Introduction
Lung cancer is the leading cause of
cancer-associated mortality, with ~1.35 million novel cases being
diagnosed each year worldwide (1). A
total of ~75–80% of lung cancer cases are non-small cell lung
cancer (NSCLC), with the overall 5-year survival rate of 10%
(2). Despite the development of
novel methods for the early diagnosis of patients as well as recent
advancements in treatment strategies, the prognosis and survival
rate of patients with NSCLC remain relatively poor (3). Between 40–50% of patients with NSCLC
eventually succumb to relapse or metastatic disease following
curative resection (4). However, a
number of prognostic biomarkers are available in clinical practice
at present, particularly regarding patients who have previously
undergone curative surgical resection (5). Therefore, it is important to identify
novel prognostic biomarkers that may aid prognosis prediction and
optimize the treatment of patients with NSCLC.
Micro (mi)RNAs (miRs) are a class of small (19–24
nucleotides in length), highly conserved non-coding RNAs that may
bind to the 3′-untranslated regions (3′-UTRs) of target mRNAs and
subsequently suppress the translation of such RNAs to proteins
(6). It has become increasingly
evident that different miRNAs may either exhibit oncogenic or
tumor-suppressive roles in numerous pathways, depending on their
target genes or cellular context (7–9). miRNAs
are not only involved in the regulation of malignant tumor cell
proliferation, but also serve an important role in tumor invasion
and metastasis (10–12). Therefore, the role of miRNAs in NSCLC
has been receiving increasing attention (13–15).
miRNAs associated with different types of cancer include oncogenic
miRNAs, tumor suppressive miRNAs and miRNA regulators (16). Previous studies have indicated that
miRNA-140-5p serves an important role in numerous tumor types
(17–20); however, studies investigated the
association between miR-140-5p expression and clinical outcomes in
patients with resected NSCLC are limited and have demonstrated
contradictory results (21,22). Therefore, the specific role of
miR-140-5p in NSCLC remains to be determined.
The aim of the present study was to identify the
target genes of miR-140-5p and to elucidate the regulatory
mechanism underlying the association between miR-140-5p and
NSCLC.
Materials and methods
Patients and tissue samples
The clinical information related to 45 NSCLC cancer
cases was collected from July 2016 to 2017 at the Qinhuangdao First
People's Hospital (Qinhuangdao, China). A total of 45 patients were
recruited in the current study [age, 31–75 years; mean age, 53±21.8
years; gender distribution (male: female), 26:19). All patients
were diagnosed with stage III or II NSCLC. Tumor and para-carcinoma
tissue samples were removed and immediately placed in liquid
nitrogen or 10% formalin for in subsequent experimentation. All
samples were confirmed by pathological examination, and no
radiotherapy or chemotherapy was performed prior to surgery. The
present study was approved by the Ethics Committee of Qinhuangdao
First People's Hospital (approval no. QDPH160503) and written
informed consent was obtained from each participant prior to the
study.
Cell culture
A549 lung cancer cells (Shanghai Institute of
Biochemistry and Cell Biology, Shanghai, China) were cultured in
Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10%
fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences,
Logan, UT, USA) and 1% penicillin/streptomycin in a 5%
CO2-humidified incubator at 37°C.
miRNA transfection
Cells were seeded into six-well plates, incubated
for 24 h and divided into the following three groups: Control
group, untreated cells; negative control (NC) group, cells treated
with 20 pmol/µl miR-NC mimics; and miR-140-5p mimics (mimics)
group, cells treated with 1.5 µl miR-140-5p mimics. The following
miRNA sequences were obtained from New England BioLabs, Inc.
(Ipswich, MA, USA): miR-mimics, 5′-ACCAUAGGGUAAAACCACUGUU-3′ and
miR-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′. Cells were transfected with
500 ng miRNA using 2.5 µl Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). After incubation for
6 h, the medium was replaced with RPMI 1640 medium supplemented
with 10% FBS. After 24 h incubation, cells were collected and used
in subsequent experimentation. The experiment was performed in
triplicate.
