Introduction
Inflammatory bowel disease (IBD) is a term mainly
used to describe two autoimmune disorders which directly affect the
gastrointestinal tract: ulcerative colitis (UC) and Crohn's disease
(1). UC is a common form IBD
characterized by bloody purulent stool, recurrent diarrhea and
abdominal pain (2). The pathogenesis
of UC is complex with numerous genetic, immune, environmental and
psychological factors suggested to be involved (3). Dysregulation of immune response in the
intestine and increased secretion of proinflammatoy cytokines serve
a critical role in the pathogenesis of IBD (4). However, the precise etiology of IBD is
unknown (4). There is an enhanced
risk of developing colorectal cancer (CRC) in patients with
long-term IBD and in particular, it has been suggested that
patients with chronic UC carry a high risk of malignant
transformation IBD (5,6). It is estimated that patients with UC
are more than 30 times more likely to develop CRC and three times
more likely to succumb to CRC compared with the general population
(6).
Recent studies revealed that complex regulatory
networks are involved in monitoring and responding to alterations
in environmental conditions and physiological states (4,7,8). Within these regulatory networks,
microRNAs (miRs) can interact with downstream target genes, some of
which have been linked to cancer progression (9). A recent study demonstrated that
increased levels of miR-132 by aryl hydrocarbon receptor attenuate
tumorigenesis associated with chronic colitis (10). Furthermore, it was revealed that
miR-141 was involved in the pathogenesis of ulcerative colitis by
targeting C-X-C motif chemokine ligand 5 (11). Emerging evidence has previously
identified miRs that can be secreted from cells into the
extracellular environment, where they are stable and resistant to
degradation by RNases (12,13). Circulating miRs are therefore
desirable candidates as both endocrine signaling molecules and
disease markers (12,13).
A previous study demonstrated that high miR-372
expression is associated with synchronous liver metastasis in
patients with CRC (14). In
addition, serum miR-372 has been suggested to be a noninvasive
biomarker for the early detection and prognosis of CRC (15). However, the involvement of
circulating miR-372 in the progression of UC remains unknown. The
present study demonstrated that the level of miR-372 in peripheral
blood is increased in patients with UC. Furthermore, receiver
operating characteristic (ROC) analysis demonstrated that levels of
miR-372 detected in blood and tissue samples could be used to
screen for patients with UC from healthy controls. These results
revealed a potential role of that circulating miR-372 as a
noninvasive biomarker to distinguish the progression of ulcerative
colitis.
Materials and methods
Human tissue and blood samples
Colonic mucosa biopsies from the sigmoid colon of 50
patients with active UC and 50 healthy patients undergoing a
screening colonoscopy were obtained from the First Affiliated
Hospital of Zhejiang Chinese Medical University (Hangzhou, China)
between December 2015 and June 2016 (Table I). This study was approved by the
Ethics Committee at The First Affiliated Hospital of Zhejiang
Chinese Medical University and written informed consent was
obtained from each patient enrolled in the study. All procedures
were conducted in compliance with the approved guidelines of the
Ethics Committee. Pathological analysis further confirmed the
diagnoses of active UC. The diagnosis of UC was confirmed by
standard parameters as previously described (16). The site of disease was defined
according to the Montreal classification (17). The clinical disease activity was
assessed by the measurement of the Mayo score for UC. Endoscopies
were performed and graded according to the ulcerative colitis
endoscopic index of severity (UCEIS) scores for UC. Patients with
infectious colitis and colorectal cancer were excluded. Individuals
who had normal height and body mass index and no history of chronic
diseases were recruited for the healthy control group. Blood
samples for the measurement of high-sensitivity C-reactive protein
(CRP) were taken 1 week prior to or after endoscopy. CRP was
measured using a nephelometric method (18). In brief, human CRP reacted with the
corresponding antisera in the liquid phase to generate
antigen-antibody complexes and produce turbidity using a CRP
immunoturbidimetric assay kit [DiaSys Diagnostic Systems (Shanghai)
Co., Ltd., Shanghai, China] according to the manufacturer's
protocol. The turbidity was associated with the antigen content and
the CRP content in the sample was calculated by comparing the
samples with PBS using Detection system 1 Hitachi 7600-020
automatic biochemical analyzer (Hitachi, Ltd., Tokyo, Japan). In
healthy people, blood CRP levels are <5 mg/l (18). To avoid bias, all gastroenterologists
performing the endoscopies were unaware of the results from the
disease activity index.
 | Table I.Clinical characteristics of patients
with UC and healthy control patients. |
Table I.
Clinical characteristics of patients
with UC and healthy control patients.
| Characteristics | Patients with UC | Healthy control
patients |
|---|
| Patients (n) | 50 | 50 |
| Males [n (%)] | 25 (50) | 25 (50) |
| Age, years (mean ±
SD) | 46±15.4 | 43±16.3 |
| Disease duration,
months [median (range)] | 65.7
(25.6–173.2) | n/a |
Sample acquisition and RNA
isolation
From each patient, a 5 ml aliquot of blood was
collected directly into anticoagulation tubes containing ethylene
diamine tetraacetic acid. Total RNA was isolated from blood or
colonic mucosa tissue samples from patients with UC or healthy
controls using RNAVzol LS (Vigorous Biotechnology Beijing Co.,
Ltd., Beijing, China), according to the manufacturer's protocol.
