Introduction
MicroRNAs (miRNAs) are a class of non-coding
endogenous single-strand RNAs, 20–23 nucleotides (nt) in length,
which negatively regulate target gene expression
post-transcriptionally to induce degradation of multiple mRNAs,
translational repression and gene silencing by pairing to the 3′
untranslated region (3′UTR) of target mRNA (1,2). miRNAs
are indicated to be involved in numerous key cellular processes,
including cell growth, differentiation and death, and in
particular, regulate the initiation, development and progression of
human cancers, including tumor growth, apoptosis, invasion and
metastasis (3). In malignancy,
miRNAs play roles as either oncogenes or tumor suppressor genes
(4,5).
Malignant tumor progression is characterized by the
process of epithelial-to-mesenchymal transition (EMT), and is
considered an essential step in tumor metastasis (6). During the EMT process, epithelial tumor
cells are induced to lose epithelial polarity and the epithelial
marker E-cadherin, and to gain mesenchymal phenotypes and increased
expression of mesenchymal markers vimentin and N-cadherin and to
exhibit increased migratory and invasive capabilities and behavior
(6,7). B-cell-specific Moloney murine leukemia
virus insertion site 1 (Bmi-1) and E2F transcription factor 3
(E2F3) are well recognized as oncogenes in cancer initiation and
progression (8,9), and are reported to promote EMT by
suppressing E-cadherin expression (10), and inhibiting the expression of cell
cycle inhibitors p14 and p16 (11,12).
miR-200 is a family of tumor suppressor miRNAs,
which includes miR-200a, miR-200b, miR-200c, miR-141 and miR-429,
and is markedly involved in tumor initiation and progression
(13). Studies have demonstrated
that expression of the miR-200 family is significantly reduced in
cells undergoing EMT (14–16). As an important member of the miR-200
family and a powerful regulator of EMT, miR-200c is indicated to
maintain the epithelial phenotype of tissues by inhibiting the
expression of E-cadherin transcription factor ZEB1 in certain types
of cancer (16,17). In a recent study, miR-200c was
reported to modulate EMT in human renal cell carcinoma (18).
Renal cancer is the most common cancer of the
urinary system worldwide, with 63,920 estimated new cases and
13,860 estimated mortalities owing to cancer in 2014 (19), and the incidence is increasing
(19–21). Due to the absence of effectiveness
and specificity, the options for therapy of metastatic renal cancer
remain limited (22); thus, the
development of new treatments to treat renal cancer in necessary.
In the present study, the effects of miR-200c were investigated by
up- and downregulating miR-200c expression in two renal cancer cell
lines (ACHN and A498) and the possible mechanisms of action were
evaluated.
Materials and methods
Cell lines and culture
Human renal cancer ACHN and A498 cells were
purchased from Guangzhou Jennio Biological Technology Co., Ltd.
(Guangzhou, China). ACHN cells were cultured in high glucose
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), and A498
cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS. The two cell lines
were cultured at 37°C under an atmosphere of 5% CO2 in
humidified air.
miRNA infection of the cells
The miR-200c and control miRNA (plasmid, pEZX-MR03)
and miR-200c inhibitor and control small interfering RNAs (siRNAs)
(plasmid, CMV-RFP-U6-miRNA inhibitor-PGK-puromycin) were purchased
from GeneCopoeia, Inc. (Guangzhou, China). Infection was performed
in accordance with the manufacturer's protocol. Briefly, ACHN and
A498 cells were cultured in medium supplemented with 10% FBS for 24
h prior to miRNA infection, and then the viral vectors were added
into the medium for infection, respectively. Cells were then
cultured in medium containing 1 µg/ml puromycin at 48 h
post-transfection.
Bioinformatic analysis
Candidate targets for miR-200c were searched for
using TargetScan Human 7.1 (http://www.targetscan.org/), a bioinformatic tool for
miRNA target screening.
Luciferase reporter assay
A luciferase reporter assay was performed with
Dual-Luciferase Reporter 1000 Assay system (Promega Corporation,
Madison, WI, USA) according to the manufacturer's protocol.
Briefly, after culturing until 70% confluence, HEK293 cells were
co-infected with miR-200c/control miRNA and the 3′UTR (wild type or
mutant) of Bmi-1 or E2F3 (GeneCopoeia, Inc.). The firefly and
Renilla luciferase activities were then analyzed.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA (enriched for miRNAs) was extracted using
an E.Z.N.A. miRNA kit (Omega Bio-Tek, Inc., Norcross, GA, USA)
according to the manufacturer's instructions. The total RNAs were
purified by treatment with gDNA Eraser from a PrimeScript RT
reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). A
RT-qPCR assay was performed using a Thermal Cycler Dice Real Time
System (TP800; Takara Biotechnology Co., Ltd.), PrimeScript miRNA
qPCR Starter kit Ver.2.0 (Takara Biotechnology Co., Ltd.) and SYBR
Premix Ex Taq II (Takara Biotechnology Co., Ltd.) according to the
manufacturers' instructions. For the miRNA expression assay,
two-step RT-qPCR was employed with specific primers for miR-200c
and RNU6B (the latter was an internal control) following the
manufacturer's protocol. The PCR was carried out at 95°C for 10
sec, followed by 40 cycles of amplification at 95°C for 5 sec and
55°C for 20 sec. For relative mRNA expression analysis, two-step
RT-qPCR was employed with specific primers for GAPDH (as internal
control), Bmi-1, E2F3, E-cadherin, N-cadherin, vimentin, p14 and
p16 following the manufacturer's protocol, and PCR was carried out
at 95°C for 30 sec, followed by 40 cycles of amplification at 95°C
for 5 sec and 56°C for 30 sec. All results were representative of
three independent assays, and the expression levels were expressed
according to the 2−ΔΔCq method (23). The designed specific primers are
listed in Table I.
 | Table I.Sequences of target gene primers for
reverse transcription-quantitative polymerase chain reaction. |
Table I.