Flow cytometry assay
Following transfection, the rate of apoptosis of
A549 cells was determined using the Annexin V-fluorescein
isothiocyanate (FITC) Apoptosis Detection kit (BD Biosciences, San
Jose, CA, USA) and flow cytometry. Briefly, the transfected cells
were seeded into 24-well plates and incubated at 37°C with 15%
CO2 for 72 h. Subsequently, cells were treated with
propidium iodide (10 µg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) and Annexin V-FITC (50 µg/ml; BD Biosciences) in the dark
for 15 min at room temperature. Apoptotic cells were subsequently
examined using a flow cytometry (FACScan™; BD Biosciences) and
Modfit software (version 3.2; Verity Software House, Inc., Topsham,
ME, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Poly(A) polymerase
was used to catalyse the addition of adenosine residues onto the
3′ends of RNA. Total RNA concentration was spectrophotometrically
determined at a wavelength of 260 nm. RNA was immediately
reverse-transcribed into cDNA using the SuperScript Reverse
Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.). qPCR
was subsequently performed using the SYBR® Green
Realtime PCR MasterMix (Toyobo Life Science, Osaka, Japan). The
following primer sequences were used for qPCR: miR-140-5p forward,
5′-ACACTCCAGCTGGGAGGCGGGGCGCCGCGGGA-3′ and reverse,
5′-CTCAACTGGTGTCGTGGA-3′; U6 forward, 5-′TGCGGGTGCTCGCTTCGGCAGC-3′
and reverse, 5′-CCAGTGCAGGGTCCGAGGT-3′; YES1 forward,
5′-ATGCTACAGTTGCCCCGAC-3′ and reverse, 5′-TCCAAAAGGAGTCACCCCTGA-3′;
Bax forward, 5′-CACCAGCTCTGAACAGATCATGA-3′ and reverse,
5′-TCAGCCCATCTTCTTCCAGATGT-3′; Bcl-2 forward,
5′-CACCCCTGGCATCTTCTCCTT-3′ and reverse,
5′-AGCGTCTTCAGAGACAGCCAG-3′; caspase-3 forward,
5′-AACTGGACTGTGGCATTGAG-3′ and reverse, 5′-ACAAAGCGACTGGATGAACC-3′;
GAPDH (352 bp) forward, 5′-AGTAGAGGCAGGGATGATGTTC-3′ and reverse,
5′-CTTTGGTATCGTGGAAGGACTC-3′. The following thermocycling
conditions were used for the qPCR: Initial denaturation at 95°C for
10 min; 50 cycles of 95°C for 15 sec and 60°C for 60 sec. Each
reaction was performed three times. The comparative cycle threshold
method was used to analyze qPCR data and 2∆∆Cq values
were used to reflect differences in the expression levels (23). GAPDH or U^ were used as the reference
genes.
Western blotting
Cells in the logarithmic growth phase were collected
in a cell culture plate and total protein was extracted using a
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Shanghai, China). Total protein was quantified using
a bicinchoninic acid assay (Thermo Fisher Scientific, Inc.) and 20
µg protein/lane was separated via SDS-PAGE on a 10% gel. The
separated proteins were transferred onto polyvinylidene fluoride
(PVDF) membranes and blocked overnight at 4°C with 5% non-fat dried
milk. The PVDF membranes were incubated with primary antibodies
against YES proto-oncogene 1 (YES1; 1:1,000; cat. no. ab109265;
Abcam, Cambridge, MA, USA); B-cell lymphoma 2 (Bcl-2; 1:1,000; cat.
no. sc-7382; Santa Cruz Biotechnology, Inc., Dallas, TX, USA);
Bcl-2 associated X (Bax; 1:1,000; sc-7480, Santa Cruz
Biotechnology, Inc.); and caspase-3 (1:1,000; cat. no. 9662; Cell
Signaling Technology, Inc., Danvers, MA, USA) for 1 h at 37°C.
GAPDH (1:1,000; cat. no. 2118; Cell Signaling Technology, Inc.) was
used as the loading control. The membranes were subsequently
incubated with a horseradish peroxidase-conjugated secondary
antibody (1:5,000; cat. no. ab6789; Abcam) at room temperature for
2 h. Subsequently, protein bands were visualized using the ECL-Plus
reagent (Santa Cruz Biotechnology, Inc.) and protein expression was
quantified using Image-Pro Plus software (version 6.0; Media
Cybernetics, Inc., Rockville, MD, USA).
Cell viability assay
Cell viability was examined using the Cell Counting
Kit-8 (CCK-8). Transfected cells were seeded in 96-well plates and
incubated for 24 h. CCK-8 reagent was subsequently added into the
culture medium and, following an additional 24-h incubation step at
37°C, the absorbance was measured at a wavelength of 450 nm using a
microplate reader.
Relative luciferase activity
assay
TargetScan bioinformatics software (www.targetscan.org/vert_71) was used to predict
potential target genes of miR-140-5p, which identified YES1. To
confirm the binding of miR-140-5p to YES1 mRNA, the 3′-UTR of YES1
mRNA was amplified by PCR and inserted into the pGL3/luciferase
vector (Promega Corporation, Madison, WI, USA). Co-transfections
with wt YES1 3′-UTR or mut YES1 3′-UTR plasmids and mimics or NC
sequences were performed using Lipofectamine® 2000.
Luciferase activity was measured 48 h after transfection using the
Dual-Luciferase® reporter assay system (Promega
Corporation). Firefly luciferase activity was normalized to
Renilla luciferase activity. Data are presented as the mean
values for triplicate experiments.
Statistical analysis
Data are presented as the mean ± standard deviation.
All statistical analyses were performed using SPSS software
(version 19.0; IBM Corp., Armonk, NY, USA). Student's t-test was
performed to analyze the difference between two groups and one-way
analysis of variance followed by Newman-Keuls post-hoc test was
used to analyze differences among multiple groups. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were performed in triplicate.