The quantity and purity of RNA were measured using a NanoDrop
spectrophotometer (ND-1000; Nanodrop Technologies; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA (1 µg) was reverse transcribed into cDNA
using the Prime-Script One-Step RT-PCR kit (cat. no. C28025-032,
Invitrogen; Thermo Scientific, Inc.), according to the
manufacturer's protocol. qPCR was subsequently performed using
SYBR® Green Supermix (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) using an iCycler iQ real-time PCR detection
system. The following thermocycling conditions were used for the
qPCR: Initial denaturation at 95°C for 10 min; 50 cycles of 95°C
for 10 sec, 55°C for 10 sec, 72°C for 5 sec, 99°C for 1 sec, 59°C
for 15 sec and 95°C for 1 sec; and then cooled to 40°C. U6 was used
as an internal control. The relative mRNA expression levels were
calculated with the 2−∆∆Cq method (19) and experiments were performed in
triplicate. The primers used in the current study were listed as
follows: miR-372-RT,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGAATA-3′; U6-RT,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATG-3′; miR-372,
forward 5′-GCGCCCTCAAATGTGGAGCAC-3′; U6, forward
5′-GCGCGTCGTGAAGCGTTC-3′; universal reverse primer,
5′-GTGCAGGGTCCGAGGT-3′.
Cell culture
Human colon cancer cell line HT-29 and 293T cells
were obtained from the Cell Bank of Chinese Academy of Sciences
(Shanghai, China) and cultured in RPMI-1640 medium (HyClone; GE
healthcare, Chicago, IL, USA). HT-29 and 293T cells were seeded at
a density of 1.5 ×104 cells/cm2 and cultured
in Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% heat-inactivated fetal calf
serum (Invitrogen; Thermo Fisher Scientific, Inc.), streptomycin
(100 mg/ml; Thermo Fisher Scientific, Inc.), and penicillin (100
U/ml; Thermo Fisher Scientific, Inc.) and maintained at 37°C in a
5% CO2-humidified incubator.
Transient transfection
HT-29 or 293 cells were seeded in the six-well plate
at a density of 106 cells/well. The cells were transfected with
miR-372 mimic (CCUCAAAUGUGGAGCACUAUUCU), miR-372 inhibitor
(AGAATAGTGCTCCACATTTAGG) or negative control (NC,
UUCUCCGAACGUGUCACGU) for 48 h using Hiperfect Transfection Reagent
(Qiagen GmbH, Hilden, Germany) according to the manufacturer's
protocol. In brief, 12 µl Hiperfect Transfection Reagent was mixed
with 100 µl serum-free DMEM (Invitrogen; Thermo Fisher Scientific,
Inc.). Additionally, 10 µl miR-372 mimic, miR-372 inhibitor or NC
was mixed with serum-free DMEM. Then, the two mixtures were mixed
and incubated at room temperature for 15 min. Then, the mixture was
added to the six-well plate at a final miR concentration of 20
nM/well. Following transfection for 48 h, the cells were collected
for subsequent experiments.
Bioinformatics analysis and
dual-luciferase reporter assay
TargetScan software 7.2 (www.targetscan.org) was used to predict the putative
target genes of miR-372. The 3′untranslated region (3′UTR) of
NLRP12 was cloned into the pmirGLO plasmid. 293T cells were
co-transfected with miR-372 mimic (or NC) and pmirGLO-NLRP12-3′UTR
plasmid (or blank pmirGLO) using Vigofect transfection reagent
(Vigorous Biotechnology Beijing Co., Ltd.), according to the
manufacturer's protocol. In brief, 293 cells were seeded in a
six-well plate at a density of 106 cells/well. Following
this, 10 µl Vigofect transfection reagent was mixed with 100 µl
serum free DMEM to create a mixture. Then 10 µl miR-372 mimic or NC
and pmirGLO-NLRP12-3′UTR plasmid was mixed with the aforementioned
mixture at room temperature for 10 min. The final mixture was added
in the six-well plate at a final miR concentration of 20 nM/well.
After 48 h, the luciferase activity was detected using a Dual
Luciferase Reporter Assay System (Promega Corporation, Madison, WI,
USA).
Western blotting
After transfection with miR-372 mimic, miR-372
inhibitor or NC for 48 h, total proteins were isolated from HT-29
using a total protein extraction kit (Beijing Solarbio Science
& Technology Co., Ltd., Beijing, China). Then, the cell lysates
were centrifuged at 12,000 × g for 30 min at 4°C. To determine the
protein concentration, a BCA protein assay kit (Pierce; Thermo
Fisher Scientific, Inc.) was applied. Following this, 20 µg protein
per lane was separated using SDS-PAGE on a 12% gel, transferred
onto polyvinylidene difluoride membranes at 300 mA for 2 h. Then,
the membranes were blocked with 5% fat-free milk at room
temperature for 2 h. The following antibodies were incubated with
membranes overnight at 4°C: Anti-NLRP12 (cat. no. ab105409;
1:1,000; Abcam, Cambridge, UK) and anti-GAPDH (cat. no. 2118;
1:5,000; Cell Signaling Technology, Inc., Danvers, MA, USA) primary
antibodies. After washing with PBST three times (5 min/wash), the
membranes were incubated with horseradish peroxidase-conjugated
goat anti-rabbit immunoglobulin G (1:5,000; cat. no. ZB-2301;
Beijing Zhongshan Golden Bridge Biotechnology Co., Beijing, China)
for 2 h at room temperature. After washing with PBST three times (5
min/wash), the protein levels were determined using enhanced
chemiluminescence (EMD Millipore, Billerica, MA, USA) according to
the manufacturer's protocol. Signals were evaluated using a Super
ECL Plus Kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) and
quantitative analysis was performed using UVP software (UVP, LLC,
Phoenix, AZ, USA). GAPDH was used as an internal control. ImageJ
1.43b software (National Institutes of Health, Bethesda, MD, USA)
was used for densitometry analysis.