Sequences of target gene primers for
reverse transcription-quantitative polymerase chain reaction.
| Gene | Primer sequence
5′-3′ | Tm (°C) |
|---|
| RNU6B | F:
CTCGCTTCGGCAGCACA | 59.42 |
|
| R:
AACGCTTCACGAATTTGCGT | 55.75 |
| miR-200c | F:
TAATACTGCCGGGTAATGATGG | 58.21 |
|
| R:
TCGTATCCAGTGCAGGGTC | 59.72 |
| GAPDH | F:
TGCACCACCAACTGCTTAG | 60.07 |
|
| R:
AGTAGAGGCAGGGATGATGTTC | 59.72 |
| Bmi-1 | F:
TGGATCGGAAAGTAAACAAAGAC | 56.60 |
|
| R:
TGCATCACAGTCATTGCTGCT | 58.01 |
| E2F3 | F:
TGCCTGACTCAATAGAGAGCCTAC | 61.97 |
|
| R:
TCCCATTGTGGTCTTGGTTGT | 58.01 |
| E-cadherin | F:
GAAAGCGGCTGATACTGACC | 59.85 |
|
| R:
CGTACATGTCAGCCGCTTC | 59.72 |
| N-cadherin | F:
GGTGGAGGAGAAGAAGACCAG | 61.92 |
|
| R:
GGCATCAGGCTCCACAGTG | 61.88 |
| Vimentin | F:
GGGAGAAATTGCAGGAGGAG | 59.85 |
|
| R:
AGGTCAAGACGTGCCAGAGAC | 61.92 |
| p14 | F:
GTTCTTGGTGACCCTCCGGATT | 61.94 |
|
| R:
ATCAGCACGAGGGCCACAG | 61.88 |
| p16 | F:
CCCAACGCACCGAATAGTTAC | 59.97 |
|
| R:
ACGGGTCGGGTGAGAGTG | 61.86 |
Western blot analysis
ACHN and A498 cells were lysed with RIPA buffer
(Beyotime Institute of Biotechnology, Shanghai, China) and total
proteins were extracted by centrifuging at 10,000 × g for 10 min at
4°C. The proteins were quantified using an Enhanced BCA Protein
Assay kit (Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. Proteins (30 µg/lane) were separated
by SDS-PAGE (10% gel) and then transferred to a PVDF membrane (EMD
Millipore, Bedford, MA, USA). The blotting membranes were incubated
overnight (16 h) at 4°C with anti-Bmi-1 antibody (40 kD; 1:20,000;
ab115251; Abcam, Cambridge, UK), anti-E2F3-1 antibody (37 kD;
1:2,000; ab50917; Abcam, Cambridge, UK), anti-E-cadherin antibody
(135 kD; 1:1,000; cat. no. 5296; Cell Signaling Technology, Inc.,
Danvers, MA, USA), anti-N-cadherin antibody (140 kD; 1:1,000;
ab18203; Abcam), anti-vimentin antibody (57 kD; 1:2,000; cat. no.
5741; Cell Signaling Technology, Inc.), anti-p14 antibody (14 kD;
1:500; cat. no. 2407; Cell Signaling Technology, Inc.), anti-p16
antibody (16 kD; 1:500; ab118459; Abcam) or anti-β-tubulin antibody
(55 kD; 1:50,000; cat. no. 70004; EarthOx Life Sciences, Millbrae,
CA, USA; loading control), respectively, and then probed with a
horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G
secondary antibody (1:10,000; E030120; EarthOx Life Sciences) or a
horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G
secondary antibody (1:10,000; E030110; EarthOx Life Sciences) for 1
h at room temperature. The bands were visualized using Luminata
Crescendo Western HRP Substrate (WBLUR0500; EMD Millipore) with
exposure to X-OMAT BT film (Carestream Health, Rochester, NY, USA).
Three replicates were performed.
Cell proliferation assays
Proliferation of ACHN and A498 cells was detected
using a CellTiter 96 AQueous One Solution Cell Proliferation Assay
(Promega Corporation) in accordance with the manufacturer's
protocol. Briefly, cells were seeded in a 96-well cell culture
cluster (Corning Incorporated, Corning, NY, USA) at a density of
3,000 cells per well with 100 µl culture medium. After 5 days, the
culture medium was removed and replaced with an equal volume of
medium containing CellTiter 96 AQueous One Solution reagent, and
the cells were then incubated at 4°C for 2 h. The absorbance was
detected at 490 nm using a 96-well plate reader.
Colony formation assay
A colony formation assay was performed according to
a slightly modified method (24).
Briefly, cells were seeded into 60-mm plastic dishes (Nest
Biotechnology, Hong Kong, China) at a density of 1,000 cells per
well, and cultured at 37°C under an atmosphere of 5% CO2
in humidified air (ACHN cells were cultured for 3 weeks and A498
cells were cultured for 2 weeks, respectively). The numbers of
colonies were counted after staining with Coomassie Brilliant
Blue.
Wound healing (migration) and
transwell (invasion) assays
ACHN and A498 cells were seeded into 12-well cell
culture plates (3×105 cells per well; Nest
Biotechnology). The wound healing assay was performed using a
sterile pipette tip to make a scratch through the confluent
monolayer when cells reached 100% confluence. Cells were washed
three times with PBS and cultured with medium supplemented with 1%
FBS. The cell migration was observed after culture for 24 h. The
percentage of wound closure was determined with 3 replications, and
five randomly chosen fields were calculated for each replicate. For
the transwell assay, 3×105 cells in 150 µl serum-free
medium supplemented with 1% FBS were placed into the upper chamber
of the insert (membrane pore size, 8 µm; Corning) with Matrigel (BD
Biosciences, Billerica, MA, USA), and 500 µl medium supplemented
with 10% FBS was added to the lower chamber of a 24-well plastic
plate. Following 24 h of culture at 37°C, the cells remaining in
the upper chamber or on the upper membrane were removed. The number
of cells adhering to the lower membrane of the inserts was counted
after staining with Crystal Violet Staining Solution (Beyotime
Institute of Biotechnology) for 10 min.