Results
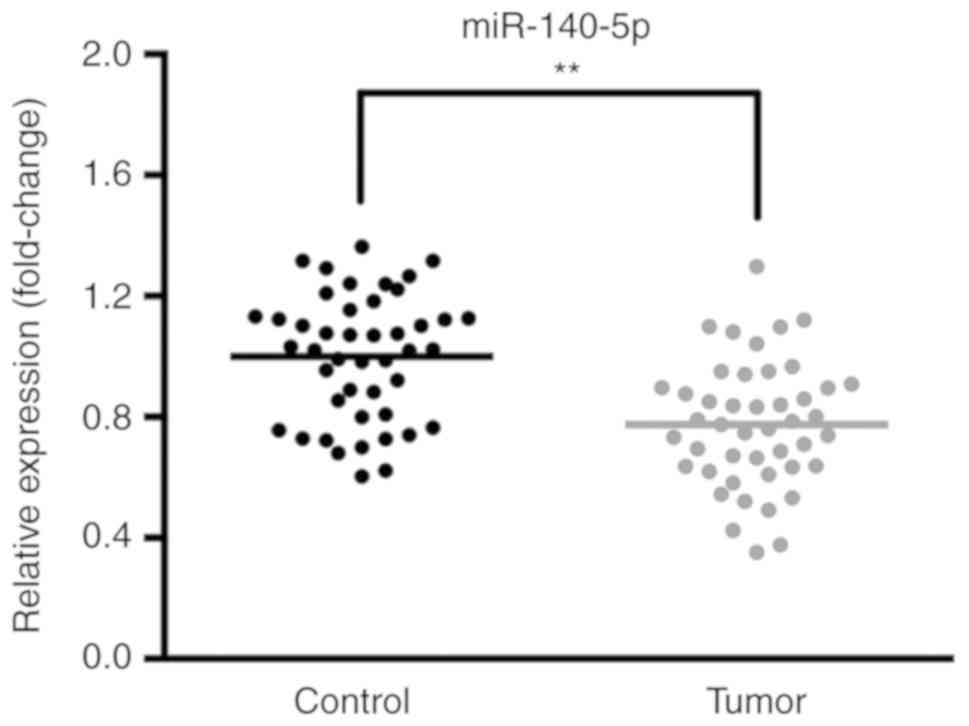
miR-140-5p is downregulated in NSCLC
tissues
The expression levels of miR-140-5p in 45 samples of
NSCLC tissues and paracancerous tissues were determined via
RT-qPCR. The results revealed that the expression of miR-140-5p was
decreased in NSCLC tissues compared with the corresponding
paracancerous tissues (Fig. 1).
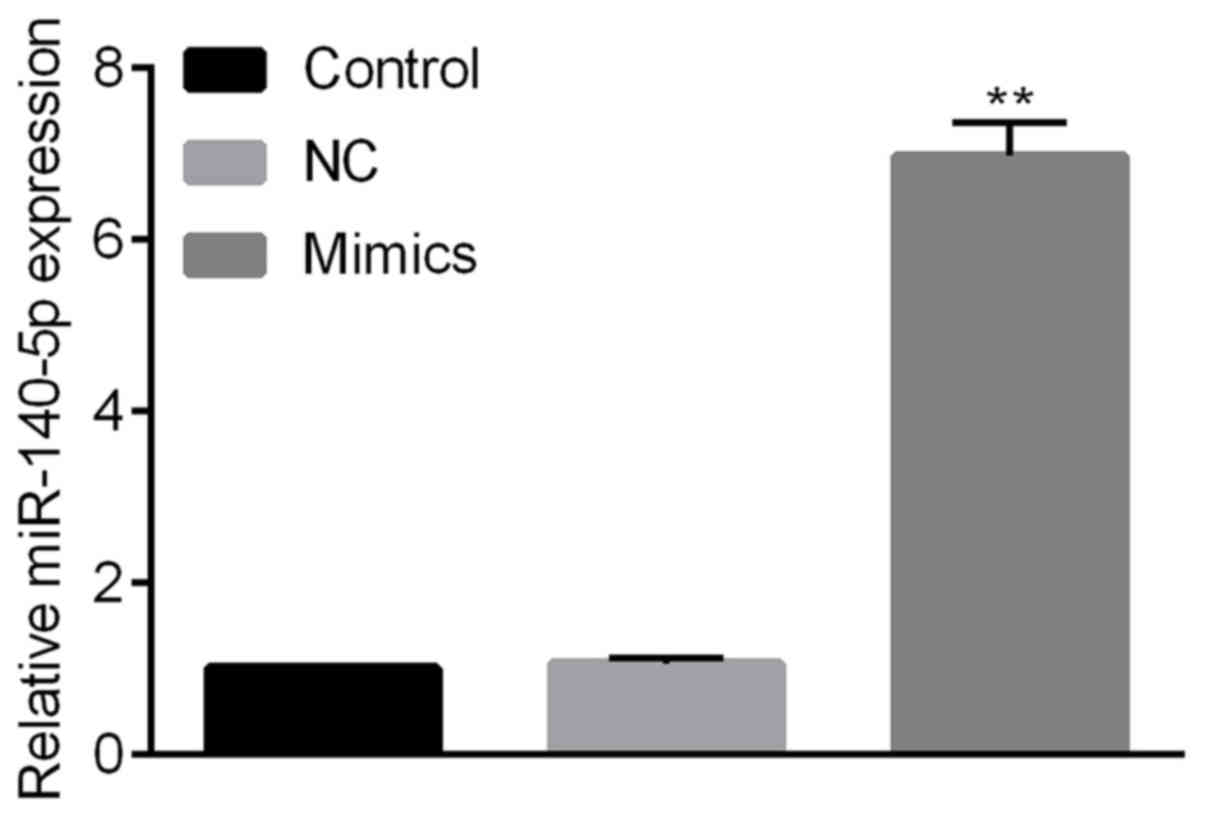
Expression of miR-140-5p following
miRNA transfection
Cells were transfected with miR-140-5p or miR-NC
mimics. The expression of miR-140-5p was significantly increased
following transfection with miR-140-5p mimics compared with the NC
group; however, there was no significant alteration between the
control group and the miR-NC group (Fig.