Statistical analysis
Data are presented as the mean ± standard deviation.
All statistical analyses were performed using SPSS software
(version 20.0; IBM Corp., Armonk, NY, USA). Student's t-test was
used for the comparisons of two groups. The use of miR-372 as a
biomarker to distinguish disease status was determined using ROC
analysis and the area under the curve was used to test
discriminative ability. Spearman's correlation coefficient was used
to measure the linear correlation between miR-372 expression and
serum CRP levels. P<0.05 was considered to indicate a
statistically significant difference.
Results
Peripheral blood miR-372 increases in
patients with UC
The expression level of miR-372 was significantly
increased in peripheral blood samples from patients with UC
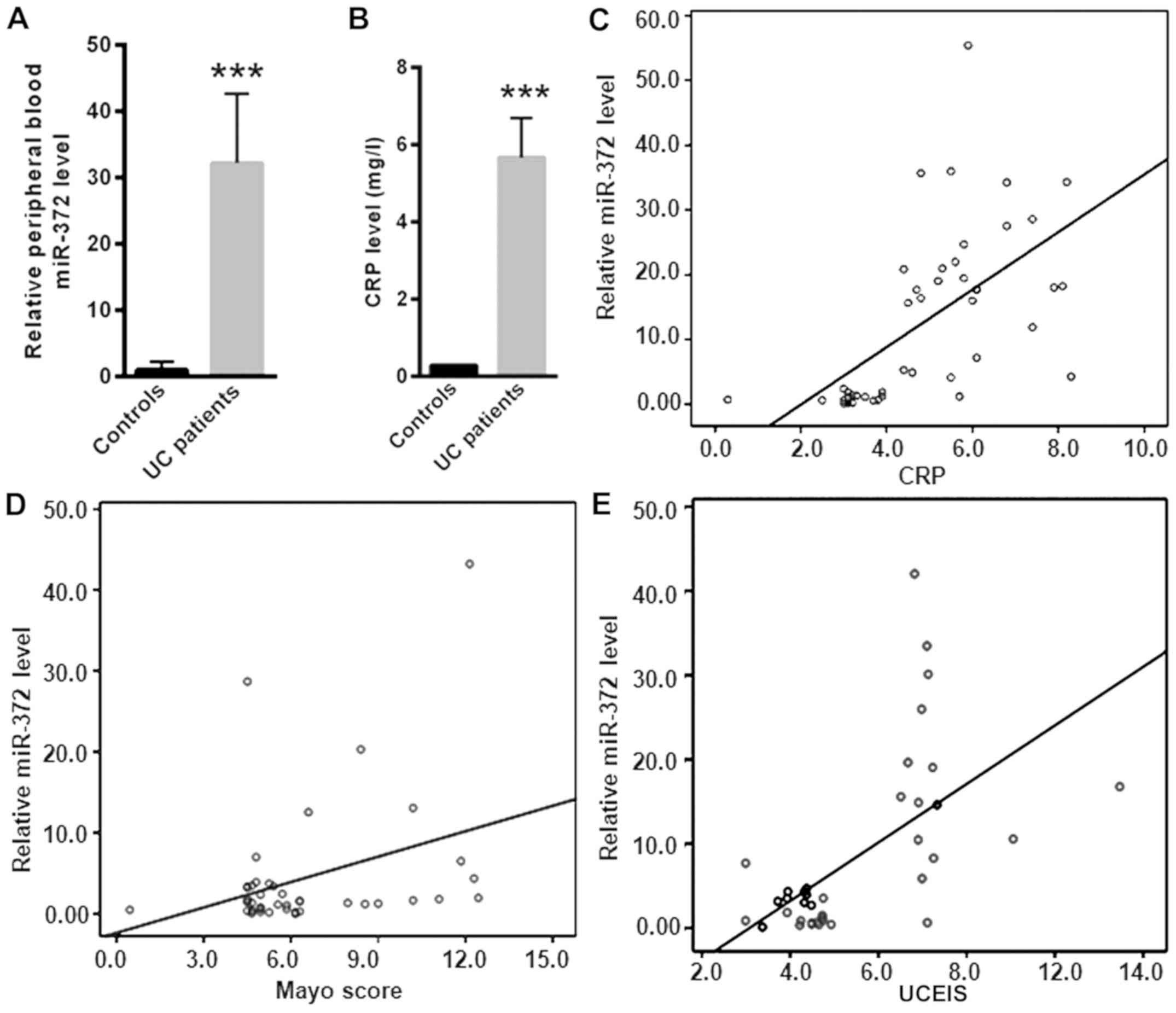
compared with healthy controls (P<0.001; Fig. 1A). In addition, serum CRP was
significantly increased in patients with UC (5.67±1.02 mg/l)
compared with healthy controls (0.28±0.03 mg/l; P<0.001;
Fig. 1B). To identify the level of
serum miR-372 expression with potential diagnostic value, the
correlation of serum miR-372 with CRP, the Mayo score and UCEIS in
patients with UC was analyzed (Fig.