Statistical analysis
The cell proliferation assay was performed in five
independent experiments and other analyses were repeated in three
independent experiments. Results are presented as mean ± standard
error of the mean. All data were analyzed using SPSS 18.0 software
(SPSS, Inc., Chicago, IL, USA) by one-way analysis of variance, and
differences between treatments were assessed using a Fisher's least
significant difference test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-200c suppressed the proliferation,
migration and invasion of renal cancer cells
To determine the biological functions of miR-200c,
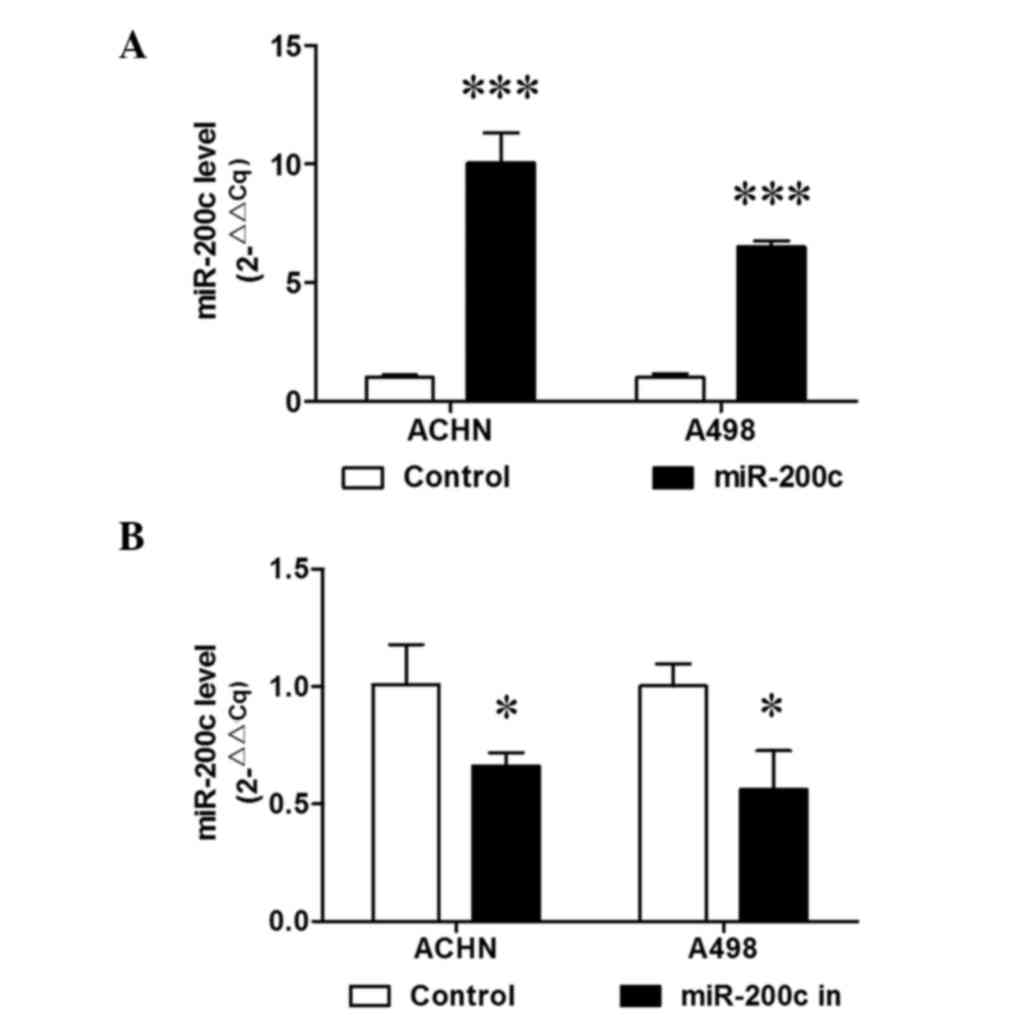
ACHN and A498 cells were stably transduced with miR-200c. RT-qPCR
analysis demonstrated that miR-200c was then stably overexpressed
(P<0.001; Fig. 1A). A cell
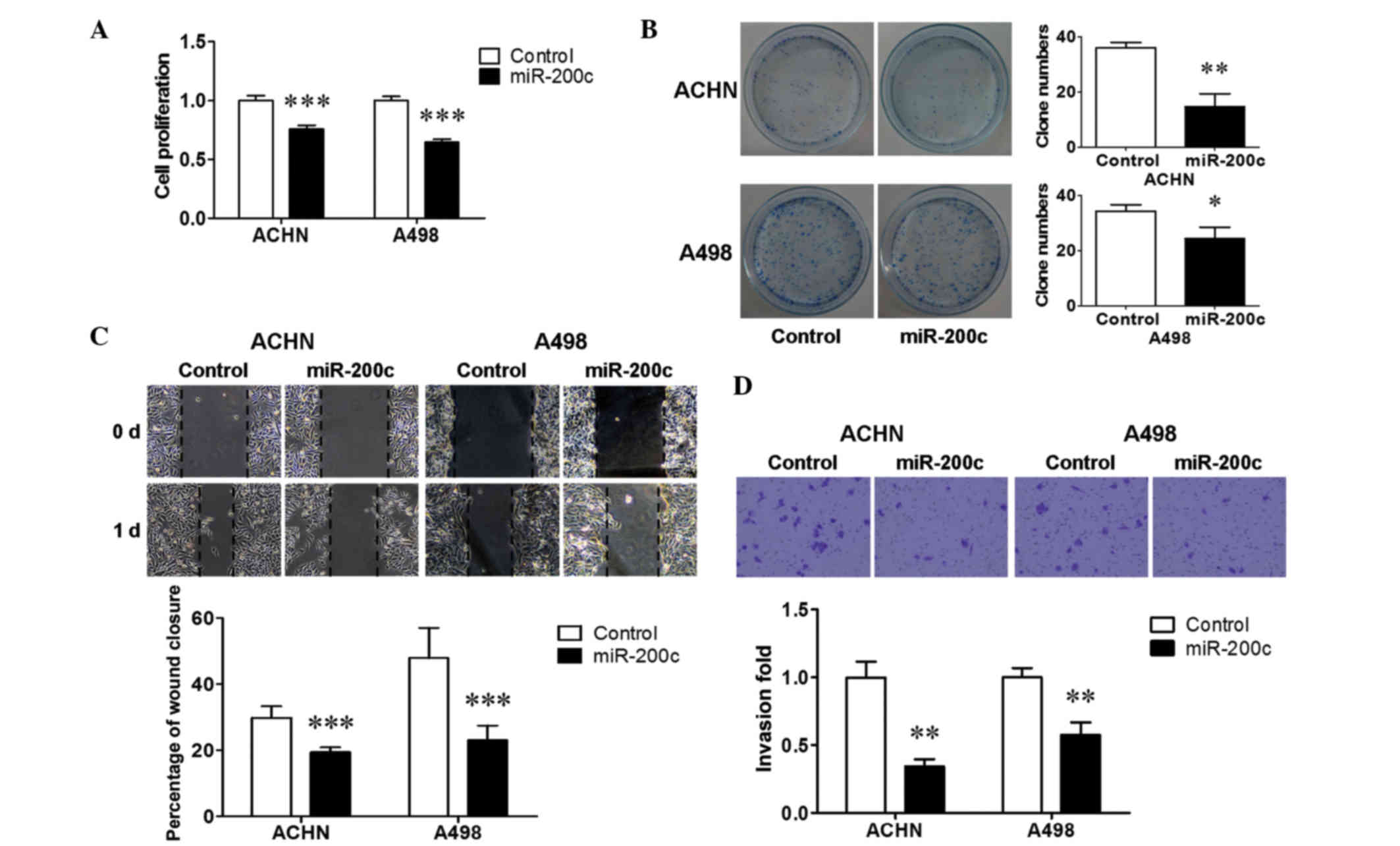
proliferation assay was performed to examine the effect of miR-200c
on renal cancer cell growth. Ectopic miR-200c induced a significant
suppression in cell proliferation in the two renal cancer cell
