Introduction
Dysfunction of pancreatic beta cells in type II
diabetes is associated with high circulatory levels of free fatty
acids and glucose. Previous results have demonstrated that levels
of free fatty acids are higher in diabetic patients than in healthy
individuals (1). High levels of
saturated fatty acids, including palmitate, lauric acid and
linoleic acids, activate signaling from toll like receptors (TLRs)
and their downstream Jun N-terminal kinase (JNK) and nuclear
factor-κB (NF-κB) pathways, thus promoting the release of
interleukin (IL)-6, tumor necrosis factor (TNF)-α and other
inflammatory cytokines from macrophages (2,3). An
increase in fat mass associated with an inflammatory response may
be involved in the development of insulin resistance, as indicated
by a previous study in obese mice, where it was observed that an
increased mass of adipose tissue led to macrophage infiltration of
adipose tissue (4). Higher levels of
circulating free fatty acids and inflammatory cytokines were also
observed, which may then act on skeletal muscle cells, adipocytes
and other cells to influence insulin signaling (5,6). In
muscle cells, the conditioned media from palmitate-stimulated
macrophages has been demonstrated to activate protein kinase C-θ
and -ε to promote inflammatory cytokine expression, thus
contributing to insulin resistance (7) Conditioned media from palmitate-treated
macrophages may also provoke insulin resistance in muscle cells at
the levels of Akt signaling, glucose transporter type 4
translocation and glucose uptake (8). Furthermore, it has been observed in a
co-culture of hepatocytes and macrophages that
lipopolysaccharide-induced TNF-α factor (LITAF) was involved in
macrophage infiltration of hepatocytes, with knockdown of LITAF
leading to improvement of insulin resistance in human hepatocytes
(9). Similarly, prolonged exposure
of beta cells to inflammatory cytokines may induce excessive
production of reactive oxygen species and endoplasmic reticulum
stress, leading to cell apoptosis and a decrease in insulin
secretion (10,11). Collectively, these data suggest that
insulin signaling and beta cell function may be altered by
inflammatory processes. However, the underlying mechanism for the
effects of macrophage-derived conditioned media on beta cell
function remains unknown.
Receptor interacting protein 140 (RIP140) was first
identified as a co-repressor of the estrogen receptor (12), with high expression of RIP140
observed in reproductive (13) and
metabolic tissues, including adipose, liver and muscular tissue
(14). RIP140 is now an established
co-repressor of various nuclear receptors, including the thyroid
receptor, peroxisome proliferator activated receptor (PPAR)
(15), liver X receptor (LXR)-α
(16), glucocorticoid receptor
(17) and retinoic acid receptor
(18), whereby it regulates the
transcription of genes involved in glucose and lipid metabolism
(16). In some instances, RIP140
also acts as a co-activator of nuclear receptors, by stimulating
histone acetylase and cyclic adenosine monophosphate-responsive
element binding protein (CBP), as the co-activator of NF-κB, thus
promoting pro-inflammatory cytokine transcription (19). Intracellular cholesterol and a high
fatty diet may increase the level of RIP140 in macrophages, whereas
microRNA miR-33 reduces the inflammatory activity of macrophages by
inhibiting RIP140 co-activation of NF-κB (20). It has been demonstrated that
macrophage treatment with a low dose of lipopolysaccharide (LPS)
stimulated phosphorylation of RIP140 by spleen tyrosine kinase,
thus stimulating RIP140 interaction with the NF-kB subunit RelA. In
turn, this triggered the degradation of RIP140, resulting in
decreased transcription of cytokine genes and a reduced
inflammatory response (21).
Additionally, mice with RIP140 knockdown in macrophages exhibit a
decreased population of circulating monocytes, along with a
reduction in inflammatory M1 (pro-inflammatory phenotype) and
expansion in M2 (anti-inflammatory phenotype) macrophages, which
subsequently improved insulin resistance induced by a high-fat diet
(22).
Co-activation of NF-κB by RIP140 mediates the
release of inflammatory cytokines. However, under lipotoxic
conditions, the influence of RIP140 on the infiltration of
pancreatic beta cells by macrophages and the subsequent effects on
beta cell function remain unknown. In the present study,
conditioned media from palmitate-stimulated macrophages was used to
stimulate beta cells and the chemotaxis of macrophages towards beta
cells was evaluated. It was determined that RIP140 promoted
inflammatory cytokine expression in macrophages under high
palmitate conditions and impaired beta cell function. In addition,
under lipotoxic conditions, RIP140 enhanced the chemotaxis of
macrophages towards pancreatic beta cells through the release of
inflammatory cytokines, thus triggering local low-grade
inflammation and impairing beta cell function.
Materials and methods
Materials and reagents
A total of 50 mM palmitate (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) was dissolved in 1 ml 99% ethanol and
mixed with 9 ml 10% bovine serum albumin (BSA; Sigma-Aldrich; Merck
KGaA), from which a 5 mM palmitate-10% BSA stock solution was
prepared, as previously described (23). Antibodies against phosphorylated
(p)-JNK (cat. no. 9255S), JNK (cat. no. 3708S), p-ERK1/2 (cat. no.
9106S) and ERK1/2 (cat. no. 4696S) were purchased from Cell
Signaling Technology, Inc. (Danvers, MA, USA). Antibodies against
RIP140 (cat. no. ab42125) and β-actin (cat. no. sc-130300) were
purchased from Abcam (Cambridge, MA, USA) and Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA), respectively. Goat
anti-rabbit immunoglobulin (Ig)-G (heavy and light chain) secondary
antibody conjugated to horseradish peroxidase (HRP; cat. no.
BS13278) was obtained from Bioworld Technology, Inc. (St. Louis
Park, MN, USA). ELISA kits for mouse TNF-α (cat. no. EK0527) and
IL-6 (cat. no. EK0411) were purchased from Wuhan Boster Biological
Technology, Ltd. (Wuhan, China).
Cell culture and treatment
Murine RAW264.7 macrophages (American Type Culture
Collection, Manassas, VA, USA) were maintained in RPMI 1640 medium
supplemented with 10% fetal bovine serum (FBS; Hyclone; GE
Healthcare, Logan, UT, USA) and 1% penicillin-streptomycin at 37°C
in a humidified atmosphere (5% CO2) for 24 h. The mouse
pancreatic beta cell line MIN6 (American Type Culture Collection)
was cultured overnight in Dulbecco's modified Eagle medium (DMEM;
Hyclone; GE Healthcare) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc. Waltham, MA, USA), 0.001%
beta-mercaptoethanol and 1% penicillin-streptomycin at 37°C in a
humidified atmosphere (5% CO2, 95% air). Macrophages
were transiently transfected with pEGFP-N1-RIP140 plasmid (or
pEGFP-N1 plasmid as a control) or RIP140 siRNA (or scramble siRNA
as a control) at 37°C for 24 h, then treated with 500 µM palmitate
at 37°C for 24 h. Supernatant was then collected in order to detect
the concentrations of TNF-α and IL-6 using the relevant ELISA kits,
according to the manufacturer's instructions. Macrophages were
washed several times with phosphate-buffered saline (PBS) and fresh
RPMI 1640 media with 10% FBS was added. Following 24 h incubation
at 37°C, palmitate-free conditioned media was collected,
centrifuged (10,000 × g, 5 min, room temperature) and added to MIN6
cells for a 24 h incubation, as previously described (7). MIN6 cells were then collected for cell
viability, gene expression and western blot analyses. For
chemotaxis assays, MIN6 cells were transiently transfected and
treated with palmitate as above and culture supernatant was
collected.
Plasmid construction and cell
transfection
A mouse RIP140 overexpression plasmid was
constructed, as previously described (24), for transfection into the RAW264.7
macrophages and MIN6 cells. Briefly, total DNA from RAW264.7
macrophages was extracted using a DNA extraction kit (cat. no.
DP304; Beijing Transgen Biotech Co., Ltd., Beijing, China) and a
fragment of RIP140 (Reference Sequence: NM 173440.2) was amplified
by polymerase chain reaction (PCR) using the following primers:
Forward, 5′-GACTCGAGGATCTTACTGACGAGAGGAGCT-3′, and reverse,
5′-ACGGTACCCAGGTACACATTTCTTCTGACTC-3′. A TransTaq High Fidelity DNA
polymerase was obtained from Beijing Transgen Biotech Co., Ltd.
(Beijing, China) and the cycling conditions were as follows: 4
cycles for 30 sec each at 94°C, 1 cycle for 30 sec at 58°C, 1 cycle
for 4 min at 72°C, 30 cycles for 30 sec each at 94°C, 1 cycle for
30 sec at 64°C and 4 min at 72°C. KpnI (GGTACC) and XhoI (CTCGAG)
restriction sites were included to facilitate ligation of the cDNA
product into a pEGFP-N1 plasmid (Biovector Science Lab, Inc.,
Beijing, China) using Kpn I, Xho I and T4 DNA ligase (all from
Fermentas Inc., Burlington, Canada), according to the
manufacturer's protocol. The sequence of pEGFP-N1-RIP140 was
verified by DNA sequencing. The sequences of RIP140 siRNA were as
follows: Forward, 5′-CGGCGUUGACAUCAAAGAAdTdT-3′ and reverse,
3′-dTdTGCCGCAACUGUAGUUUCUU-5′. Transient transfection was conducted
with Lipofectamine™ 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to manufacturer's protocol.
Lipofectamine 2000 and 250 µl optimum-minimal essential medium
(Opti-MEM; Gibco; Thermo Fisher Scientific, Inc.) were mixed,
incubated at room temperature for 5 min, then 5 µg plasmid or 200
pmol/l siRNA was added to a separate 250 µl Opti-MEM. The two
solutions were mixed and incubated for 20 min at room temperature,
then added to serum-free medium-cultured RAW264.7 cells. The
mixture was replaced with fresh medium after 6 h incubation at
37°C.