1C-E). This study revealed that the level of serum miR-372 had
a positive correlation with CRP (r=0.592; P<0.001), the Mayo
score (r=0.604; P<0.001) and UCEIS (r=0.338; P<0.001) in
patients with UC. The positive correlation with these indicators of
disease activity in UC suggests that the serum miR-372 level is
associated with UC disease severity.
miR-372 increased in colonic mucosa
tissue samples from patients with UC
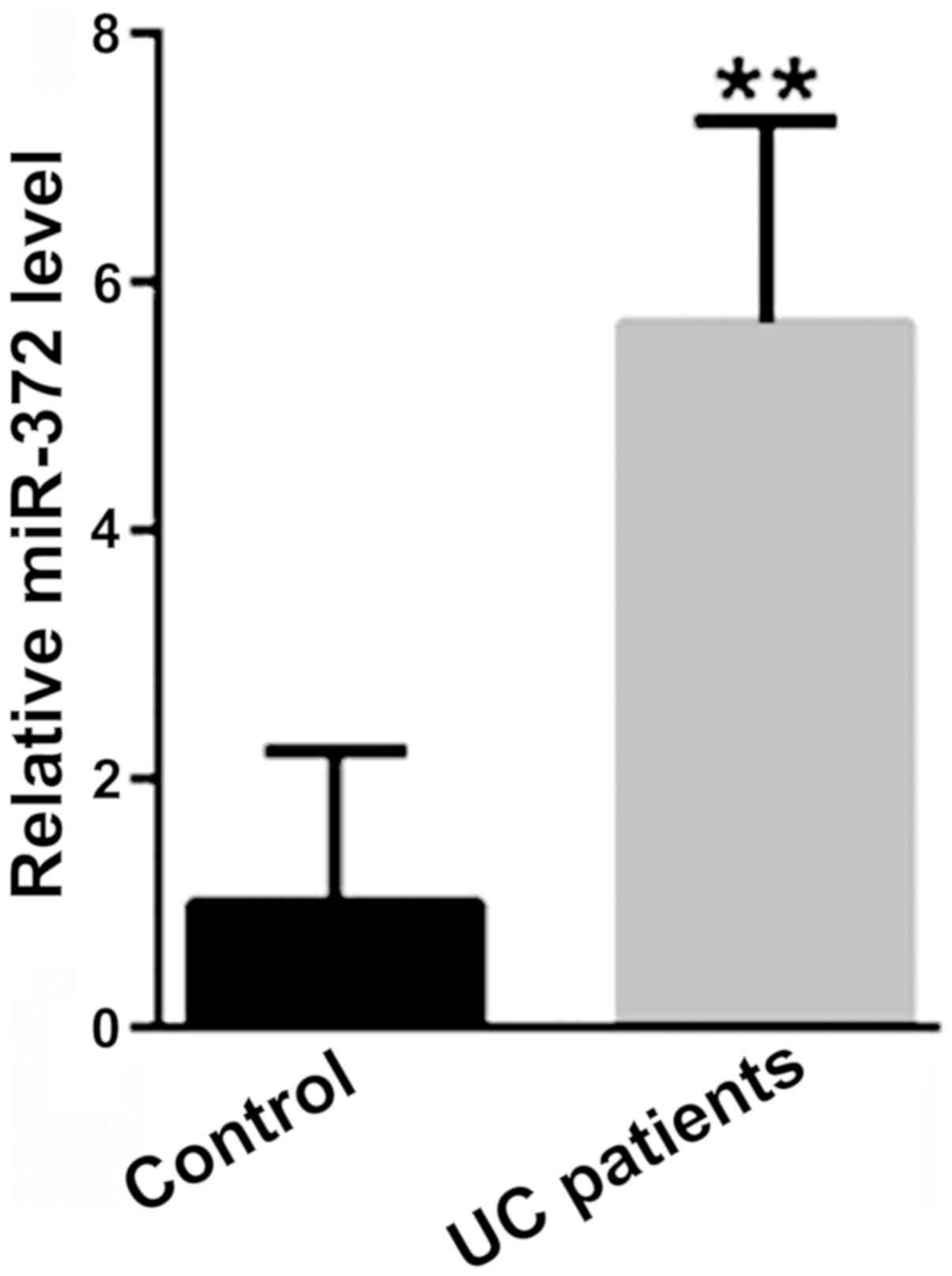
The mRNA expression level of miR-372 was
significantly increased in colonic mucosa tissue samples from
patients with UC, compared with tissue samples from healthy
controls (P<0.01; Fig. 2).