2).
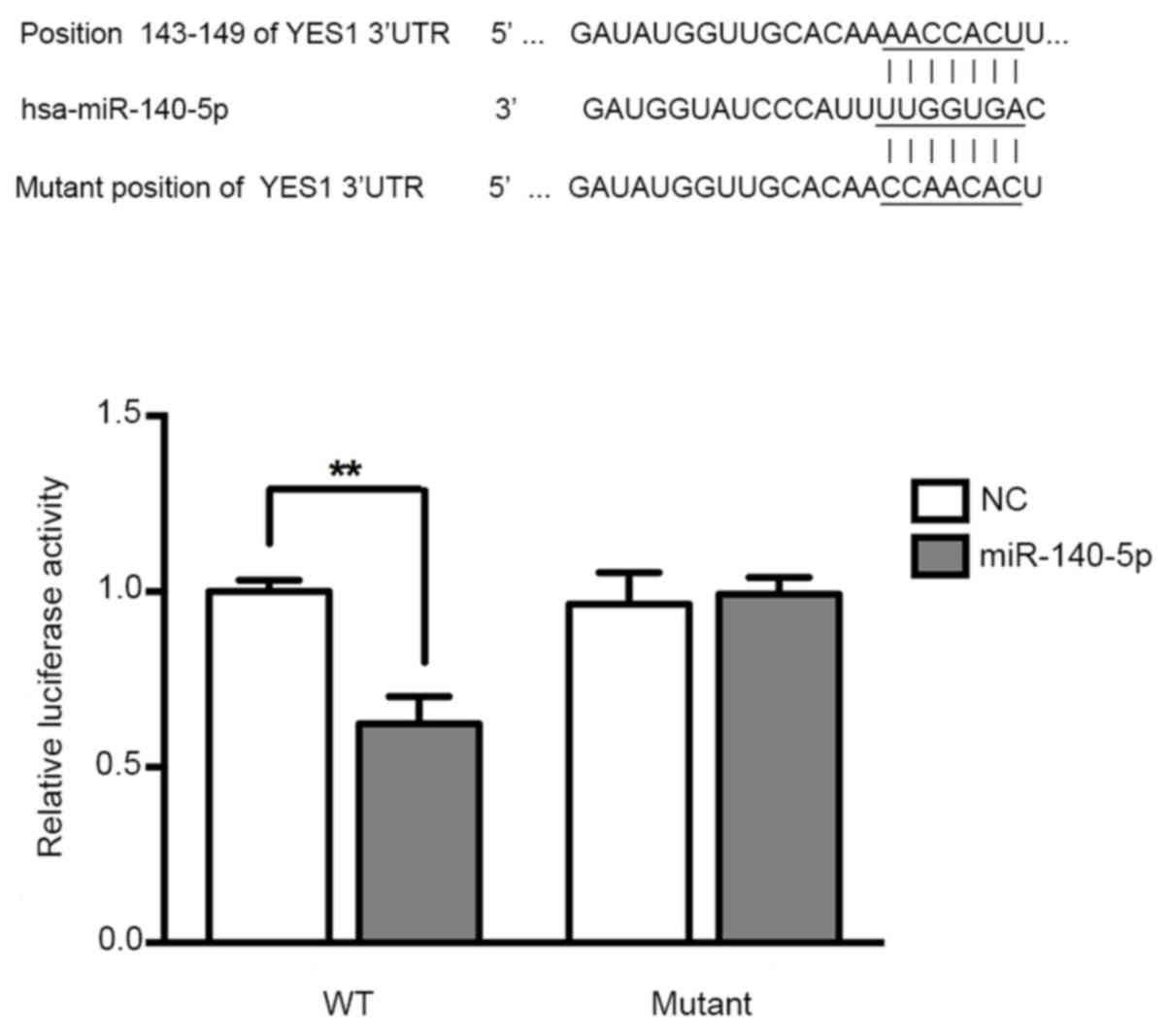
miR-140-5p directly targets YES1 in
A549 cells
To determine the function of miR-140-5p in NSCLC,
target genes of miR-140-5p were predicted using Targetscan
software, and the results revealed that YES1 is a potential target
gene of miR-140-5p. To investigate the potential interaction
between the YES1 3′-UTR and miR-140-5p, luciferase reporter assays
were performed, and the results indicated that the luciferase
activity was significantly inhibited following co-transfection with
pre-miR-140-5p and vectors carrying the wide type 3′-UTR of YES1
(Fig. 3).