Total RNA extraction and reverse
transcription (RT)-PCR
Total RNA was extracted with a Spin Column RNA
Extraction kit (Sangon Biotech Co., Ltd., Shanghai, China) and
transcribed to cDNA using Moloney Murine Leukemia Virus reverse
transcriptase (Promega Corporation, Madison, WI, USA). PCR for
RIP140 was performed using a SYBR-Green PCR Master Mix kit (Takara
Bio, Inc., Otsu, Japan), according to the manufacturer's protocol.
The cycling conditions were as follows: An initial denaturation
step at 95°C for 5 min, followed by 40 PCR cycles for 5 sec each at
95°C, 1 cycle for 30 sec at 58°C and 1 cycle for 30 sec at 72°C.
Semiquantitative RT-PCR was performed using Taq DNA polymerase
(Takara Bio, Inc.) for comparison of the cDNAs developed, under the
following conditions: An initial denaturation at 95°C for 5 min,
followed by 30 PCR cycles for 5 sec each at 95°C, 1 cycle for 30
sec at 56°C and 1 cycle for 30 sec at 72°C. Semiquantification was
performed using mouse β-actin as an internal standard. Levels of
target bands in the agar gel were analyzed by AlphaEaseFC 4.0
software (Alpha Innotech Corporation; ProteinSimple, San Jose, CA,
USA), The primers of all genes quantified are listed in Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Genes (accession
no.) | Primer sequences
(5′ to 3′) |
|---|
| RIP140 (NM
73440.2) |
|
|
Forward |
GGCAGCAAACCTGAATTCGGC |
|
Reverse |
CTCACCGGGCACGGCACATC |
| β-actin (NM
0007393.3) |
|
|
Forward |
TCTACAATGAGCTGCGTGTG |
|
Reverse |
GGTGAGGATCTTCATGAGGT |
| TNF-α (NM
013693.2) |
|
|
Forward |
CCACCACGCTCTTCTGTCTAC |
|
Reverse |
TGGGCTACAGGCTTGTCACT |
| IL-6 (NM
031168.1) |
|
|
Forward |
CCACTTCACAAGTCGGAGGCTTA |
|
Reverse |
GCAAGTGCATCATCGTTGTTCATAC |
| PGC-1α (NM
008904.2) |
|
|
Forward |
GTGTTCCCGATCACCATATTCC |
|
Reverse |
CGGTGTCTGTAGTGGCTTGATTC |
| PEPCK (NM
011044.2) |
|
|
Forward |
ATCTTTGGTGGCCGTAGACCT |
|
Reverse |
GCCAGTGGGCCAGGTATTT |
| PDX-1 (NM
008814.3) |
|
|
Forward |
GATGAAATCCACCAAAGCTCA |
|
Reverse |
AGAATTCCTTCTCCAGCTCCA |
| PCNA (NM
011045.2) |
|
|
Forward |
TCGACACATACCGCTGCGACC |
|
Reverse |
GCAAATTCACCCGACGGCATC |
| UCP-2 (NM
011671.4) |
|
|
Forward |
GCATTGGCCTCTACGACTCT |
|
Reverse |
CTGGAAGCGGACCTTTACC |
| iNOS1 (NM
010927.3) |
|
|
Forward |
AATGGCAACATCAGGTCGGCCATCACT |
|
Reverse |
GCTGTGTGTCACAGAAGTCTCGAACTC |
Western blotting
MIN6 cells were washed three times with PBS and
lysed on ice in radioimmunoprecipitation assay buffer containing 50
mM Tris hydrochloride (pH 7.4), 150 mM sodium chloride (NaCl), 1%
NP-40 lysis buffer, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride
and 1 mM phosphatase inhibitor (Applygen Technologies Inc, Beijing,
China) for 30 min, then centrifuged at 10,000 × g at 4°C for 10
min. The supernatant was collected and protein concentration was
determined using a BCA Protein Assay kit (Applygen Technologies,
Inc., Beijing, China), according to the manufacturer's protocol.
SDS-PAGE, trans-blotting and subsequent immunodetection were
performed, as previously described (25). Briefly, blots were incubated with
primary antibodies (anti-RIP40, 1:1,000; anti-p-JNK, 1:250; and
anti-JNK, anti-p-ERK and anti-ERK, 1:500) at 4°C overnight, then
with HRP-conjugated secondary IgG (1:5,000) at 37 °C for 1 h.
β-actin was used as an internal control. Levels of target protein
within bands were determined using AlphaEaseFC 4.0 software.
Cell viability
MIN6 cells (5×103 cells/per well) were
cultured in 96 well plates in complete DMEM at 37°C. After 24 h of
culture, cells were treated with 100 µl conditioned media from the
following RAW264.7 cells: i) Treated with BSA, ii) treated with 500
µM palmitate, iii) transfected with pEGFP-N1 control plasmid, iv)
transfected with pEGFP-N1-RIP140 plasmid, v) transfected with
scramble siRNA and vi) transfected with RIP140 siRNA. Groups 3–6
were treated with 500 µM palmitate for 24 h post-transfection. Cell
viability was then measured by a
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. Specifically, 5 mg/ml MTT solution (Sigma-Aldrich; Merck
KGaA) was added to each well prior to 4 h incubation at 37°C. The
resulting formazan precipitate was dissolved by adding 100 µl
dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) for 10 min at room
temperature. Color intensity was then detected at 570 nm by a
microplate reader (Thermo Labsystems, Beverly, MA, USA). Three
independent experiments were performed.
Glucose-stimulated insulin secretion
assay
MIN6 cells were exposed to the conditioned media of
RAW264.7 cells for 24 h. MIN6 cells were then washed twice with
PBS, incubated in Krebs-Ringer bicarbonate
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer
(KRBH buffer: 115 mM NaCl, 24 mM sodium bicarbonate, 5 mM potassium
chloride, 1 mM magnesium chloride, 25 mM HEPES, 0.5% BSA, pH 7.4)
with 2.8 mM glucose for 40 min. Supernatant was then collected for
measurement of basal insulin secretion (BIS). Fresh KRBH buffer
containing 25 mM glucose was added to MIN6 cells and cells were
incubated at 37°C for 60 min. Supernatant was then collected to
measure glucose-stimulated insulin secretion (GSIS). Insulin
concentrations were measured with an ELISA kit (cat. no.
CSB-E05071m; Cusabio Biotech Co., Ltd., Wuhan China), according to
the manufacturer's protocol. The experiment was performed three
times.
Chemotaxis assays
Chemotaxis assays were performed on Transwell plates
(24-well format, 8 µm; Corning Incorporated, Corning, NY, USA). A
total of 500 µl conditioned media from MIN6 cells was added to the
lower chamber. RAW264.7 cells were washed with PBS three times and
suspended in RPMI 1640 medium containing 0.5% FBS, then seeded into
the upper chamber (104 cells in total). Chambers were
incubated for 6 h at 37°C. Cells that remained in the upper chamber
were removed while cells in the lower chamber were fixed with 4%
paraformaldehyde for 20 min at room temperature, followed by
incubation with 1% crystal violet (Sigma-Aldrich; Merck KGaA) for
10 min at room temperature. Stained cells were manually counted
under a light microscope in 6 randomly selected fields. The
experiment was performed three times.
Chemotaxis of murine peritoneal
macrophages was also measured
Peritoneal macrophages were isolated from 3 male
c57BL/6 mice obtained from the Animal Center of Wuhan University,
Wuhan, China (mean age, 5.0±0.5 weeks; mean weight, 19.0±0.5 g).
The research protocol was approved by Medical Ethical Committee of
Zhongnan Hospital (Wuhan, China). The mice were sacrificed by
cervical dislocation and soaked in 75% ethyl alcohol for 5 min,
injected intraperitoneally with 5 ml serum-free RPMI 1640 medium,
then placed in a supine position for 5 min. The abdominal cavity
was opened and 5 ml peritoneal fluid was aspirated and centrifuged
at 1,000 × g at 4°C for 5 min. Isolated cells were maintained in
RPMI1640 medium supplemented with 10% FBS at 37°C for 24 h. After
adhering to the culture dishes, macrophages were harvested and
added to the upper Transwell chamber and conditioned media from
MIN6 cells was added to the lower chamber. Chambers were incubated
for 6 h at 37°C and chemotaxis assays were performed as before.
Statistical analysis
Data are presented as the mean ± standard deviation.
When two groups were being compared, differences between mean
values were determined by Independent Sample t tests. One-way
analysis of variance followed by a Turkey test was used to
determine differences between the mean values of >2 groups.
Statistical analysis was performed using SPSS 17.0 (SPSS, Inc.
Chicago, IL, USA) and P<0.05 was considered to indicate a
statistically significant difference.