miR-372 as a potential biomarker for
UC
To examine the feasibility of using miR-372 as a
diagnostic marker, blood and tissue samples were collected from
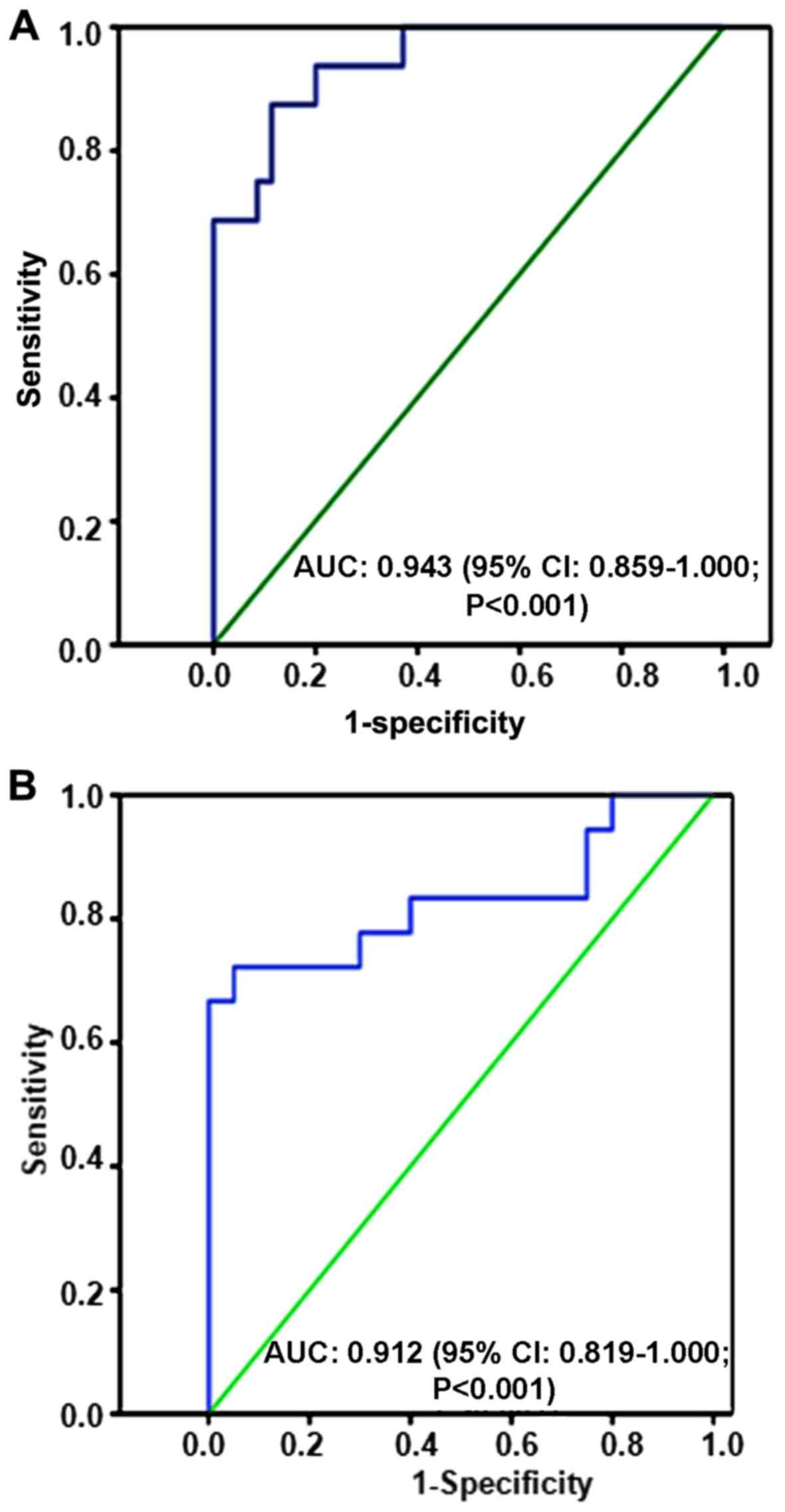
patients with UC and healthy controls. ROC analyses indicated that
the serum miR-372 level had a predictive power of 0.943 (95%
confidence interval: 0.859–1.000; P<0.001; Fig. 3A) for distinguishing potential
patients with UC from the healthy control group. Furthermore, ROC
analyses indicated that the tissue miR-372 level had a predictive
power of 0.912 (95% confidence interval: 0.819–1.000; P<0.001;
Fig. 3B) for distinguishing UC
patients from the healthy control group.
NLRP12 is a novel target gene of
miR-372
To investigate miR-372 and its involvement in the
progression of UC, TargetScan was used to identify potential
targets of miR-372. TargetScan identified a conserved miR-372
binding site in the 3′UTR of NLRP12 (Fig. 4A), which was previously suggested to
attenuate the progression of UC. The dual luciferase assay
demonstrated that miR-372 overexpression significantly suppressed
the relative luciferase activity of pmirGLO-NLRP12-3′UTR compared
with control pmirGLO (P<0.001; Fig.
4B). The effect of miR-153 on RUNX2 expression was examined in
human colon cancer cell line HT-29 following transfection with
either miR-372 mimic or miR-NC. The mRNA expression level of
miR-372 significantly increased in human colon cancer cells
transfected with miR-372 mimic compared with NC (P<0.001;
Fig. 4C). Overexpression of miR-372
significantly decreased the protein expression level of NLRP12
(P<0.01; Fig. 4D). To further
understand the effect of miR-372, HT-29 cells were transfected with
either miR-372 inhibitor or miR-NC. The mRNA expression level of
miR-372 significantly decreased in human colon cancer cells
transfected with miR-372 inhibitor compared with NC (P<0.001;
Fig. 4E). Knockdown of miR-372
significantly increased the protein expression level of NLRP12
(P<0.01; Fig. 4F).