lines (P<0.001; Fig. 2A). A
colony formation assay was then performed, and it was found that
upregulation of miR-200c reduced colony formation by ACHN cells
(P<0.01; Fig. 2B) and A498 cells
(P<0.05; Fig. 2B), consistent
with the results of the cell proliferation assay. Furthermore, a
wound healing assay and transwell assay were performed to evaluate
the effect of miR-200c on renal cancer cell metastasis. Ectopic
miR-200c significantly decreased the extent of wound healing (%
wound closure) in the two cell lines (P<0.001; Fig. 2C). Transwell assays with Matrigel
demonstrated that over-expressed miR-200c significantly inhibited
the invasive capacity of the two cell lines (P<0.01; Fig. 2D). These results indicated that
miR-200c inhibited the cell growth and metastasis of renal cancer
cells.
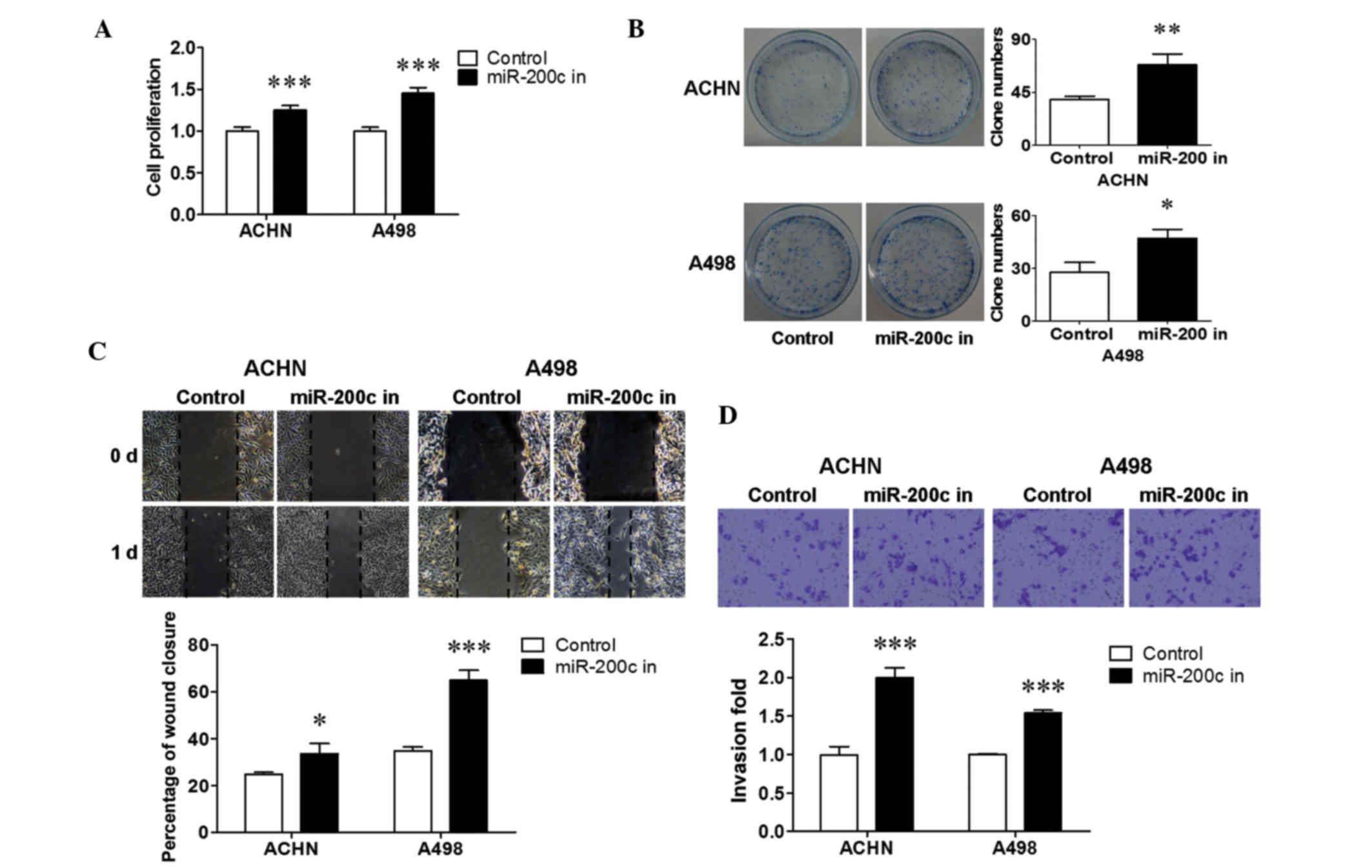
Downregulated endogenous miR-200c
increased renal cancer cell growth and metastasis
miR-200c was stably inhibited in ACHN and A498 cells
to reveal the biological significance of endogenous miR-200c.