Results
TNF-α and IL-6 levels are elevated by
RIP140 in palmitate-treated macrophages
Expression of RIP140, TNF-α and IL-6 at the mRNA and
protein level in palmitate-treated RAW264.7 macrophages was
determined by semiquantitative RT-PCR and ELISA, respectively. It
was observed that the level of RIP140 mRNA expression in RAW264.7
cells was significantly increased by 500 µM palmitate, compared
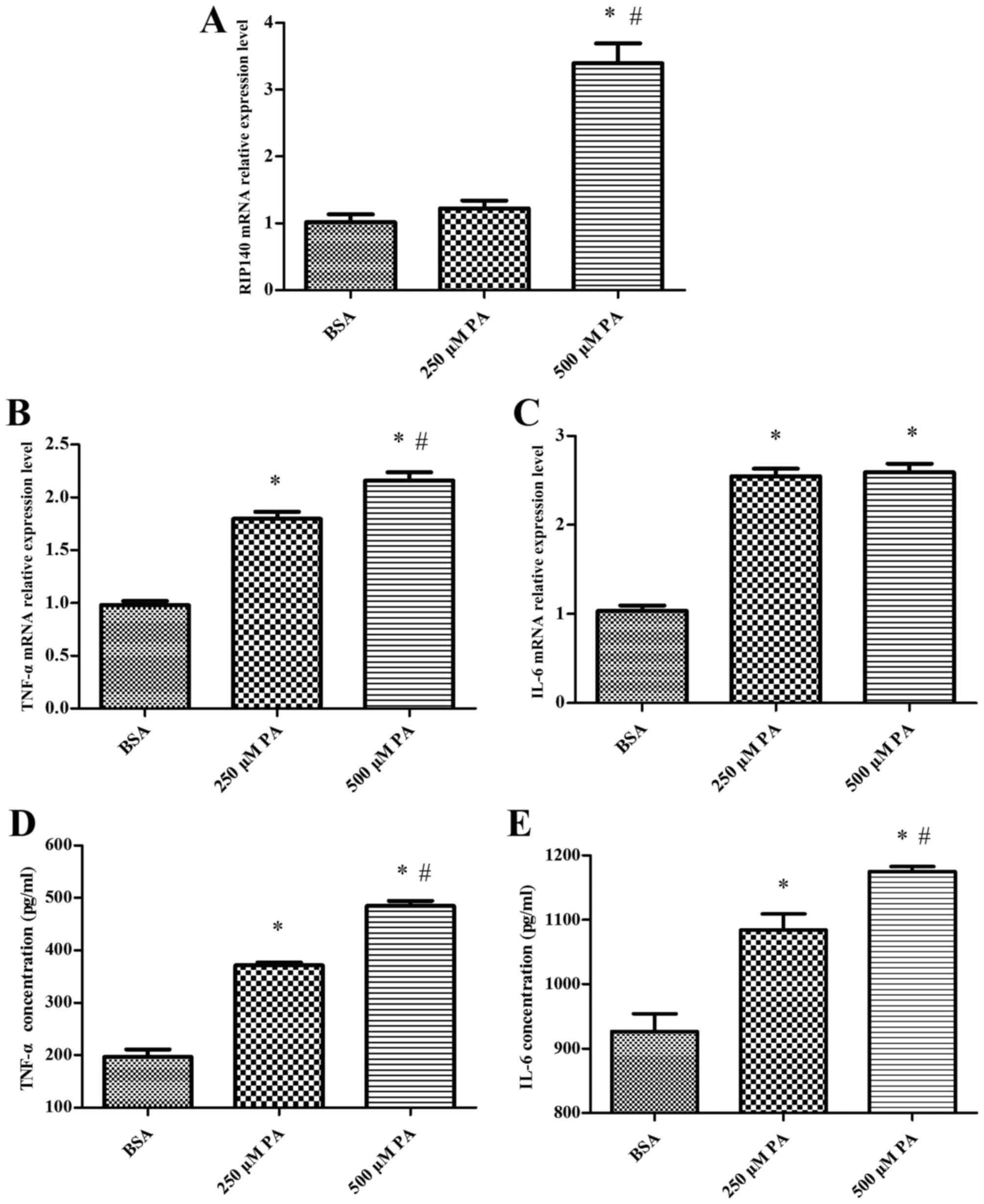
with control cells treated with BSA alone (P<0.05; Fig. 1A). Expression of the inflammatory
cytokines TNF-α and IL-6 was also significantly increased at the
mRNA and protein levels by 250 µM and 500 µM palmitate, relative to
control cells. Relative to 250 µM palmitate, expression of TNF-α
and IL-6 was also significantly increased at the mRNA and protein
levels by 500 µM palmitate (P<0.05; Fig. 1B-E). The level of RIP140 expression
was subsequently regulated by transfection of RAW264.7 macrophages
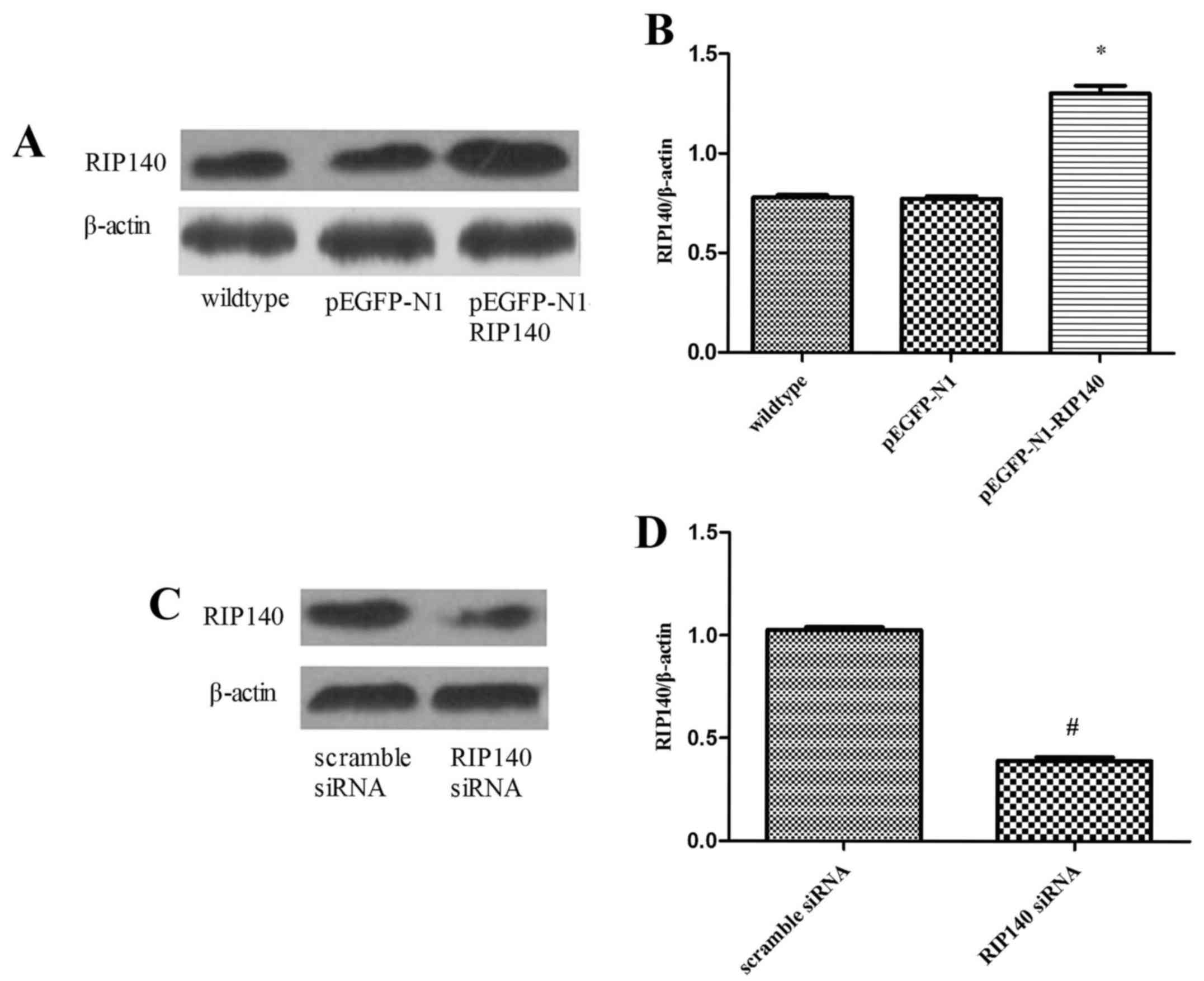
with RIP140 plasmid or siRNA. As demonstrated by western blot
analysis, the level of RIP140 protein expression was significantly
increased in cells transfected with pEGFP-N1-RIP140 plasmid,
relative to wild type control cells (P<0.05; Fig. 2A and B). In addition, transfection
with RIP140 siRNA significantly decreased RIP140 mRNA, relative to
control cells transfected with scramble siRNA (P<0.05; Fig. 2C and D). The influence of these
transfections on inflammatory cytokine expression in
palmitate-treated macrophages was subsequently evaluated. In
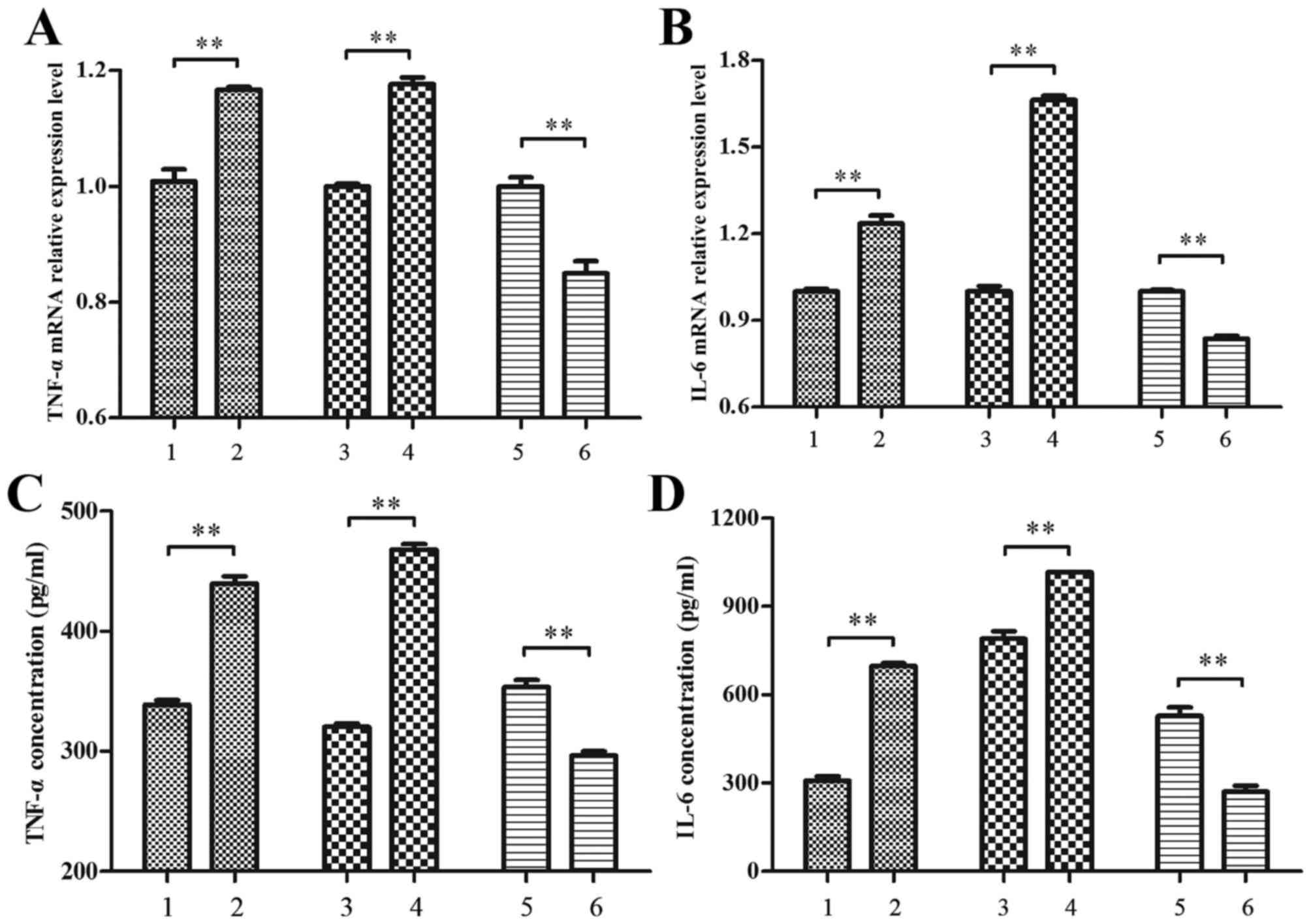
RAW264.7 macrophages treated with 500 µM palmitate, it was observed
that the expression of TNF-α and IL-6 were significantly increased
at the mRNA and protein levels by the RIP140 overexpression
plasmid, relative to control cells transfected with pEGFP-N1
plasmid alone (P<0.01; Fig. 3).