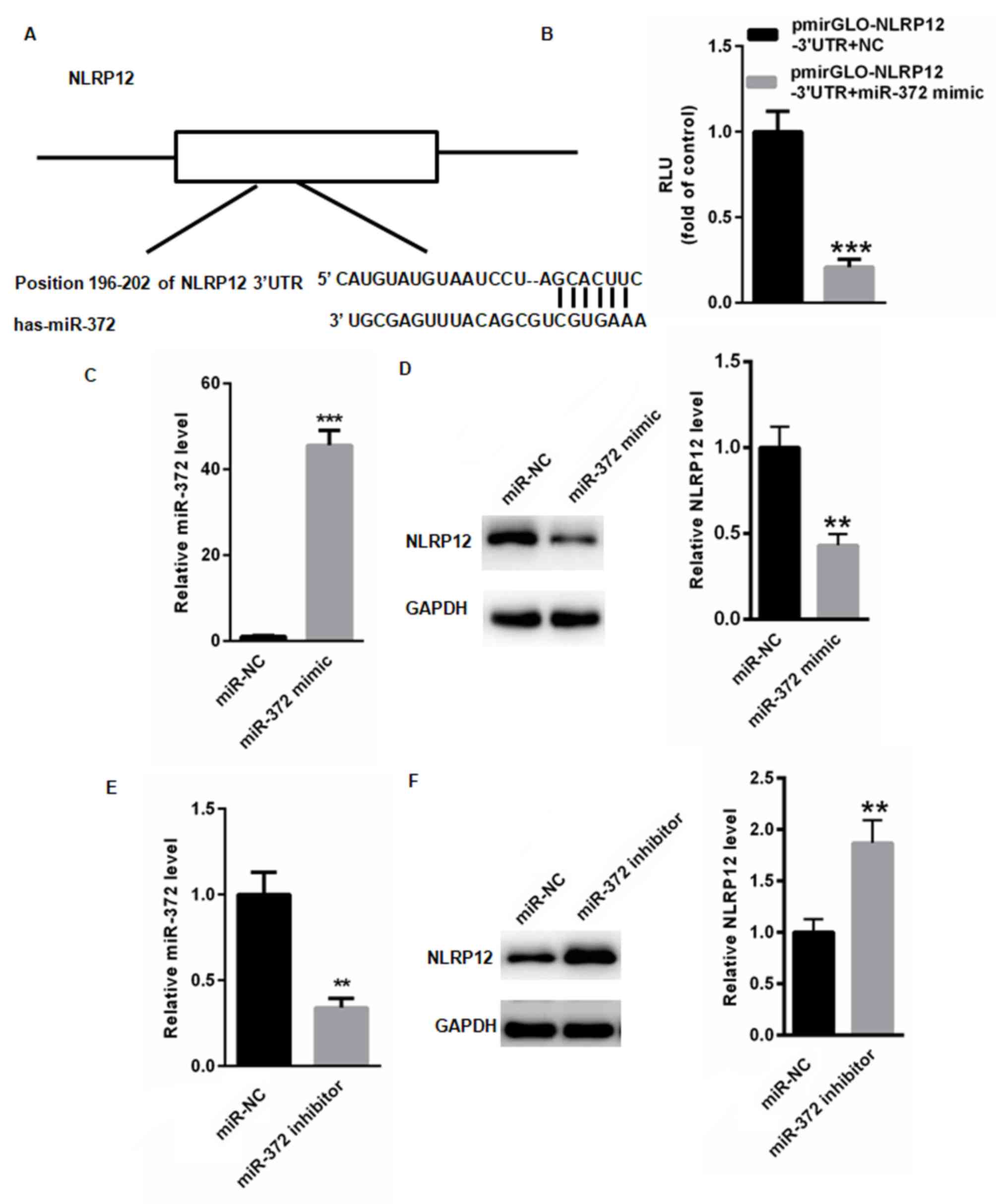 | Figure 4.NLRP12 is a novel target gene of
miR-372. (A) Bioinformatics analysis was used to identify a
conserved miR-372 binding site in NLRP12. The structure represents
the 3′UTR region of NLPR12 and possible binding bases between
miR-372 and the 3′UTR of NLPR12. (B) miR-372 mimic and pmirGLO or
pmirPLO-NLRP12-3′UTR were transiently transfected into 293T cells,
respectively, and the relative luciferase activity of
pmirGLO-NLRP12-3′UTR was measured relative to control. miR-372
mimic and miR-NC were transiently transfected into HT-29 cells,
respectively. (C) The mRNA expression level of miR-372 was detected
by RT-qPCR. (D) The protein expression level of NLRP12 was
determined by western blotting. miR-372 inhibitor and miR-NC were
transiently transfected into HT-29 cells, respectively. (E) The
mRNA expression level of miR-372 was detected by RT-qPCR. (F) The
protein expression level of NLRP12 was determined by western
blotting. **P<0.01 and ***P<0.001 vs. control. NLRP12, NLR
family pyrin domain containing 12; HT-29, human colon cancer cell
line; miR-NC, HT-29 cells transfected with scramble miR; miR-372
mimic, HT-29 cells transfected with miR-372 mimic; miR-NC, HT-29
cells transfected with miR-NC; miR-372 inhibitor, HT-29 cells
transfected with miR-372 inhibitor; miR-372 + pmirGLO, 293T cells
co-transfected with miR-372 mimic and pmirGLO; miR-372 +
pmirGLO-NLRP12-3′UTR, 293T cells co-transfected with miR-372 mimic
and pmirGLO-NLRP12-3′UTR; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; miR,
microRNA. |
Discussion
High sensitivity CRP is a common non-specific marker
of inflammation that can be elevated in patients with active IBD
(20,21). However, CRP can be challenging to use
as a biomarker due to its low specificity and high expression
heterogeneity (22,23). It is difficult to screen and monitor
the progression of UC and therefore necessary to invest the use of
other novel noninvasive biomarkers for patients with UC (24). Increasing evidence suggests that miRs
are involved in the progression of a number of diseases, which
include cancer and inflammatory diseases (25,26).