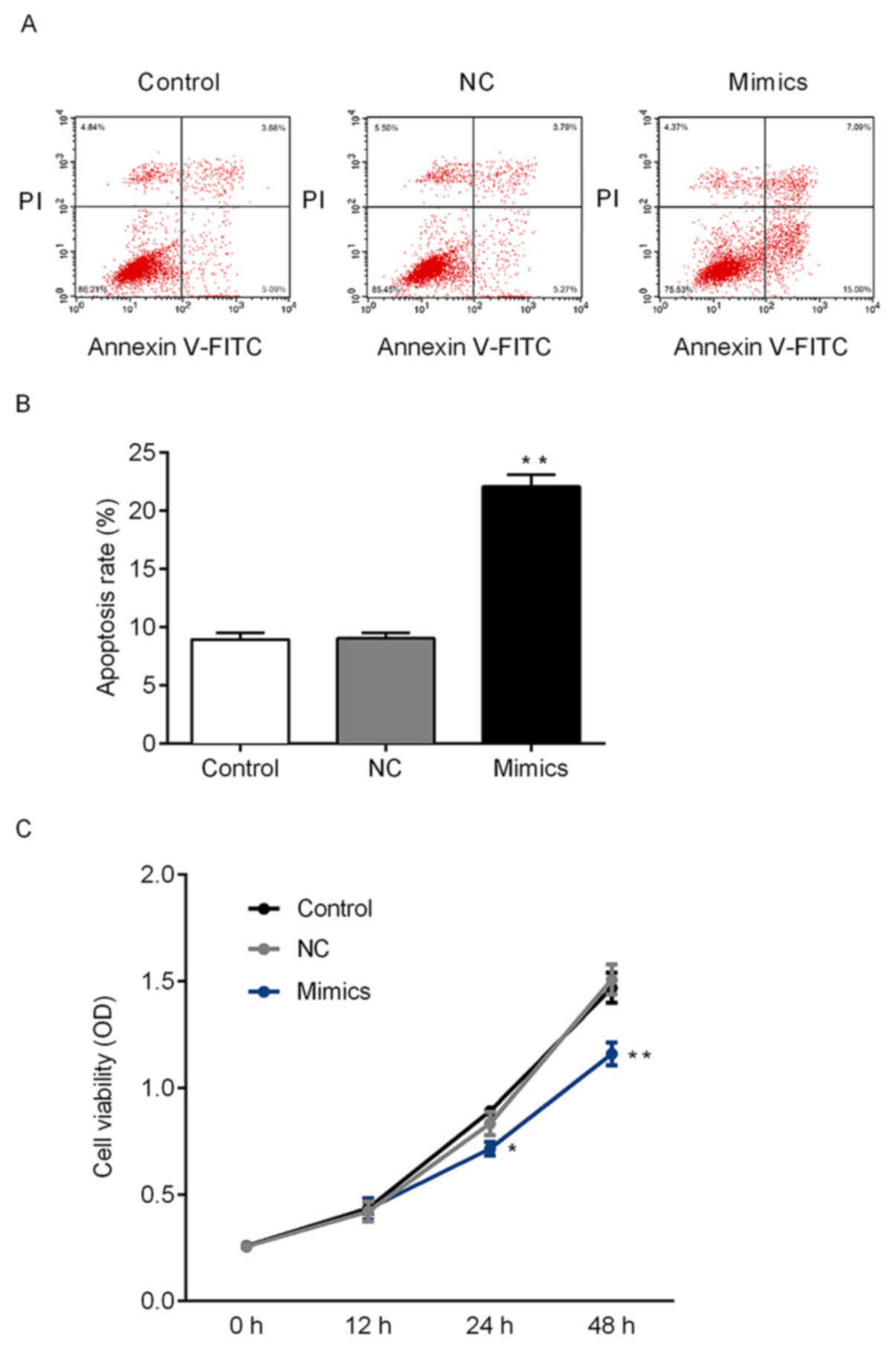
miR-140-5p regulate the cell viability
and apoptosis of NSCLC cells via targeting YES1
As demonstrated above, miR-140-5p may target YES1.
Therefore, the effects of miR-140-5p on the growth of tumor cells
were further investigated. CCK-8 and flow cytometry assays were
performed to determine the cell viability and apoptosis of A549
cells in vitro. The rate of apoptosis of A549 cells
following transfection with miR-140-5p mimics significantly
increased compared with the NC group, and transfection with
miR-140-5p mimics significantly suppressed cell viability in A549
cells compared with he NC group (Fig.
4). Therefore, miR-140-5p expression may suppress the
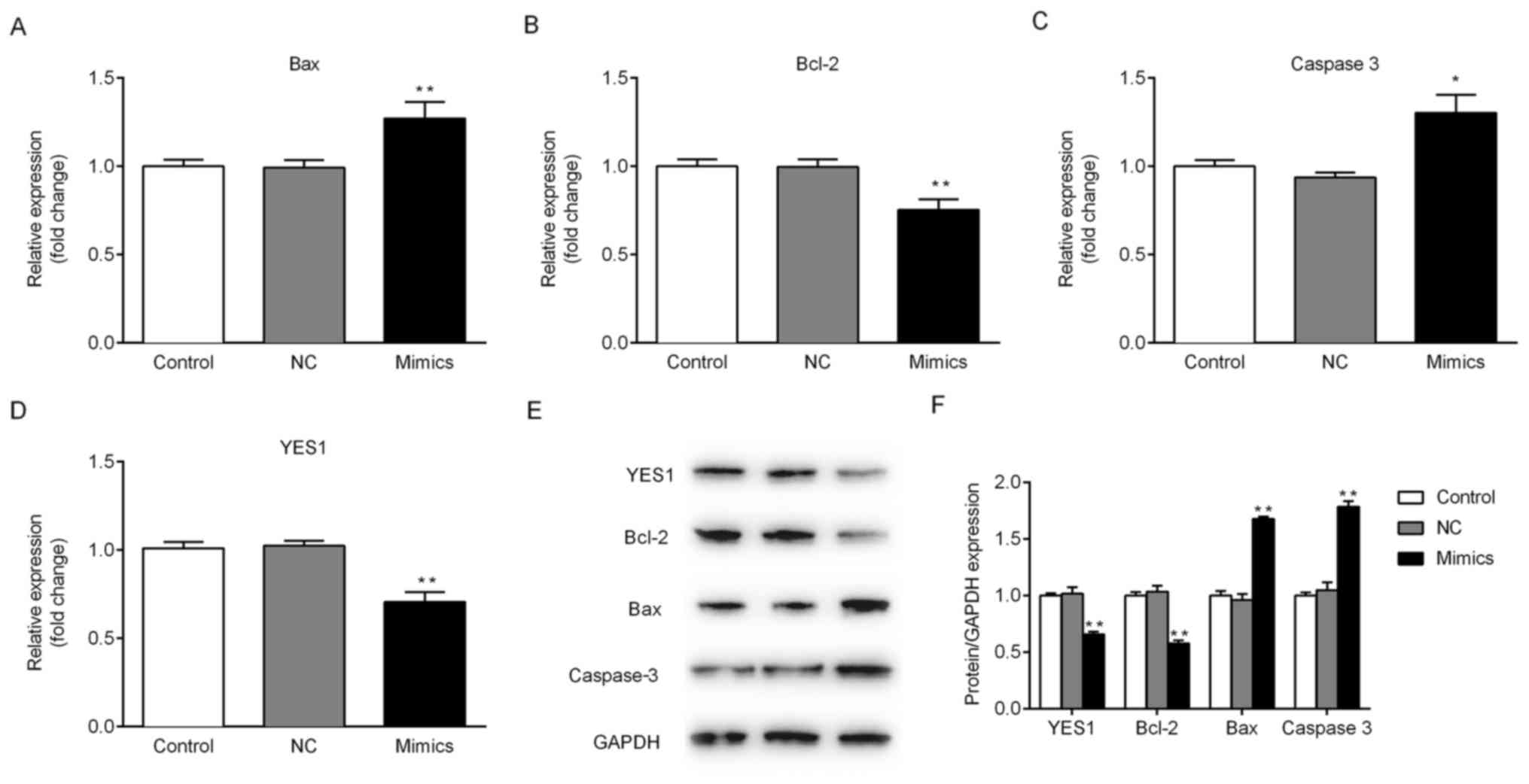
proliferation of NSCLC cells. To further investigate whether
miR-140-5p regulates the expression of YES1 and other
apoptosis-associated proteins, mRNA and protein expression levels
were determined. Ectopic expression of miR-140-5p in A549 cells
significantly decreased the expression of YES1 and Bcl-2 compared
with the NC group at the mRNA and protein level. Furthermore, the
expression of Bax and caspase-3 was significantly increased in the
mimics group compared with control group (Fig. 5). The aforementioned results
suggested that miR-140-5p may regulate cell viability in
vitro by targeting of YES1.
Discussion
Lung cancer is the only malignant tumor for which
morbidity and mortality rates have been increasing in recent years
(1). In addition, lung cancer has
become the leading cause of cancer-associated mortality worldwide
(1). Lung cancer is pathologically
categorized as either small cell lung cancer or NSCLC (22). NSCLC accounts for ~80% of patients
with lung cancer, and the majority of patients with NSCLC are
diagnosed during the mid-late stages of the disease, and thus may
no longer be subjected to operative therapy (3). Radiotherapy, chemotherapy and molecular