In addition, TNF-α and IL-6 expression was significantly decreased
by RIP140 siRNA, relative to scramble siRNA control cells
(P<0.01; Fig. 3). These data
suggest that RIP140 promotes TNF-α and IL-6 expression in
macrophages in high palmitate conditions.
Conditioned media from macrophages
modifies cell viability
Conditioned media collected from palmitate-treated
RAW264.7 macrophages (500 µM palmitate) was added to MIN6
pancreatic beta cells and cell viability was measured by an MTT
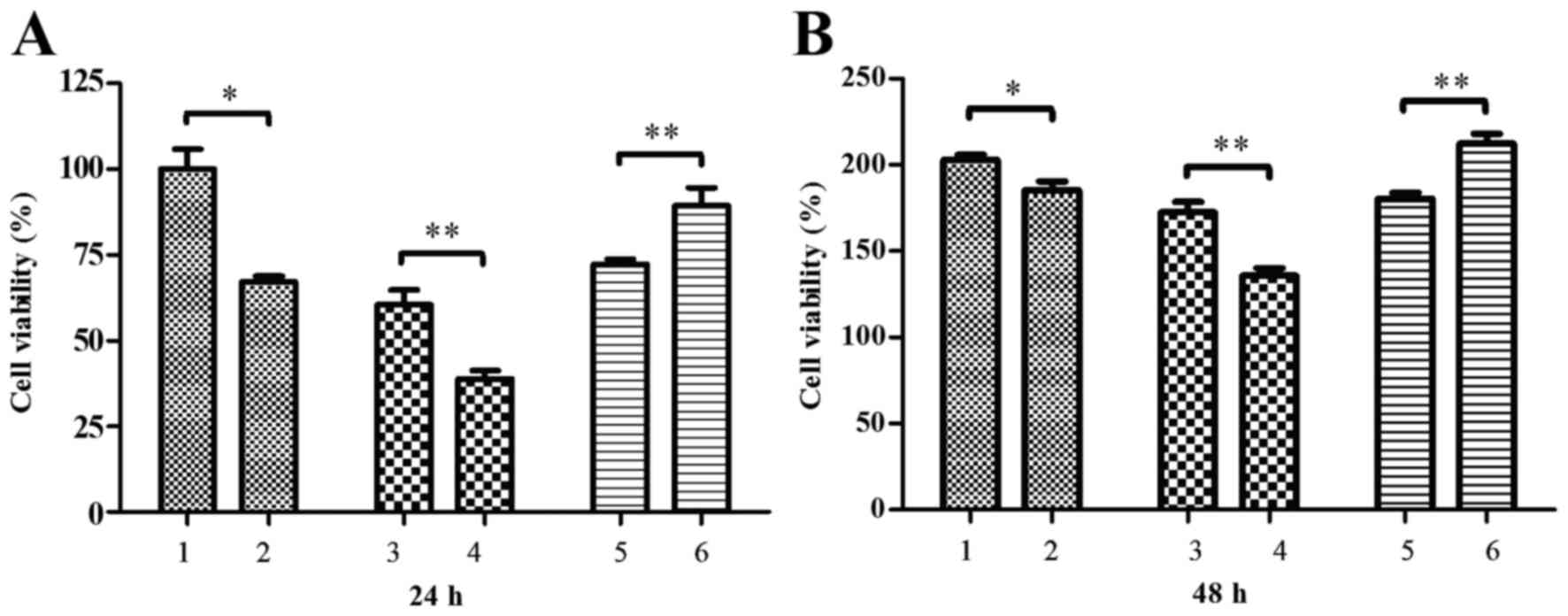
assay. It was observed that cell viability was significantly
reduced by conditioned media from palmitate-treated RAW264.7 cells
at 24 and 48 h post-media treatment, relative to media from control
BSA-treated RAW264.7 cells (P<0.05; Fig. 4). It was also demonstrated that the
viability of MIN6 cells was significantly decreased by conditioned
media from palmitate-treated RAW264.7 cells transfected with RIP140
overexpression plasmid at 24 and 48 h post-treatment, relative to
media of palmitate-treated RAW264.7 cells transfected with pEGFP-N1
plasmid alone (P<0.01; Fig. 4).
By contrast, conditioned media of palmitate-treated RAW264.7 cells
transfected with RIP140 siRNA significantly increased MIN6 cell
viability at 24 and 48 h post-media treatment, relative to media
from control palmitate-treated RAW264.7 cells transfected with
scramble siRNA (P<0.01; Fig.
4).
Conditioned media modifies insulin
secretion in MIN6 cells
Levels of GSIS and BIS in MIN6 cells treated with
conditioned media from RAW264.7 macrophages prior to palmitate
treatment were measured by ELISA. As depicted in Fig. 5, it was observed that GSIS was
significantly decreased by conditioned media from palmitate-treated
RAW264.7 cells, relative to the media of control RAW264.7 cells
(P<0.01; Fig. 5A). However, the
insulin secretion index (ISI; defined as GSIS/BIS) did not differ
significantly (P=0.121; Fig. 5B).
This suggests that pancreatic beta cells may increase insulin
secretion with short-term increases in levels of free fatty acids
by a compensatory mechanism.
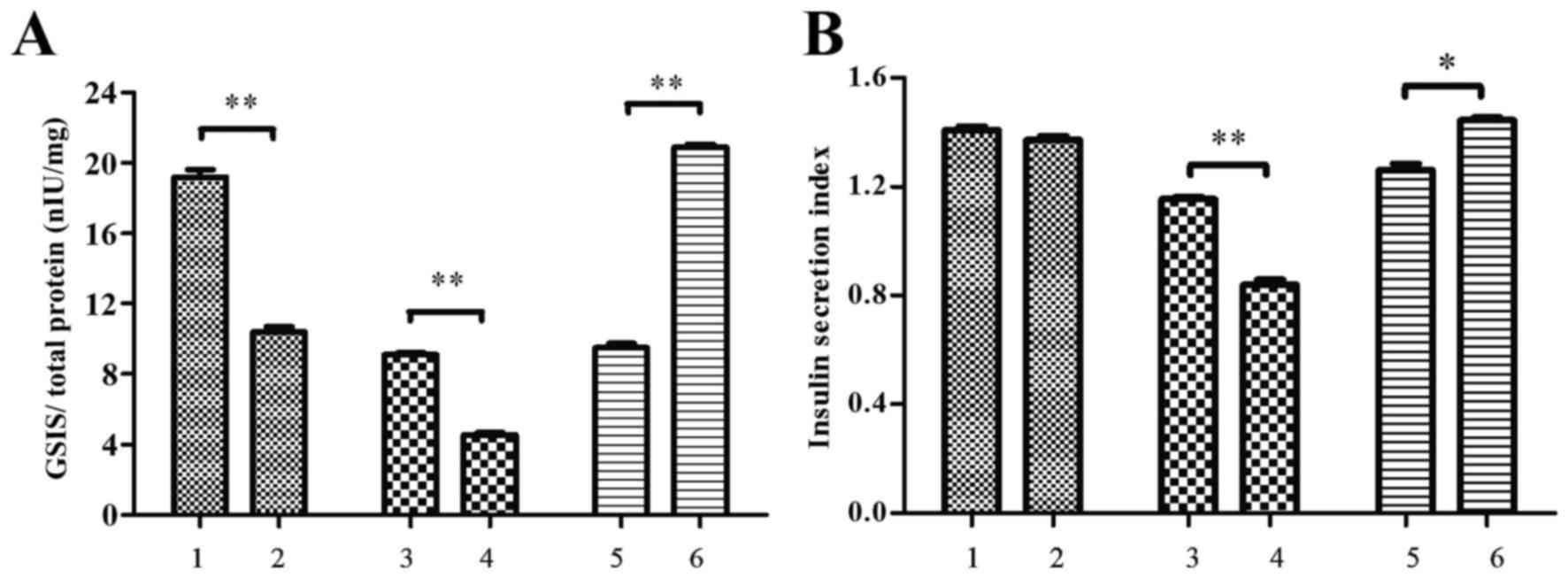 | Figure 5.Conditioned media altered insulin
secretion in MIN6 cells. MIN6 cells were incubated with conditioned
media from RAW264.7 macrophage cultures and the level of insulin
secretion (nIU/ml) in response to 2.8 and 25 mM glucose (BIS and
GSIS, respectively) was determined by ELISA. Total protein
concentration (mg/ml) was used as a control. The ISI was derived as
GSIS/BIS. Measurements of (A) GSIS and (B) ISI are shown. MIN6
cells were incubated with conditioned media obtained from the
following: i) RAW264.7 cells treated with bovine serum albumin, ii)
RAW264.7 cells treated with 500 µM palmitate, iii) RAW264.7 cells
transfected with pEGFP-N1 control plasmid, iv) RAW264.7 cells
transfected with pEGFP-N1-RIP140, v) RAW264.7 cells transfected
with scramble siRNA and vi) RAW264.7 cells transfected with RIP140
siRNA. Groups 3–6 were treated with 500 µM palmitate
post-transfection. Data are presented as the mean ± standard
deviation of three independent experiments. *P<0.05,
**P<0.01. MIN6 cells, murine pancreatic beta cell line; BIS,
basal insulin secretion; GSIS, glucose-stimulated insulin
secretion; ISI, insulin secretion index; siRNA, small interfering
RNA. |
It was also demonstrated that GSIS and ISI in MIN6
cells were significantly decreased by conditioned media from
RAW264.7 cells transfected with RIP140 overexpression plasmid,
relative to media of RAW264.7 cells transfected with pEGFP-N1
plasmid alone (P<0.01; Fig. 5).