The present study demonstrated that levels of
miR-372 were significantly increased in the peripheral blood and
colonic mucosa tissue samples from patients with UC, compared with
healthy controls. Furthermore, the level of miR-372 in circulation
was positively correlated with serum CRP levels. ROC analysis
demonstrated that levels of miR-372 detected in both peripheral
blood and colonic mucosa tissue samples could be used to screen for
patients with UC from healthy controls. These results demonstrated
a potential role of miR-372 as a diagnostic marker and therapeutic
target for patients with UC.
The abnormal activation of inflammatory responses is
a hallmark of UC (27,28). The nucleotide-binding domain
leucine-rich repeat proteins are important regulators of
inflammatory and innate immune response, exerting pro- or
anti-inflammatory functions in the development and progression of
UC (29,30). Studies have revealed that NLRP12
negatively regulates inflammatory signaling by suppressing both
canonical and non-canonical NF-κB signaling pathways, as well as
regulating gut microbial communities (31–33).
IBD-profiling studies indicated that NLRP12 expression is
negatively correlated with active UC (33,34).
Furthermore, an imbalance in the intestinal microbiota, or
dysbiosis serves a key role in IBD pathogenesis (35–37).
There is a complex cause-effect association between intestinal
microbial diversity and human disease (38). In human metabolic and inflammatory
disorders including IBD, a reduction in gut microbiome richness and
diversity can be used as a biomarker for disease (39,40). A
recent study demonstrated that NLRP12 attenuates excessive
inflammatory cytokine production to limit intestinal inflammation
by maintaining colonic microbial diversity and promoting protective
commensal bacterial growth (33).
The current study identified NLRP12 as a novel
target gene of miR-372. Dual luciferase assay demonstrated that
overexpression of miR-372 significantly reduced the relative
luciferase activity of pmirGLO-NLRP12-3′UTR compared with control
pmirGLO. In addition, western blot analysis indicated that
overexpression of miR-372 significantly decreased the protein
expression level of NLRP12. Therefore, it was hypothesized that
miR-372 may promote the progression of UC by suppressing NLRP12
expression, thereby inducing the production of excessive
inflammatory cytokines.
In conclusion, the current study demonstrated that
levels of miR-372 detected in peripheral blood were significantly
increased in patients with UC compared with healthy controls.
Furthermore, circulating miR-372 was positively correlated with
serum CRP levels. High levels of miR-372 detected in peripheral
blood samples may serve as a potential biomarker to screen patients
with UC from healthy control patients.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the First
Affiliated Hospital of Zhejiang Chinese Medical University
(Hangzhou, China; grant no. ZJCMU-201609).
Availability of data and materials
All datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MS performed the experiments and analyzed the data.
LM designed the study, analyzed the data and gave final approval
for the version to be published.
Ethical approval and consent to
participate
The current study was approved by The Ethics
Committee at the First Affiliated Hospital of Zhejiang Chinese
Medical University (Hangzhou, China).
Patient consent for publication
Informed consent for participation in the study or
use of their tissue was obtained from all participants and all
patients were consent for publication of this study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bernstein CN, Blanchard JF, Kliewer E and
Wajda A: Cancer risk in patients with inflammatory bowel disease: A
population-based study. Cancer. 91:854–862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haberman Y, Karns R, Dexheimer PJ,
Schirmer M, Somekh J, Jurickova I, Braun T, Novak E, Bauman L,
Collins MH, et al: Ulcerative colitis mucosal transcriptomes reveal
mitochondriopathy and personalized mechanisms underlying disease
severity and treatment response. Nat Commun. 10:382019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hoving JC: Targeting IL-13 as a
Host-Directed Therapy Against Ulcerative Colitis. Front Cell Infect
Microbiol. 8:3952018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen JH, Andrews JM, Kariyawasam V, Moran
N, Gounder P, Collins G, Walsh AJ, Connor S, Lee TW, Koh CE, et al:
Review article: Acute severe ulcerative colitis-evidence-based
consensus statements. Aliment Pharmacol Ther. 44:127–144. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Akao Y, Nakagawa Y and Naoe T: let-7
microRNA functions as a potential growth suppressor in human colon
cancer cells. Biol Pharm Bull. 29:903–906. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kanaan Z, Rai SN, Eichenberger MR, Barnes
C, Dworkin AM, Weller C, Cohen E, Roberts H, Keskey B, Petras RE,
et al: Differential microRNA expression tracks neoplastic
progression in inflammatory bowel disease-associated colorectal
cancer. Hum Mutat. 33:551–560. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hutchings HA, Alrubiay L, Watkins A,
Cheung WY, Seagrove AC and Williams JG: Validation of the Crohn's
and Ulcerative Colitis questionnaire in patients with acute severe
ulcerative colitis. United European Gastroenterol J. 5:571–578.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Leake I: IBD: Treatment for acute severe
ulcerative colitis. Nat Rev Gastroenterol Hepatol. 13:4362016.