targeted therapy represent current therapeutic strategies for the
treatment of patients with NSCLC, however, the use of such
strategies has not significantly improved the survival rate of
patients with lung cancer (18).
miRNAs are widely and stably expressed in human body
fluids (24). Aberrant levels of
circulating miRNAs represent potential biomarkers for the early
diagnosis of tumors as well as for the prediction of therapeutic
responses (25). miRNAs exhibit
important regulatory roles by targeting mRNAs for protein cleavage
or translational repression (3).
Considering that >50% of miRNAs are located in cancer-associated
genomic regions or in fragile sites, miRNAs may serve an important
role in cancer pathogenesis (26).
Furthermore, aberrant miRNA expression has been demonstrated in
lung cancer, which contributes to carcinogenesis and cancer
development by either promoting oncogene expression or inhibiting
tumor suppressor genes (27).
miR-140-5p was found to serve a role in carcinogenesis of NSCLC
(21,22). Target genes associated with
miR-140-5p may serve as biomarkers and aid in the diagnosis and
prognostic prediction of NSCLC (28,29). The
miR-140-5p/monocyte to macrophage differentiation-associated (MMD)
axis could affect cell proliferation and invasion of lung cancer
cells by regulating the Erk signaling pathway (21,22). In
the present study, miR-140-5p expression was demonstrated to be
markedly decreased in NSCLC tissues compared with normal adjacent
tissues.
YES1 is an important oncogene belonging to the Src
protein-tyrosine kinase family (30). Increasing evidence has indicated that
YES1 is overexpressed in numerous types of cancer and promotes the
proliferation and metastasis of tumor cells, which may have
potential therapeutic applications regarding the clinical diagnosis
and treatment of tumors (31–34).
However, whether miR-140-5p may inhibit proliferation and induce
apoptosis of NSCLC cells via targeting of YES1 remains unclear.