By contrast, GSIS and ISI were significantly increased by
conditioned media from RIP140-knockdown RAW264.7 cells (P<0.01
and P<0.05, respectively), relative to media of RAW264.7 cells
transfected with scramble siRNA (Fig.
5).
Conditioned media modifies specific
gene expression in MIN6 cells
To identify the effects of the conditioned media
from palmitate-treated RAW264.7 macrophages on pancreatic beta cell
function, the levels of mRNA for oxidative stress-related genes,
namely PPARγ coactivator 1α (PGC-1α), uncoupling protein 2 (UCP-2),
nitric-oxide synthase 1 (iNOS1), insulin secretion related gene
pancreatic and duodenal homeobox 1 (PDX-1), hepatic
gluconeogenesis-related phosphoenulpyruvate carboxykinase (PEPCK)
and proliferating cell nuclear antigen (PCNA) were determined by
semiquantitative RT-PCR. It was observed that treatment with
conditioned media from palmitate-treated RAW264.7 cells
overexpressing RIP140 led to a significant downregulation in the
levels of PGC-1α (P<0.05), PEPCK (P<0.05) and PCNA
(P<0.01) mRNA in MIN6 cells, while significant upregulation in
the mRNA of UCP-2 (P<0.01), iNOS1 (P<0.01) and PDX-1
(P<0.05) was observed. However, in MIN6 cells treated with media
from palmitate-treated, RIP140-knockdown RAW264.7 cells,
significant decreases were observed in the levels of PGC-1α
(P<0.05), PEPCK (P<0.05) and PCNA (P<0.01) mRNA were
significantly increased, while levels of UCP-2 (P<0.01), iNOS1
(P<0.05) and PDX-1 (P<0.05) mRNA (Fig. 6). These results suggest that
conditioned media from palmitate-treated macrophages influences the
transcription of genes associated with oxidative stress, insulin
secretion and glucose metabolism in pancreatic beta cells.
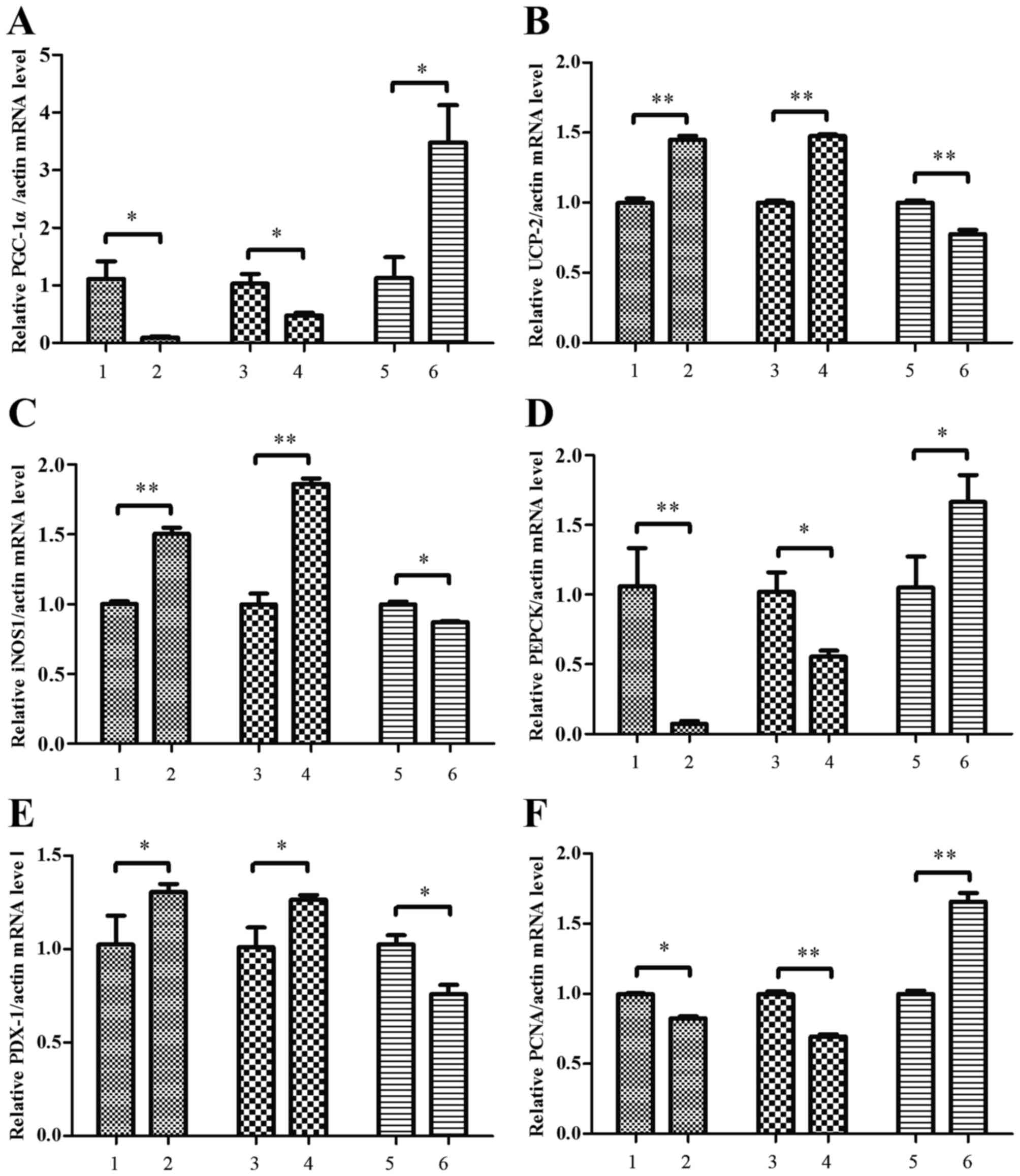 | Figure 6.Conditioned media altered the
expression of oxidative-stress related genes in MIN6 cells. MIN6
cells were treated with conditioned media from RAW264.7 macrophage
cultures and the levels of (A) PGC-1α, (B) UCP-2, (C) iNOS-1, (D)
PEPCK, (E) PDX-1 and (F) PCNA mRNA expression in MIN6 cells were
determined by semiquantitative reverse transcription polymerase
chain reaction. MIN6 cells were incubated with conditioned media
obtained from the following: i) RAW264.7 cells treated with bovine
serum albumin, ii) RAW264.7 cells treated with 500 µM palmitate,
iii) RAW264.7 cells transfected with pEGFP-N1 control plasmid, iv)
RAW264.7 cells transfected with pEGFP-N1-RIP140 plasmid, v)
RAW264.7 cells transfected with scramble siRNA and vi) RAW264.7
cells transfected with RIP140 siRNA. Groups 3–6 were treated with
500 µM palmitate post-transfection. Data are presented as the mean
± standard deviation of three independent experiments. *P<0.05,
**P<0.01. MIN6, murine pancreatic beta cell line; PPARγ
coactivator 1α; UCP-2, uncoupling protein 2; iNOS1, nitric-oxide
synthase 1; PEPCK, phosphoenulpyruvate carboxykinase; PDX-1,
pancreatic and duodenal homeobox 1; PCNA, proliferating cell
nuclear antigen; siRNA, small interfering RNA. |
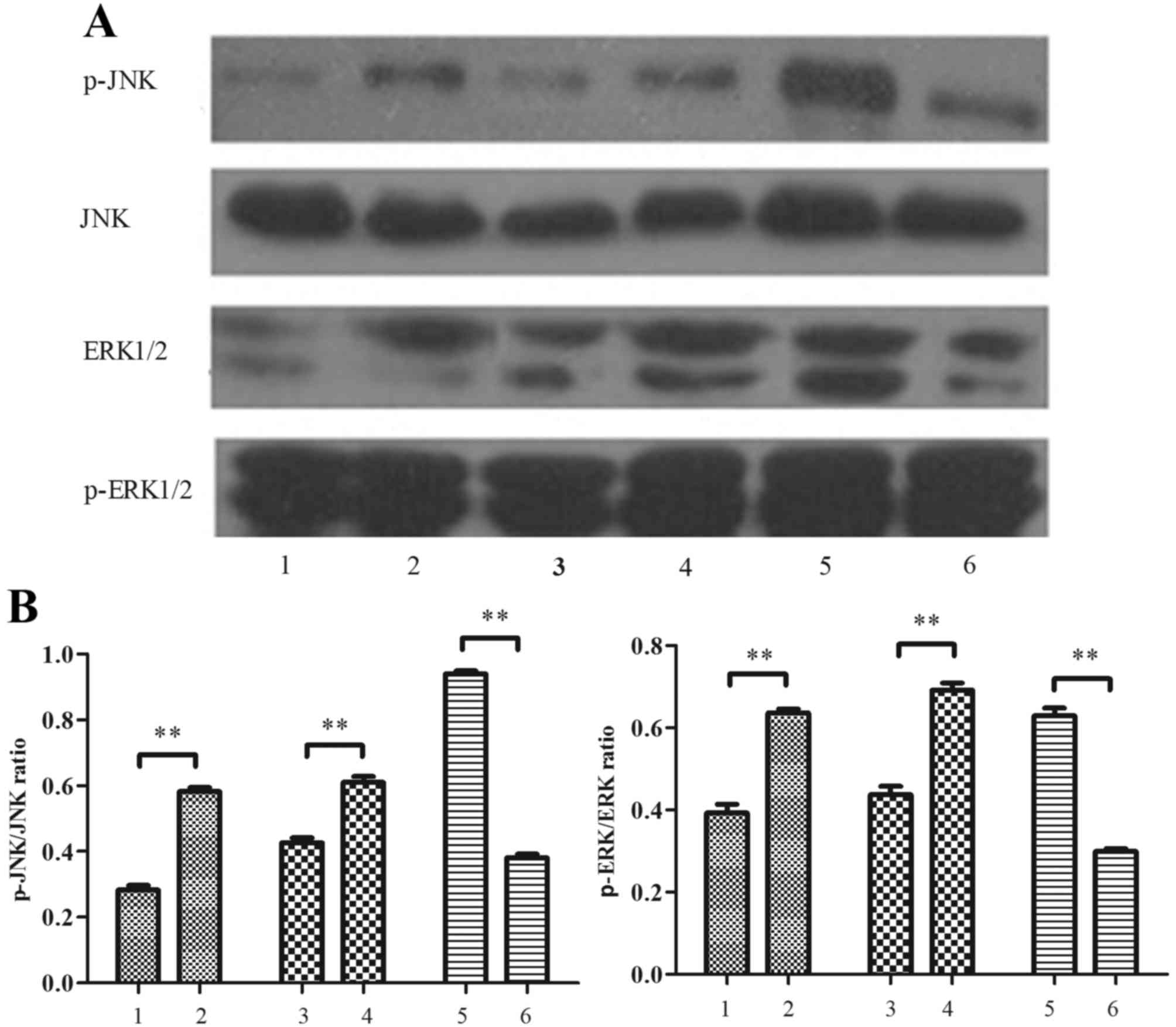
Conditioned media modifies JNK and
ERK1/2 phosphorylation in MIN6 cells
Levels of p-JNK and p-ERK1/2 in MIN6 cells treated
with conditioned media were measured by western blotting. As
depicted in Fig. 7, it was
demonstrated that conditioned media from palmitate-treated
macrophages stimulated a significant increase in JNK and ERK1/2