RT-qPCR demonstrated that miR-200c was stably downregulated in the
two cell lines (P<0.05; Fig. 1B).
To determine the influence of downregulated miR-200c on renal
cancer cell growth, a cell proliferation and colony formation assay
was performed. It is shown in Fig. 3
that the inhibition of endogenous miR-200c resulted in an increase
in cell proliferation (P<0.001; Fig.
3A), and a promotion in colony formation in ACHN (P<0.01;
Fig. 3B) and A498 (P<0.05;
Fig. 3B) cells. The changes in renal
cancer cell metastasis were then evaluated using a wound healing
assay and a transwell assay. The results revealed that the
downregulation of endogenous miR-200c significantly stimulated
migration (P<0.05 for ACHN cells; P<0.001 for A498 cells;
Fig. 3C) and invasion (P<0.001;
Fig. 3D) in both cell lines compared
with that in the control. Together, these results suggest that
downregulated endogenous miR-200c promoted cell growth and
metastasis in renal cancer cells.
miR-200c regulated cell proliferation
and metastasis by directly targeting of oncogenes Bmi-1 and
E2F3
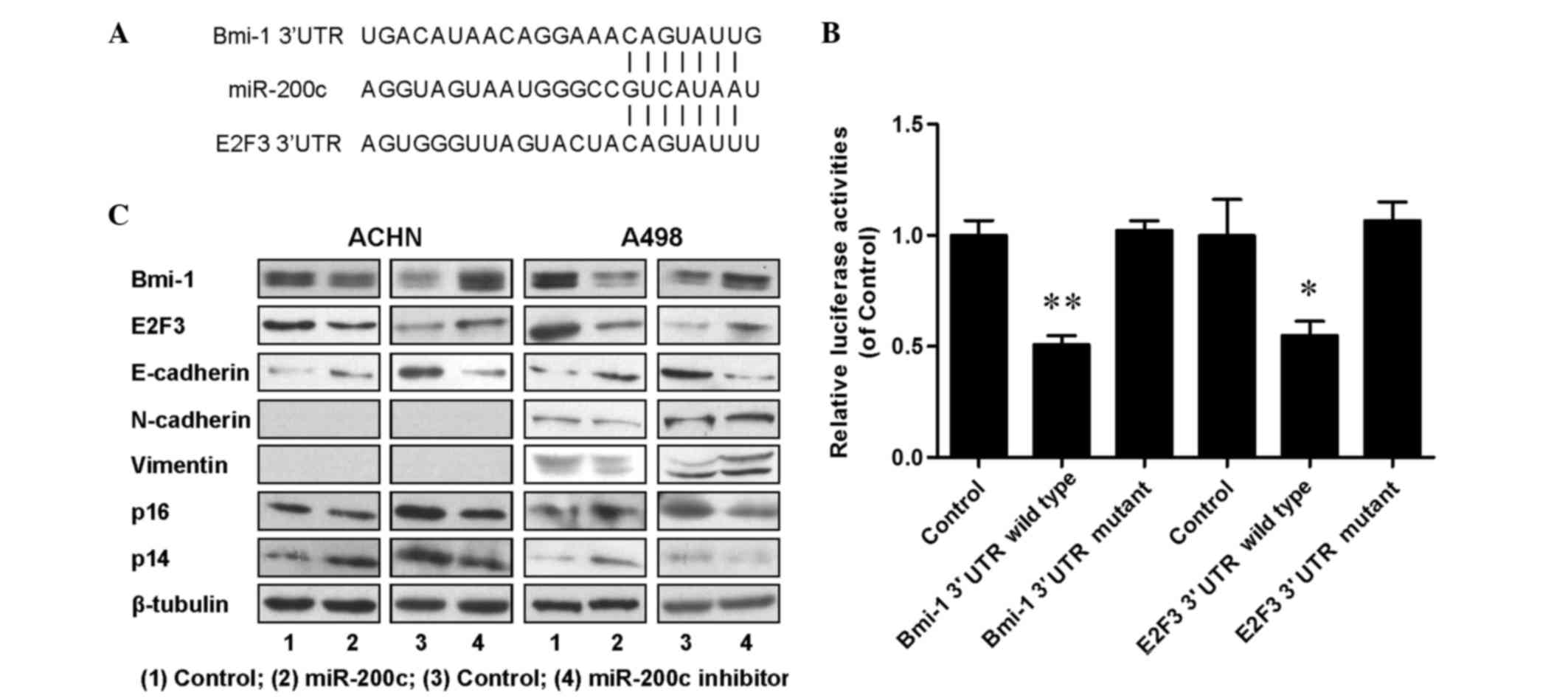
To dissect the molecular mechanism by which miR-200c
acts as tumor suppressor in renal cancer cells, candidate targets
for miR-200c were searched for using TargetScan, a bioinformatic
tool for miRNA target screening. It was found that oncogenes Bmi-1
and E2F3 were putative targets of miR-200c (Fig. 4A). A miR-200c-based luciferase assay
in HEK293 cells was then performed to measure if miR-200c directly
binds to the 3′UTRs of Bmi-1 and E2F3, and the results demonstrated
that miR-200c directly binds to the 3′UTRs of Bmi-1 (P<0.01;
Fig. 4B) and E2F3 (P<0.05;
Fig. 4B), as significantly decreased
luciferase activities were observed for the wild type but not for
the mutant 3′UTR. A western blot assay was performed to evaluate
the expression levels of Bmi-1 and E2F3, and their downstream
proteins including E-cadherin, N-cadherin, vimentin, p14 and p16.
