddanna P: Chebulagic acid
(CA) attenuates LPS-induced inflammation by suppressing NF-kappaB
and MAPK activation in RAW 264.7 macrophages. Biochem Biophys Res
Commun. 381:112–117. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wong PM, Chugn SW and Sultzer BM: Genes,
receptors, signals and responses to lipopolysaccharide endotoxin.
Scand J Immunol. 51:123–127. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang F, Sun X, Beech W, Teter B, Wu S,
Sigel J, Vinters HV, Frautschy SA and Cole GM: Antibody to
caspase-cleaved actin detects apoptosis in differentiated
neuroblastoma and plaque-associated neurons and microglia in
Alzheimer's disease. Am J Pathol. 152:379–389. 1998.PubMed/NCBI
|
|
29
|
Radhakrishnan VM and Martinez JD:
14-3-3gamma induces oncogenic transformation by stimulating MAP
kinase and PI3K signaling. PLOS One. 5:114332010. View Article : Google Scholar
|
|
30
|
Samuel T, Weber HO, Rauch P, Verdoodt B,
Eppel JT, McShea A, Hermeking H and Funk JO: The G2/M regulator
14-3-3sigma prevents apoptosis through sequestration of Bax. J Biol
Chem. 276:45201–45206. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Obsilova V, Silhan J, Boura E, Teisinger J
and Obsil T: 14-3-3 proteins: A family of versatile molecular
regulators. Physiol Res. 57(Suppl 3): S11–S21. 2008.PubMed/NCBI
|
|
32
|
Boutinaud M, Guinard-Flamenta J and Jammes
H: The number and activity of mammary epithelial cells, determining
factors for milk production. Reprod Nutr Dev. 44:499–508. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ametaj BN, Emmanuel DG, Zebeli Q and Dunn
SM: Feeding high proportions of barley grain in a total mixed
ration perturbs diurnal patterns of plasma metabolites in lactating
dairy cows. J Dairy Sci. 92:1084–1091. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zebeli Q, Dunn SM and Ametaj BN:
Perturbations of plasma metabolites correlated with the rise of
rumen endotoxin in dairy cows fed diets rich in easily degradable
carbohydrates. J Dairy Sci. 94:2374–2382. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mao SY, Zhang RY, Wang DS and Zhu WY:
Impact of subacute ruminal acidosis (SARA) adaptation on rumen
microbiota in dairy cattle using pyrosequencing. Anaerobe.
24:12–19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kleen JL, Hooijer GA, Rehage J and
Noordhuizen JP: Subacute ruminal acidosis (SARA): A review. J Vet
Med A Physiol Pathol Clin Med. 50:406–414. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zu L, He J, Jiang H, Xu C, Pu S and Xu G:
Bacterial endotoxin stimulates adipose lipolysis via toll-like
receptor 4 and extracellular signal-regulated kinase pathway. J
Biol Chem. 284:5915–5926. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ling B and Alcorn J: LPS-induced
inflammation downregulates mammary gland glucose, fatty acid, and
L-carnitine transporter expression at different lactation stages.
Res Vet Sci. 89:200–202. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
López-Soriano FJ and Williamson DH: Acute
effects of endotoxin (lipopolysaccharide) on tissue lipid
metabolism in the lactating rat. The role of delivery of intestinal
glucose. Mol Cell Biochem. 141:113–120. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pekala PH, Kawakami M, Angus CW, Lane MD
and Cerami A: Selective inhibition of synthesis of enzymes for de
novo fatty acid biosynthesis by an endotoxin-induced mediator from
exudate cells. Proc Natl Acad Sci USA. 80:2743–2747. 1983.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bionaz M and Loor JJ: Gene networks
driving bovine milk fat synthesis during the lactation cycle. BMC
Genomics. 9:3662008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang Q and Li Y: Roles of PPARs on
regulating myocardial energy and lipid homeostasis. J Mol Med
(Berl). 85:697–706. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wan Y, Saghatelian A, Chong LW, Zhang CL,
Cravatt BF and Evans RM: Maternal PPAR gamma protects nursing
neonates by suppressing the production of inflammatory milk. Genes
Dev. 21:1895–1908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Anderson SM, Rudolph MC, McManaman JL and
Neville MC: Key stages in mammary gland development. Secretory
activation in the mammary gland: It's not just about milk protein
synthesis! Breast Cancer Res. 9:2042007.
|