phosphorylation, relative to conditioned media from control
RAW264.7 macrophages (P<0.01). It was also demonstrated that
levels of p-JNK and p-ERK 1/2 were significantly increased in MIN6
cells by the conditioned media of palmitate-treated RAW264.7 cells
transfected with RIP140 overexpression plasmid, relative to the
media of RAW264.7 cells transfected with pEGFP-N1 plasmid alone
(P<0.01; Fig. 7). By contrast,
levels of p-JNK and p-ERK 1/2 were significantly decreased by the
conditioned media of palmitate-treated, RIP140-knockdown RAW264.7
cells, relative to the media of control RAW264.7 cells transfected
with scramble siRNA (P<0.01; Fig.
7).
Macrophages chemotax towards MIN6
cells
Chemotaxis of murine macrophages (RAW264.7 and
peritoneal) towards MIN6 cells following treatment with conditioned
media from palmitate-treated MIN6 cells (500 µM palmitate), was
determined by Transwell assays (Fig. 8A
and B). For RAW264.7 macrophages, it was observed that cell
chemotaxis towards MIN6 cells was significantly increased by
conditioned media from palmitate-treated MIN6 cells, relative to
media from control BSA-treated MIN6 cells (P<0.01; Fig. 8C). It was also demonstrated that
RAW264.7 cell chemotaxis was significantly increased by conditioned
media from palmitate-treated MIN6 cells transfected with RIP140
overexpression plasmid, relative to media from palmitate-treated
MIN6 cells transfected with pEGFP-N1 plasmid alone (P<0.01;
Fig. 8C). By contrast, RAW264.7 cell
chemotaxis was significantly decreased by the conditioned media of
palmitate-treated, RIP140 siRNA-knockdown MIN6 cells, relative to
the media of palmitate-treated MIN6 cells transfected with scramble
siRNA (P<0.01; Fig. 8C).
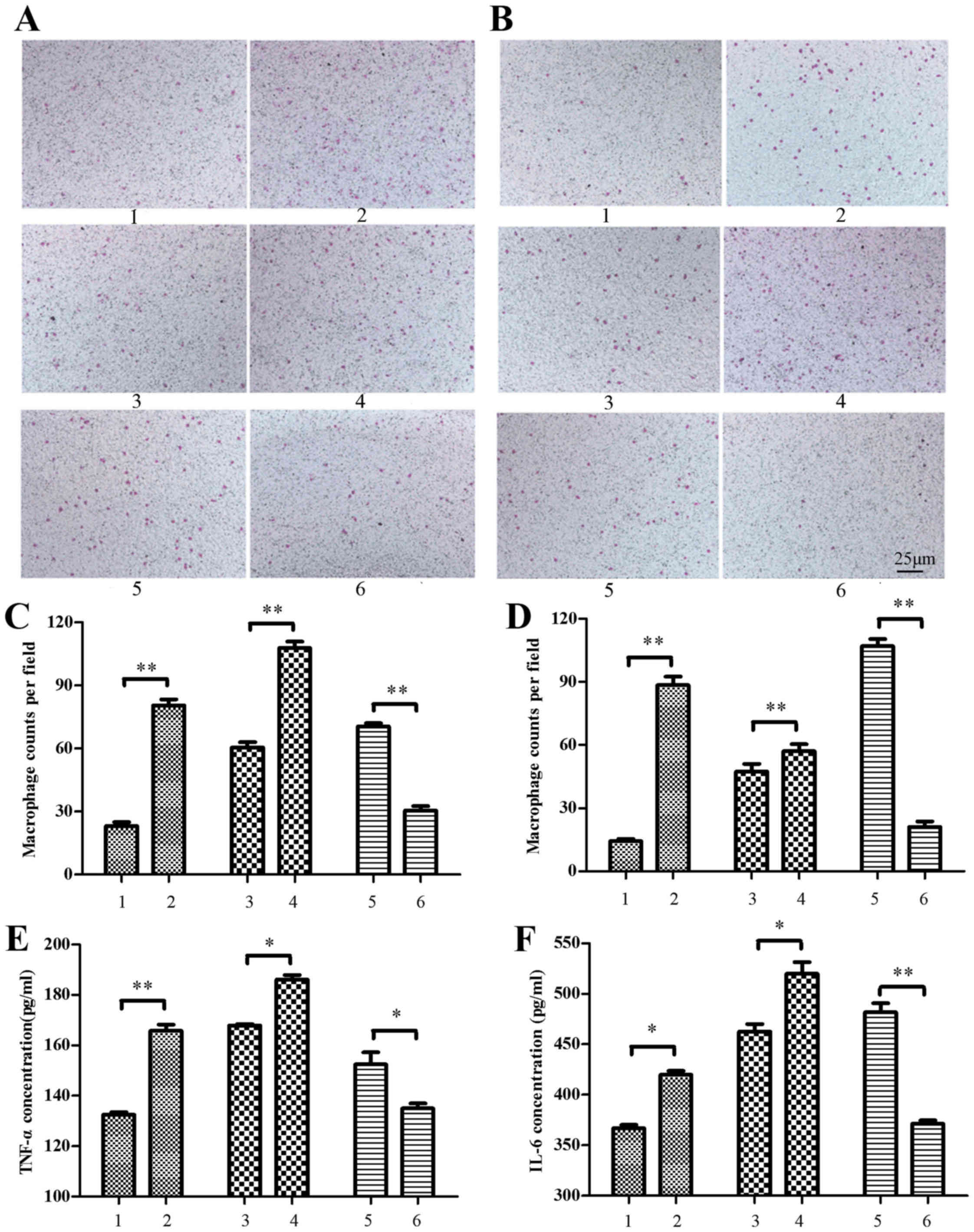 | Figure 8.Macrophages chemotax towards MIN6
cells. Murine macrophages were treated with MIN6 cell culture
supernatant for chemotaxis assays. (A) The RAW264.7 murine
macrophage cell line and (B) peritoneal macrophages isolated from
c57BL/6 mice were stained with crystal violet and viewed under a
light microscope (magnification, ×200). (C) RAW264.7 and (D)
peritoneal macrophages were counted in six randomly selected
fields. The concentration of (E) TNF-α and (F) IL-6 in the culture
supernatant was also measured by ELISA. Supernatant for macrophage
treatment was obtained from the following: i) MIN6 cells treated
with bovine serum albumin, ii) MIN6 cells treated with 500 µM
palmitate, iii) MIN6 cells transfected with pEGFP-N1 control
plasmid, iv) MIN6 cells transfected with pEGFP-N1-RIP140 plasmid,
v) MIN6 cells transfected with scramble siRNA and vi) MIN6 cells
transfected with RIP140 siRNA. Groups 3–6 were treated with 500 µM
palmitate post-transfection. Data are presented as the mean ±
standard deviation of three independent experiments. *P<0.05,
**P<0.01. MIN6, murine pancreatic beta cell line; TNF-α, tumor
necrosis factor-α; IL-6, interleukin-6; siRNA, small interfering
RNA. |
Similar results were observed for the peritoneal
macrophages isolated from c57BL/6 mice. Cell chemotaxis towards
MIN6 cells was significantly increased by conditioned media from
palmitate-treated MIN6 cells, with and without RIP140
overexpression, while chemotaxis was significantly decreased by
conditioned media from palmitate-treated MIN6 cells with knockdown
of RIP140, relative to their respective control groups (P<0.01;
Fig. 8D).