The results (Fig. 4C) indicated that
the expression of Bmi-1, E2F3 and N-cadherin was downregulated, and
the expression of E-cadherin, p14 and p16 was upregulated in
miR-200c overexpressing cells; and increased expression levels of
Bmi-1, E2F3 and N-cadherin, and decreased expression levels of
E-cadherin, p14 and p16 were detected in endogenous miR-200c
downregulated cells. An RT-qPCR assay was also performed and
similar tendencies were detected for the associated mRNAs (Fig. 5). All the aforementioned data
demonstrated that miR-200c regulated renal cancer cell growth and
metastasis by directly targeting the 3′UTR of Bmi-1 and E2F3 and
inducing changes in the expression levels of Bmi-1 and E2F3 and
their downstream genes in cell proliferation and EMT.
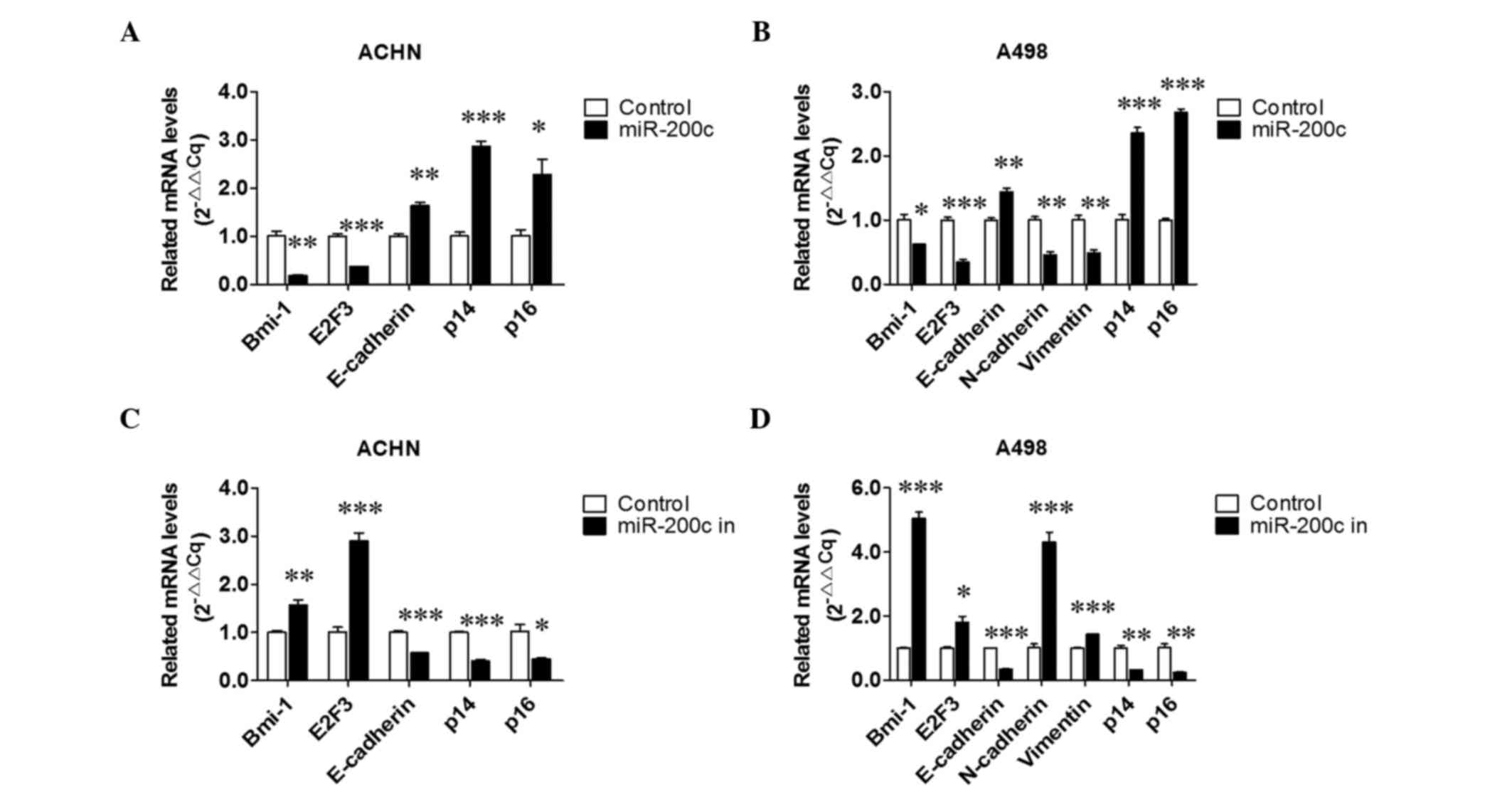 | Figure 5.Effect of over- and downexpressed
miR-200c on relative mRNA levels. Effects of miR-200c
overexpression on relative mRNA levels in (A) ACHN and (B) A498
cells. Effects of downexpressed miR-200c on Bmi-1, E2F3,
E-cadherin, N-cadherin, vimentin, p14 and p16 expression in (C)
ACHN and (D) A498 cells. *P<0.05, **P<0.01 and **P<0.01
vs. the control group. miR, microRNA; Bmi-1, B-cell-specific
Moloney murine leukemia virus insertion site 1; E2F3, E2F
transcription factor 3; 3′UTR, 3′ untranslated region. |
Discussion
MicroRNAs have been demonstrated to play critical
roles as oncogenes or tumor suppressors in human cancers in
previous studies (25). miR-200 is a
family of tumor suppressor miRNAs consisting of five members
significantly involved in the inhibition of EMT (13), and as a member of the miR-200 family,
miR-200c was reported to modulate EMT in human renal cell carcinoma
in a recent study (18). In the
present study, the effects of up- and downregulation of the
expression of miR-200c on cell proliferation, migration and
invasion in renal cancer cells, and the mechanisms of miR-200c
regulation, were investigated. The results demonstrated that
ectopic miR-200c significantly inhibited cell growth, migration and
invasion, and a reduction of endogenous miR-200c expression
recovered cell proliferation and metastasis in renal cancer cells.