Differential expression of miR-140-5p has been revealed in numerous
signaling pathways associated with the regulation of cell apoptosis
(21,22). A previous study demonstrated that the
YES1 is a target gene of miR-140-5p (35). In the current study, target genes of
miR-140-5p were predicted using TargetScan bioinformatics software,
and the results revealed that the YES1 oncogene represented a
potential target of miR-140-5p. Furthermore, the results of the
present study revealed that ectopic expression of miR-140-5p
suppressed NSCLC viability and promoted apoptosis.
Apoptosis serves an important role in
anti-carcinogenesis (36). The Bcl-2
protein family and the caspase family are currently the most
frequently investigated genes (37,38).
Bcl-2 and Bax are the most important regulatory genes in the
apoptotic pathway with opposing regulatory functions (37). The anti-apoptotic gene Bcl-2 was the
first anti-apoptotic gene discovered in biomedical research
(39). Bax is a pro-apoptotic gene
opposing the activity of apoptosis-inhibiting proteins, including
Bcl-2, to prevent the anti-apoptotic effects (40). Caspase-3 is a physiological enzymatic
protease of Bcl-2 involved in the process of apoptosis (41). In the current study, overexpression
of miR-140-5p in A549 cells markedly inhibited YES1 and Bcl-2
expression; however, the protein expression levels of Bax and
caspase-3 were upregulated following transfection with miR-140-5p,
compared with the NC group. The apoptosis rate of A549 cells
following transfection with miR-140-5p mimics increased compared
with the NC group, and transfection with miR-140-5p mimics
suppressed the level of cell viability. These results may be used
for the development of a novel diagnostic or therapeutic strategy
and may indicate potential therapeutic targets for NSCLC. The
current study was limited by the use of only one cell line, and in
future studies, multiple cell lines will be used to verify these
results.
In conclusion, the present study revealed a novel
target gene of miR-140-5p associated with the regulation of lung
cancer cell apoptosis and viability.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WZ contributed to the analysis and interpretation of
data, conception and design. XW and DY were responsible for the
acquisition of data and writing of the manuscript. LX, HW and ZM
contributed to the acquisition, analysis and interpretation of
data. ZW and YS performed statistical analysis. QZ provided
technical support, and contributed to the analysis and
interpretation of data. ZZ contributed to the conception and design
of the study, interpretation of data and revision of the
manuscript. YY was responsible for the conception, design and
supervision of the study, and the revision of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Qinhuangdao First People's Hospital (Qinhuangdao,
China) and written informed consent was obtained from each
participant prior to the study.
Patient consent for publication
All patients provided consent for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu P, Pu J, Zhang J, Chen Z, Wei K and
Shi L: Bioinformatic analysis of miR-4792 regulates Radix
Tetrastigma hemsleyani flavone to inhibit proliferation, invasion,
and induce apoptosis of A549 cells. Onco Targets Ther.
12:1401–1412. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pope CA III, Burnett RT, Thun MJ, Calle
EE, Krewski D, Ito K and Thurston GD: Lung cancer, cardiopulmonary
mortality, and long-term exposure to fine particulate air
pollution. JAMA. 287:1132–1141. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sui A, Zhang X and Zhu Q: Diagnostic value
of serum miR197 and miR145 in non-small cell lung cancer. Oncol
Lett. 17:3247–3252. 2019.PubMed/NCBI
|
|
4
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carvalho PE, Antonãngelo L, Bernardi FD,
Leão LE, Rodrigues OR and Capelozzi VL: Useful prognostic panel
markers to express the biological tumor status in resected lung
adenocarcinomas. Jpn J Clin Oncol. 30:478–486. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dews M, Homayouni A, Yu D, Murphy D,
Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT and
Thomas-Tikhonenko A: Augmentation of tumor angiogenesis by a
Myc-activated microRNA cluster. Nat Genet. 38:1060–1065. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mitra R, Edmonds MD, Sun J, Zhao M, Yu H,
Eischen CM and Zhao Z: Reproducible combinatorial regulatory
networks elucidate novel oncogenic microRNAs in non-small cell lung
cancer. RNA. 20:1356–1368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wozniak MB, Scelo G, Muller DC, Mukeria A,
Zaridze D and Brennan P: Circulating microRNAs as non-invasive
biomarkers for early detection of non-small-cell lung cancer. PLoS
One. 10:e01250262015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yao Q, Zhang AM, Ma H, Lin S, Wang XX, Sun
JG and Chen ZT: Novel molecular beacons to monitor microRNAs in
non-small-cell lung cancer. Mol Cell Probes. 26:182–187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Goga A and Benz C: Anti-oncomir
suppression of tumor phenotypes. Mol Interv. 7:199–202, 180. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan X, Zhu Z, Xu S, Yang LN, Liao XH,
Zheng M, Yang D, Wang J, Chen D, Wang L, et al: MicroRNA-140-5p
inhibits hepatocellular carcinoma by directly targeting the unique
isomerase Pin1 to block multiple cancer-driving pathways. Sci Rep.
7:459152017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou CW, Zhao WJ, Zhu YG and Zhao XD:
MiR-185 inhibits tumor growth and enhances chemo-resistance via
targeting SRY-related high mobility group box transcription factor
13 in non-small-cell carcinoma. Am J Transl Res. 10:2600–2609.