TNF-α and IL-6 concentration in the supernatant of
MIN6 cells were also measured by ELISA following 500 µM palmitate
treatment. It was observed that levels of TNF-α and IL-6 were
significantly increased in the supernatant of palmitate-treated
MIN6 cells (P<0.01 and P<0.05, respectively) and in that of
palmitate-treated cells overexpressing RIP140 (both P<0.05),
relative to their respective control groups. By contrast, TNF-α and
IL-6 were significantly decreased in the supernatant of
palmitate-treated, RIP140-knockdown MIN6 cells, relative to control
cells (P<0.05 and P<0.01, respectively; Fig. 8E and F).
Discussion
Inflammation is involved in insulin resistance
within adipose and muscular tissue, thus promoting the onset of
type II diabetes (26). In addition,
pancreatic beta cell dysfunction serves a key role in
pathophysiology of type II diabetes (27). However, whether crosstalk between
macrophages and beta cells, with involvement of nuclear receptor
cofactor RIP140, contributes to the onset of diabetes remains
unknown. Previous results suggest that loss of RIP140 impairs
inflammatory responses mediated by TLRs. RIP140 is considered to
provide a platform for the stabilization of the NF-κB cofactors,
RelA, and CBP, thus regulating the expression of pro-inflammatory
cytokines, including TNF-α, IL-1β and IL-6 (19). In turn, high levels of inflammatory
cytokines may disturb beta cell function and proliferation
(28). The results of the present
study suggested that the level of TNF-α and IL-6 expression in
macrophages was increased by upregulation of RIP140 in the presence
of palmitate, and that higher levels of TNF-α and IL-6 triggered an
inflammatory response, thus contributing to decreased cell
viability and insulin secretion of beta cells. These findings
indicate the effects of macrophage-derived cytokines on beta cell
function and the involvement of RIP140. Specifically, RIP140 may
promote the release of inflammatory cytokines under lipotoxic
conditions, thus accelerating beta cell failure.
In the present study, under high palmitate
conditions, conditioned media derived from macrophages with and
without RIP140 overexpression were able to activate JNK and ERK1/2
signaling. Phosphorylation of JNK and ERK1/2 serve important roles
in cell signal transduction events, including gene transcription,
proliferation, apoptosis, differentiation and insulin signaling
(29). In addition, it has been
observed that palmitate activates the phosphorylation of ERK1/2 and
p38 mitogen-activated protein kinase pathways and endoplasmic
reticulum stress in cell such as skeletal muscle and mesenchymal
stem cells (30). A number of
inflammatory cytokines, including IL-1β, IFN-γ and TNF-α, are
considered to be activators of JNK and ERK1/2 signaling, which may
lead to an increase in the levels of reactive oxygen species and
compensatory generation of nitric oxide (31). Collectively, these data suggest that
the high levels of inflammatory mediators identified in the
conditioned media of palmitate-treated and RIP140-upregulated
macrophages may have disturbed beta cell signal transduction and
proliferation, thus contributing to a decrease in insulin
secretion. However, the mechanistic involvement of JNK and ERK1/2
is not fully understood and studies evaluating their
pharmacological inhibition are warranted.
High levels of inflammatory cytokines may also
affect the transcription of specific genes, including PGC-1α,
UCP-2, iNOS1, PEPCK, PDX-1 and PCNA. As a transcriptional
activator, PGC-1α interacts with RIP140 and upregulates the
expression of genes associated with mitochondrial metabolism
(32). UCP-2 is a ubiquitously
expressed mitochondrial carrier protein that uncouples the
respiratory chain from ATP synthesis (33), with increases in UCP-2 possibly
involved in reduced insulin secretion during oxidative stress
(34). iNOS1 stimulates L-arginine
to generate NO, inducing an inflammatory reaction (35). In the present study, it was
demonstrated that conditioned media from palmitate-treated and
RIP140-upregulated macrophages, consisting of high levels of TNF-α
and IL-6, induced a significant decrease in the level of PGC-1α
mRNA and a significant increase in the levels of UCP-2 and iNOS1
mRNA. These results indicate that RIP140 in macrophages induces
cytokine secretion, which may then alter beta cell function and
increase oxidative reaction stress. It has been demonstrated that
RIP140 is necessary for LXR-dependent expression of PEPCK (16), as a rate-limiting enzyme in
gluconeogenesis. In the current study, conditioned media containing
high TNF-α and IL-6 was found to decrease PEPCK expression,
suggesting that inflammatory cytokines may contribute to
carbohydrate metabolism disorder. Moore et al (36) demonstrated that palmitate reduces
insulin secretion by downregulating the expression of the insulin
transcription factors PDX-1 and MafA. In addition, it has been
observed that LPS represses PDX-1 expression and insulin secretion
by pathways involving TLR4 and NF-κB (37). In the present study, conditioned
media from macrophages overexpressing RIP140 prior to palmitate
exposure significantly decreased GSIS and ISI, by upregulating
PDX-1 mRNA expression. Fontes et al (38) suggested that Per-ARNT-Sim kinase may
be independent from the ERK1/2 pathway and have no effect on MafA
expression, suggesting that ≥3 independent signaling pathways may
contribute to reduced insulin gene expression. Inflammatory
cytokines may also affect PDX-1 expression by other pathways. In
the current study, increasing levels of PDX-1 expression may have
been a compensatory mechanism that occurred during stressful
conditions, though insulin secretion did not increase accordingly
and further study is warranted to verify this.
Macrophage infiltration of adipose and muscular
tissue and hepatocytes may lead to peripheral insulin resistance
(7–9). In the present study, chemotaxis assays
were performed using Transwell plates. The conditioned media from
MIN6 beta cells was added to the lower chamber and murine
macrophages (peritoneal or RAW264.7) were added to the upper
chamber. In assays of each cell type, cells migrated across the
Transwell membrane, possibly due to the presence of a
chemoattractant gradient. This is in accordance with the results of
a previous study, in which the effects of a chemoattractant
gradient were demonstrated by counting the number of macrophages on
the lower surface of a Transwell membrane (39). In the present study, levels of TNF-α
and IL-6 in the supernatant of MIN6 beta cells were significantly
decreased by downregulation of RIP140 under high palmitate
conditions, and the chemotaxis of RAW264.7 macrophages towards beta
cells was reduced. By contrast, upregulation of RIP140 in beta
cells under high palmitate conditions led to enhanced RAW264.7
macrophage chemotaxis and levels of TNF-α and IL-6 in the
supernatant were elevated. These results were analogous for
peritoneal macrophages. Collectively, these findings suggest that
RIP140 mediates inflammatory cytokine secretion in the presence of
palmitate, which subsequently increases macrophage chemotaxis
towards beta cells. However, macrophages are divided into two
phenotypic categories: i) Anti-inflammatory (M2), which produce
anti-inflammatory cytokines such as IL-10 and ii) pro-inflammatory
(M1), which produce inflammatory cytokines such as IL-6 and TNF-α
(40). Therefore, a range of pro-
and anti-inflammatory cytokines may be secreted in vivo.
Therefore, the cumulative effects of inflammatory cytokines in
vivo warrant further study.
In conclusion, the present study indicated that in
high concentrations of palmitate, expression of RIP140 in
macrophages stimulates the release of inflammatory cytokines, which
may subsequently suppress beta cell viability and insulin
secretion. The underlying mechanism remains unknown, though
possibly involves regulation of specific gene expression and
activation of JNK and ERK signaling. The present results also
suggest that RIP140-mediated cytokine secretion under lipotoxic
conditions may modulate macrophage chemotaxis towards beta cells.
This may promote macrophage infiltration and local inflammation,
ultimately leading to beta cell dysfunction.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81170769).
References
|
1
|
Xue J, Zhao H, Shang G, Zou R, Dai Z, Zhou
D, Huang Q and Xu Y: RIP140 is associated with subclinical
inflammation in type 2 diabetic patients. Exp Clin Endocrinol
Diabetes. 121:37–42. 2013.PubMed/NCBI
|
|
2
|
Solinas G, Vilcu C, Neels JG,
Bandyopadhyay GK, Luo JL, Naugler W, Grivennikow S, Wynshaw-Boris
A, Scadeng M, Olefsky JM and Karin M: JNK1 in hematopoietically
derived cells contributes to diet-induced inflammation and insulin
resistance without affecting obesity. Cell Metab. 6:386–397. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nguyen MT, Favelyukis S, Nguyen AK,
Reichart D, Scott PA, Jenn A, Liu-Bryan R, Glass CK, Neels JG and
Olefsky JM: A subpopulation of macrophages infiltrates hypertrophic
adipose tissue and is activated by free fatty acids via Toll-like
receptors 2 and 4 and JNK-dependent pathways. J Biol Chem.