They also indicated that miR-200c suppressed renal cancer cell
growth and metastasis by directly targeting oncogenes Bmi-1 and
E2F3, and then modulating the expression of downstream
proteins.
The miR-200 family has been demonstrated to be
downregulated during tumor progression and acts as a critical
suppressor in the regulation of epithelial-to-mesenchymal
transition (EMT), cancer stem cell (CSs) self-renewal and
differentiation, cell division and apoptosis, and reversal of
chemoresistance (13). miR-200c is
an important member of the miR-200 family, and downregulation of
its expression level has been observed in several tumors of the
urinary system (26–30). A recent study of renal cell carcinoma
demonstrated that miR-200c was downregulated in renal cell
carcinoma, and restoration of miR-200c markedly suppressed the
migration and invasion of SN12-PM6 and 786–0 cells (18); these results suggest that miR-200c
acted as a tumor suppressor in renal cell carcinoma by modulating
EMT. In the present study, the over- and downregulation of miR-200c
expression was enforced in two renal cancer cell lines and the
results indicated that ectopically expressed miR-200c significantly
inhibited the metastatic ability of ACHN and A498 cells, and
suppressed expression of endogenous miR-200c promoted the
metastatic capacity of the two renal cancer cell lines. These
results were consisted with the aforementioned studies, and
indicate that miR-200c functions as a tumor suppressor miRNA in
renal cancer by inhibiting the EMT process. Yu et al
(31) investigated the influence of
miR-200c on the proliferation of pancreatic cancer cells and
indicated that upregulation of miR-200c could stimulate the
proliferation of pancreatic cancer cells. However, the results of
the present study are inconsistent with this, as they indicated
that miR-200c significantly decreased renal cancer cell
proliferation. It is suggested the different results may be due to
the difference in organ type.
Bmi-1 plays a key role in cell cycle regulation,
cell immortalization and cell senescence as a polycomb gene family
member (8,32). It is well recognized that Bmi-1 is
frequently upregulated in various human cancers, and plays
important roles as an oncogene in cancer initiation and progression
(8,33). E2F3 is another oncogene that is
overexpressed in bladder and prostate cancers (34,35), and
participated in controlling the cell cycle progression and
proliferation of cancer cells (9).
In the present study, Bmi-1 and E2F3 were predicted as functional
targets of miR-200c by TargetScan analysis, and the effects of
miR-200c on the 3′UTRs and expression of Bmi-1 and E2F3 were then
measured. It was found that miR-200c directly bound to the 3′UTRs
of Bmi-1 and E2F3, and decreased the expression levels of Bmi-1 and
E2F3 mRNAs and proteins. These results are consistent with the
results for cell functions in this study, and suggest that Bmi-1
and E2F3 are oncogenes in renal cancer that are inhibited by
miR-200c.
EMT is a critical process driving cancer metastasis,
which is characterized by the loss of epithelial marker E-cadherin
and a stimulation of the mesenchymal markers vimentin and
N-cadherin, followed by an induction of increases in migratory and
invasive behavior (7). Bmi-1 is
reported to induce EMT by repressing E-cadherin expression
(10). In the present study, the
expression levels of E-cadherin and N-cadherin were examined, and
it was demonstrated that the ectopic expression of miR-200c
increased E-cadherin expression and decreased N-cadherin
expression, whereas the downregulated expression of endogenous
miR-200c decreased E-cadherin expression and increased N-cadherin
expression. These results demonstrated that miR-200c regulated
Bmi-1 expression and then modulated EMT by stimulating E-cadherin
expression and suppression of N-cadherin expression. Furthermore,
Bmi-1 is reported as an oncogene, which acts by regulating the
expression of cell cycle inhibitors p14 and p16 (11,12,36,37). P14
is reported to prevent the degradation and inactivation of p53 by
binding to E3 ubiquitin-protein ligase (38), and p16 is recognized as an inhibitor
of cyclin-dependent kinases 4 and 6 (35), which arrests the cell cycle in the
G1/S phase (39). In the present
study, the expression levels of p14 and p16 were also measured and
the results indicated that overexpressed miR-200c increased the
expression of p14 and p16, and the downregulation of miR-200c
suppressed the expression of p14 and p16. These results suggest
that miR-200c regulates Bmi-1 expression and then modulates cell
proliferation by increasing the expression of p14 and p16.
In summary, the present study investigated the
functions and mechanisms of miR-200c in the regulation of renal
cancer cell proliferation and metastasis. The results demonstrated
that miR-200c acted as a tumor suppressor by inhibiting cell
proliferation, migration and invasion in renal cancer cells. In
addition, downregulated endogenous miR-200c recovered renal cancer
cell growth and metastasis, and miR-200c possessed the potency to
suppress renal cancer cell growth and metastasis via the
downregulation of the oncogenes Bmi-1 and E2F3. These findings
provide important basic information relevant to renal cancer
intervention, prevention and therapy.
Acknowledgements
This study was supported in part by the following
grants: Science and Technology Planning Project of Guangdong
Province (grant no. 2012B031800221) and The National Natural
Science Funds (grant no. 81272833) of China.