2018.PubMed/NCBI
|
|
19
|
Yang H, Fang F, Chang R and Yang L:
MicroRNA-140-5p suppresses tumor growth and metastasis by targeting
transforming growth factor β receptor 1 and fibroblast growth
factor 9 in hepatocellular carcinoma. Hepatology. 58:205–217. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang W, Zou C, Pan L, Xu Y, Qi W, Ma G,
Hou Y and Jiang P: MicroRNA-140-5p inhibits the progression of
colorectal cancer by targeting VEGFA. Cell Physiol Biochem.
37:1123–1133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dong C, Liu X, Wang H, Li J, Dai L, Li J
and Xu Z: Hypoxic non-small-cell lung cancer cell-derived exosomal
miR-21 promotes resistance of normoxic cell to cisplatin. Onco
Targets Ther. 12:1947–1956. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Flamini V, Jiang WG and Cui Y: Therapeutic
Role of MiR-140-5p for the treatment of non-small cell lung cancer.
Anticancer Res. 37:4319–4327. 2017.PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arcà B, Colantoni A, Fiorillo C, Severini
F, Benes V, Di Luca M, Calogero RA and Lombardo F: MicroRNAs from
saliva of anopheline mosquitoes mimic human endogenous miRNAs and
may contribute to vector-host-pathogen interactions. Sci Rep.
9:29552019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mirzaei H, Khataminfar S, Mohammadparast
S, Sales SS, Maftouh M, Mohammadi M, Simonian M, Parizadeh SM,
Hassanian SM and Avan A: Circulating microRNAs as potential
diagnostic biomarkers and therapeutic targets in gastric cancer:
Current status and future perspectives. Curr Med Chem.
23:4135–4150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Long XB, Sun GB, Hu S, Liang GT, Wang N,
Zhang XH, Cao PP, Zhen HT, Cui YH and Liu Z: Let-7a microRNA
functions as a potential tumor suppressor in human laryngeal
cancer. Oncol Rep. 22:1189–1195. 2009.PubMed/NCBI
|
|
27
|
Cheng TF, Choudhuri S and Muldoon-Jacobs
K: Epigenetic targets of some toxicologically relevant metals: A
review of the literature. J Appl Toxicol. 32:643–653. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang K, Chen M and Wu W: Analysis of
microRNA (miRNA) expression profiles reveals 11 key biomarkers
associated with non-small cell lung cancer. World J Surg Oncol.
15:1752017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu L, Ai J, Long H, Liu W, Wang X, Zuo Y,
Li Y, Wu Q and Deng Y: Integrative microRNA and gene profiling data
analysis reveals novel biomarkers and mechanisms for lung cancer.
Oncotarget. 7:8441–8454. 2016.PubMed/NCBI
|
|
30
|
Xiao X, Mruk DD, Lee WM and Cheng CY:
c-Yes regulates cell adhesion at the blood-testis barrier and the
apical ectoplasmic specialization in the seminiferous epithelium of
rat testes. Int J Biochem Cell Biol. 43:651–665. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bilal E, Alexe G, Yao M, Cong L, Kulkarni
A, Ginjala V, Toppmeyer D, Ganesan S and Bhanot G: Identification
of the YES1 kinase as a therapeutic target in basal-like breast
cancers. Genes Cancer. 1:1063–1073. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Seki T, Fujii G, Mori S, Tamaoki N and
Shibuya M: Amplification of c-yes-1 proto-oncogene in a primary
human gastric cancer. Jpn J Cancer Res. 76:907–910. 1985.PubMed/NCBI
|
|
33
|
Liu L, Yang J, Zhu X, Li D, Lv Z and Zhang
X: Long noncoding RNA H19 competitively binds miR-17-5p to regulate
YES1 expression in thyroid cancer. FEBS J. 283:2326–2339. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee SA, Kim JS, Park SY, Kim HJ, Yu SK,
Kim CS, Chun HS, Kim J, Park JT, Go D and Kim DK: miR-203
downregulates Yes-1 and suppresses oncogenic activity in human oral
cancer cells. J Biosci Bioeng. 120:351–358. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fang Z, Yin S, Sun R, Zhang S, Fu M, Wu Y,
Zhang T, Khaliq J and Li Y: miR-140-5p suppresses the
proliferation, migration and invasion of gastric cancer by
regulating YES1. Mol Cancer. 16:1392017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mohamed MS, Bishr MK, Almutairi FM and Ali
AG: Inhibitors of apoptosis: Clinical implications in cancer.
Apoptosis. 22:1487–1509. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Warren CFA, Wong-Brown MW and Bowden NA:
BCL-2 family isoforms in apoptosis and cancer. Cell Death Dis.
10:1772019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Julien O and Wells JA: Caspases and their
substrates. Cell Death Differ. 24:1380–1389. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Adams CM, Clark-Garvey S, Porcu P and
Eischen CM: Targeting the Bcl-2 family in B cell lymphoma. Front
Oncol. 8:6362019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lin L, Cheng K, Xie Z, Chen C, Chen L,
Huang Y and Liang Z: Purification and characterization a
polysaccharide from Hedyotis diffusa and its apoptosis inducing
activity toward human lung cancer cell line A549. Int J Biol
Macromol. 122:64–71. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu P, Cai X, Zhang W, Li Y, Qiu P, Lu D
and He X: Flavonoids of Rosa roxburghii Tratt exhibit
radioprotection and anti-apoptosis properties via the
Bcl-2(Ca(2+))/Caspase-3/PARP-1 pathway. Apoptosis. 21:1125–1143.
2016. View Article : Google Scholar : PubMed/NCBI
|