282:35279–35292. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nov O, Shapiro H, Ovadia H, Tarnovscki T,
Dvir I, Shemesh E, Kovsan J, Shelef I, Carmi Y, Voronov E, et al:
Interleukin-1β regulates fat-liver crosstalk in obesity by
auto-paracrine modulation of adipose tissue inflammation and
expandability. PLoS One. 8:e536262013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lumeng CN, Deyoung SM and Saltiel AR:
Macrophages block insulin action in adipocytes by altering
expression of signaling and glucose transport proteins. Am J
Physiol Endocrinol Metab. 292:E166–E174. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Varma V, Yao-Borengasser A, Rasouli N,
Nolen GT, Phanavanh B, Starks T, Gurley C, Simpson P, McGehee RE
Jr, Kern PA and Peterson CA: Muscle inflammatory response and
insulin resistance: Synergistic interaction between macrophages and
fatty acids leads to impaired insulin action. Am J Physiol
Endocrinol Metab. 296:E1300–E1310. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kewalramani G, Fink LN, Asadi F and Klip
A: Palmitate-activated macrophages confer insulin resistance to
muscle cells by a mechanism involving protein kinase C θ and ε.
PLoS One. 6:e269472011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Samokhvalov V, Bilan PJ, Schertzer JD,
Antonescu CN and Klip A: Palmitate- and
lipopolysaccharide-activated macrophages evoke contrasting insulin
responses in muscle cells. Am J Physiol Endocrinol Metab.
296:E37–E46. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang Y, Liu J, Xu Y, Dai Z and Alves MH:
Reduction of insulin resistance in HepG2 cells by knockdown of
LITAF expression in human THP-1 macrophages. J Huazhong Univ Sci
Technolog Med Sci. 32:53–58. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cunha DA, Hekerman P, Ladrière L,
Bazarra-Castro A, Ortis F, Wakeham MC, Moore F, Rasschaert J,
Cardozo AK, Bellomo E, et al: Initiation and execution of lipotoxic
ER stress in pancreatic beta-cells. J Cell Sci. 121:2308–2318.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fonseca SG, Gromada J and Urano F:
Endoplasmic reticulum stress and pancreatic beta-cell death. Trends
Endocrinol Metab. 22:266–274. 2011.PubMed/NCBI
|
|
12
|
Cavailles V, Dauvois S, L'Horset F, Lopez
G, Hoare S, Kushner PJ and Parker MG: Nuclear factor RIP140
modulates transcriptional activation by the estrogen receptor. EMBO
J. 14:3741–3751. 1995.PubMed/NCBI
|
|
13
|
Docquier A, Garcia A, Savatier J,
Boulahtouf A, Bonnet S, Bellet V, Busson M, Margeat E, Jalaguier S,
Royer C, et al: Negative regulation of estrogen signaling by ERb
and RIP140 in ovarian cancer cells. Mol Endocrinol. 27:1429–1441.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Leonardsson G, Steel JH, Christian M,
Pocock V, Milligan S, Bell J, So PW, Medina-Gomez G, Vidal-Puig A,
White R and Parker MG: Nuclear receptor corepressor RIP140
regulates fat accumulation. Proc Natl Acad Sci USA. 101:pp.
8437–8442. 2004; View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Debevec D, Christian M, Morganstein D,
Seth A, Herzog B, Parker M and White R: Receptor interacting
protein 140 regulates expression of uncoupling protein 1 in
adipocytes through specific peroxisome proliferator activated
receptor isoforms and estrogen-related receptor alpha. Mol
Endocrinol. 21:1581–1592. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Herzog B, Hallberg M, Seth A, Woods A,
White R and Parker MG: The nuclear receptor cofactor,
receptor-interacting protein 140, is required for the regulation of
hepatic lipid and glucose metabolism by liver X receptor. Mol
Endocrinol. 21:2687–2697. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Windahl SH, Treuter E, Ford J, Zilliacus
J, Gustafsson JA and McEwan IJ: The nuclear-receptor interacting
protein (RIP) 140 binds to the human glucocorticoid receptor and
modulates hormone-dependent transactivation. J Steroid Biochem Mol
Biol. 71:93–102. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee CH and Wei LN: Characterization of
receptor-interacting protein 140 in retinoid receptor activities. J
Biol Chem. 274:31320–31326. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zschiedrich I, Hardeland U, Krones-Herzig
A, Diaz M Berriel, Vegiopoulos A, Müggenburg J, Sombroek D, Hofmann
TG, Zawatzky R, Yu X, et al: Coactivator function of RIP140 for
NFkappaB/RelA-dependent cytokine gene expression. Blood.
112:264–276. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ho PC, Chang KC, Chuang YS and Wei LN:
Cholesterol regulation of receptor-interacting protein 140 via
microRNA-33 in inflammatory cytokine production. FASEB J.
25:1758–1766. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ho PC, Tsui YC, Feng X, Greaves DR and Wei
LN: NF-κB-mediated degradation of the coactivator RIP140 regulates
inflammatory responses and contributes to endotoxin tolerance. Nat
Immunol. 13:379–386. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu PS, Lin YW, Lee B, McCrady-Spitzer SK,
Levine JA and Wei LN: Reducing RIP140 expression in macrophage
alters ATM infiltration, facilitates white adipose tissue browning,
and prevents high fat diet-induced insulin resistance. Diabetes.
63:4021–4031. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dimopoulos N, Watson M, Sakamoto K and
Hundal HS: Differential effects of palmitate and palmitoleate on
insulin action and glucose utilization in rat L6 skeletal muscle
cells. Biochem J. 399:473–481. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gupta P, Ho PC, Huq MD, Khan AA, Tsai NP
and Wei LN: PKCepsilon stimulated arginine methylation of RIP140
for its nuclear-cytoplasmic export in adipocyte differentiation.
PLoS One. 3:e26582008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kusminski CM, da Silva NF, Creely SJ,
Fisher FM, Harte AL, Baker AR, Kumar S and McTernan PG: The in
vitro effects of resistin on the innate immune signaling pathway in
isolated human subcutaneous adipocytes. J Clin Endocrinol Metab.
92:270–276. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Olefsky JM and Glass CK: Macrophages,
inflammation, and insulin resistance. Annu Rev Physiol. 72:219–246.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cernea S and Dobreanu M: Diabetes and beta
cell function: From mechanisms to evaluation and clinical
implications. Biochem Med (Zagreb). 23:266–280. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang S and Kim KH: TNF-alpha inhibits
glucose-induced insulin secretion in a pancreatic beta-cell line
(INS-1). FEBS Lett. 377:237–239. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tanti JF and Jager J: Cellular mechanisms
of insulin resistance: Role of stress-regulated serine kinases and
insulin receptor substrates (IRS) serine phosphorylation. Curr Opin
Pharmacol. 9:753–762. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lu J, Wang Q, Huang L, Dong H, Lin L, Lin
N, Zheng F and Tan J: Palmitate causes endoplasmic reticulum stress
and apoptosis in human mesenchymal stem cells: Prevention by AMPK
activator. Endocrinology. 153:5275–5284. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Seidelin JB and Nielsen OH: Continuous
cytokine exposure of colonic epithelial cells induces DNA damage.
Eur J Gastroenterol Hepatol. 17:363–369. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen Y, Wang Y, Chen J, Chen X, Cao W,
Chen S, Xu S, Huang H and Liu P: Roles of transcriptional
corepressor RIP140 and coactivator PGC-1α in energy state of
chronically infarcted rat hearts and mitochondrial function of
cardiomyocytes. Mol Cell Endocrinol. 362:11–18. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang CY, Baffy G, Perret P, Krauss S,
Peroni O, Grujic D, Hagen T, Vidal-Puig AJ, Boss O, Kim YB, et al:
Uncoupling protein-2 negatively regulates insulin secretion and is
a major link between obesity, beta cell dysfunction, and type 2
diabetes. Cell. 105:745–755. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Y, Maedler K, Shu L and Haataja L:
UCP-2 and UCP-3 proteins are differentially regulated in pancreatic
beta-cells. PLoS One. 3:e13972008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chatterjee A, Black SM and Catravas JD:
Endothelial nitric oxide (NO) and its pathophysiologic regulation.
Vascul Pharmacol. 49:134–140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Moore PC, Ugas MA, Hagman DK, Parazzoli SD
and Poitout V: Evidence against the involvement of oxidative stress
in fatty acid inhibition of insulin secretion. Diabetes.
53:2610–2616. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Amyot J, Semache M, Ferdaoussi M, Fontés G
and Poitout V: Lipopolysaccharides impair insulin gene expression
in isolated islets of Langerhans via Toll-Like Receptor-4 and NF-κB
signalling. PLoS One. 7:e362002012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fontes G, Semache M, Hagman DK, Tremblay
C, Shah R, Rhodes CJ, Rutter J and Poitout V: Involvement of
Per-Arnt-Sim Kinase and extracellular-regulated kinases-1/2 in
palmitate inhibition of insulin gene expression in pancreatic
beta-cells. Diabetes. 58:2048–2058. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bruggeman LA, Drawz PE, Kahoud N, Lin K,
Barisoni L and Nelson PJ: TNFR2 interposes the proliferative and
NF-κB-mediated inflammatory response by podocytes to TNF-α. Lab
Invest. 91:413–425. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Antuna-Puente B, Feve B, Fellahi S and
Bastard JP: Adipokines: The missing link between insulin resistance
and obesity. Diabetes Metab. 34:2–11. 2008. View Article : Google Scholar : PubMed/NCBI
|