References
|
1
|
Brown M, Suryawanshi H, Hafner M, Farazi
TA and Tuschl T: Mammalian miRNA curation through next-generation
sequencing. Front Genet. 4:1452013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
3
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang B, Pan X, Cobb GP and Anderson TA:
MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pavelic S Kraljevic, Sedic M, Bosnjak H,
Spaventi S and Pavelic K: Metastasis: New perspectives on an old
problem. Mol Cancer. 10:222011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang L, Li J and Song L: Bmi-1, stem
cells and cancer. Acta Biochim Biophys Sin (Shanghai). 41:527–534.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Olsson AY, Feber A, Edwards S, Te Poele R,
Giddings I, Merson S and Cooper CS: Role of E2F3 expression in
modulating cellular proliferation rate in human bladder and
prostate cancer cells. Oncogene. 26:1028–1037. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu
LJ, Kong QL, Xu LH, Zhang X, Liu WL, et al: The polycomb group
protein Bmi-1 represses the tumor suppressor PTEN and induces
epithelial-mesenchymal transition in human nasopharyngeal
epithelial cells. J Clin Invest. 119:3626–3636. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim JH, Yoon SY, Kim CN, Joo JH, Moon SK,
Choe IS, Choe YK and Kim JW: The Bmi-1 oncoprotein is overexpressed
in human colorectal cancer and correlates with the reduced
p16INK4a/p14ARF proteins. Cancer Lett. 203:217–224. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Silva J, Garcia JM, Peña C, García V,
Domínguez G, Suárez D, Camacho FI, Espinosa R, Provencio M, España
P and Bonilla F: Implication of polycomb members Bmi-1, Mel-18 and
Hpc-2 in the regulation of p16INK4a, p14ARF, h-TERT, and c-Myc
expression in primary breast carcinomas. Clin Cancer Res.
12:6929–6936. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feng X, Wang Z, Fillmore R and Xi Y:
MiR-200, a new star miRNA in human cancer. Cancer Lett.
344:166–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mongroo PS and Rustgi AK: The role of the
miR-200 family in epithelial-mesenchymal transition. Cancer Biol
Ther. 10:219–222. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Korpal M and Kang Y: The emerging role of
miR-200 family of microRNAs in epithelial-mesenchymal transition
and cancer metastasis. RNA Biol. 5:115–119. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang X, Chen X, Wang R, Xiao P, Xu Z, Chen
L, Hang W, Ruan A, Yang H and Zhang X: MicroRNA-200c modulates the
epithelial-to-mesenchymal transition in human renal cell carcinoma
metastasis. Oncol Rep. 30:643–650. 2013.PubMed/NCBI
|
|
19
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stadler WM: Maturing of renal cancer
therapeutics. J Clin Oncol. 32:722–724. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang H, Fang F, Chang R and Yang L:
MicroRNA-140-5p suppresses tumor growth and metastasis by targeting
transforming growth factor β receptor 1 and fibroblast growth
factor 9 in hepatocellular carcinoma. Hepatology. 58:205–217. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wiklund ED, Bramsen JB, Hulf T, Dyrskjøt
L, Ramanathan R, Hansen TB, Villadsen SB, Gao S, Ostenfeld MS,
Borre M, et al: Coordinated epigenetic repression of the miR-200
family and miR-205 in invasive bladder cancer. Int J Cancer.
128:1327–1334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Castro-Vega LJ, Jouravleva K, Liu WY,
Martinez C, Gestraud P, Hupé P, Servant N, Albaud B, Gentien D, Gad
S, et al: Telomere crisis in kidney epithelial cells promotes the
acquisition of a microRNA signature retrieved in aggressive renal
cell carcinomas. Carcinogenesis. 34:1173–1180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nakada C, Matsuura K, Tsukamoto Y,
Tanigawa M, Yoshimoto T, Narimatsu T, Nguyen LT, Hijiya N, Uchida
T, Sato F, et al: Genome-wide microRNA expression profiling in
renal cell carcinoma: Significant down-regulation of miR-141 and
miR-200c. J Pathol. 216:418–427. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kong D, Li Y, Wang Z, Banerjee S, Ahmad A,
Kim HR and Sarkar FH: MiR-200 regulates PDGFDmediated
epithelial-mesenchymal transition, adhesion, and invasion of
prostate cancer cells. Stem Cells. 27:1712–1721. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kong D, Banerjee S, Ahmad A, Li Y, Wang Z,
Sethi S and Sarkar FH: Epithelial to mesenchymal transition is
mechanistically linked with stem cell signatures in prostate cancer
cells. PloS One. 5:e124452010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu J, Ohuchida K, Mizumoto K, Sato N,
Kayashima T, Fujita H, Nakata K and Tanaka M: MicroRNA,
hsa-miR-200c, is an independent prognostic factor in pancreatic
cancer and its upregulation inhibits pancreatic cancer invasion but
increases cell proliferation. Mol Cancer. 9:1692010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shimono Y, Zabala M, Cho RW, Lobo N,
Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al:
Downregulation of miRNA-200c links breast cancer stem cells with
normal stem cells. Cell. 138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu X, Jiang X, Li H, Guo L, Jiang W and Lu
SH: MiR-203 inhibits the proliferation and self-renewal of
esophageal cancer stem-like cells by suppressing stem renewal
factor Bmi-1. Stem Cells Dev. 23:576–585. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feber A, Clark J, Goodwin G, Dodson AR,
Smith PH, Fletcher A, Edwards S, Flohr P, Falconer A, Roe T, et al:
Amplification and overexpression of E2F3 in human bladder cancer.
Oncogene. 23:1627–1630. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Foster CS, Falconer A, Dodson AR, Norman
AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar
S, et al: Transcription factor E2F3 overexpressed in prostate
cancer independently predicts clinical outcome. Oncogene.
23:5871–5879. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lukas J, Parry D, Aagaard L, Mann DJ,
Bartkova J, Strauss M, Peters G and Bartek J:
Retinoblastoma-protein-dependent cell-cycle inhibition by the
tumour suppressor p16. Nature. 375:503–536. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Molofsky AV, He S, Bydon M, Morrison SJ
and Pardal R: Bmi-1 promotes neural stem cell self-renewal and
neural development but not mouse growth and survival by repressing
the p16Ink4a and p19Arf senescence pathways. Genes Dev.
19:1432–1437. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Y, Xiong Y and Yarbrough WG: ARF
promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus
deletion impairs both the Rb and p53 tumor suppression pathways.
Cell. 92:725–734. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nilsson K and Landberg G: Subcellular
localization, modification and protein complex formation of the
cdk-inhibitor p16 in Rb-functional and Rb-inactivated tumor cells.
Int J Cancer. 118:1120–1125. 2006. View Article : Google Scholar : PubMed/NCBI
|