View Article : Google Scholar
|
|
9
|
Tili E, Michaille JJ, Piurowski V, Rigot B
and Croce CM: MicroRNAs in intestinal barrier function,
inflammatory bowel disease and related cancers-their effects and
therapeutic potentials. Curr Opin Pharmacol. 37:142–150. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alzahrani AM, Hanieh H, Ibrahim HM,
Mohafez O, Shehata T, Bani Ismail M and Alfwuaires M: Enhancing
miR-132 expression by aryl hydrocarbon receptor attenuates
tumorigenesis associated with chronic colitis. Int Immunopharmacol.
52:342–351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai M, Chen S and Hu W: MicroRNA-141 Is
Involved in Ulcerative Colitis Pathogenesis via Aiming at CXCL5. J
Interferon Cytokine Res. 37:415–420. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matsuzaki J and Suzuki H: Circulating
microRNAs as potential biomarkers to detect transformation of
Barrett's oesophagus to oesophageal adenocarcinoma. BMJ Open
Gastroenterol. 4:e0001602017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Matsuzaki J and Ochiya T: Circulating
microRNAs and extracellular vesicles as potential cancer
biomarkers: A systematic review. Int J Clin Oncol. 22:413–420.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamashita S, Yamamoto H, Mimori K, Nishida
N, Takahashi H, Haraguchi N, Tanaka F, Shibata K, Sekimoto M, Ishii
H, et al: MicroRNA-372 is associated with poor prognosis in
colorectal cancer. Oncology. 82:205–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu J, Jin L, Jiang L, Gao L, Zhou J, Hu Y,
Li W, Zhi Q and Zhu X: Serum miR-372 is a diagnostic and prognostic
biomarker in patients with early colorectal cancer. Anticancer
Agents Med Chem. 16:424–431. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Conrad K, Roggenbuck D and Laass MW:
Diagnosis and classification of ulcerative colitis. Autoimmun Rev.
13:463–466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kocsis D, Toth Z, Csontos AA, Miheller P,
Pák P, Herszényi L, Tóth M, Tulassay Z and Juhász M: Prevalence of
inflammatory bowel disease among coeliac disease patients in a
Hungarian coeliac centre. BMC Gastroenterol. 15:1412015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hanafy AS, Monir MH, Abdel Malak H and
Desoky Aiad M: A simple noninvasive score predicts disease activity
and deep remission in ulcerative colitis. Inflamm Intest Dis.
3:16–24. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Asadzadeh-Aghdaee H, Shahrokh S,
Norouzinia M, Hosseini M, Keramatinia A, Jamalan M, Naghibzadeh B,
Sadeghi A, Jahani Sherafat S and Zali MR: Introduction of
inflammatory bowel disease biomarkers panel using protein-protein
interaction (PPI) network analysis. Gastroenterol Hepatol Bed
Bench. 9 (Suppl 1):S8–S13. 2016.PubMed/NCBI
|
|
21
|
Barnes EL and Burakoff R: New biomarkers
for diagnosing inflammatory bowel disease and assessing treatment
outcomes. Inflamm Bowel Dis. 22:2956–2965. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pepys MB and Hirschfield GM: C-reactive
protein: A critical update. J Clin Invest. 111:1805–1812. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fengming Y and Jianbing W: Biomarkers of
inflammatory bowel disease. Dis Markers. 2014:7109152014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Starr AE, Deeke SA, Ning Z, Chiang CK,
Zhang X, Mottawea W, Singleton R, Benchimol EI, Wen M, Mack DR and
Stintzi A: Proteomic analysis of ascending colon biopsies from a
paediatric inflammatory bowel disease inception cohort identifies
protein biomarkers that differentiate Crohn's disease from UC. Gut.
66:1573–1583. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang H, Chao K, Ng SC, Bai AH, Yu Q, Yu J,
Li M, Cui Y, Chen M, Hu JF and Zhang S: Pro-inflammatory miR-223
mediates the cross-talk between the IL23 pathway and the intestinal
barrier in inflammatory bowel disease. Genome Biol. 17:582016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pierdomenico M, Cesi V, Cucchiara S,
Vitali R, Prete E, Costanzo M, Aloi M, Oliva S and Stronati L: NOD2
is regulated By Mir-320 in physiological conditions but this
control is altered in inflamed tissues of patients with
inflammatory bowel disease. Inflamm Bowel Dis. 22:315–326. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krugliak Cleveland N, Rubin DT, Hart J,
Weber CR, Meckel K, Tran AL, Aelvoet AS, Pan I, Gonsalves A,
Gaetano JN, et al: Patients With Ulcerative Colitis and Primary
Sclerosing Cholangitis Frequently Have Subclinical Inflammation in
the Proximal Colon. Clin Gastroenterol Hepatol. 16:68–74. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nairn RC, Savvas R, Hocking G, Kovala M
and Rolland JM: Ulcerative colitis. Animal model: Immunoreactive
inflammation in fetal colon implants in syngeneic adult rats. Am J
Pathol. 96:647–650. 1979.PubMed/NCBI
|
|
29
|
Al Nabhani Z, Montcuquet N, Roy M,
Dussaillant M, Hugot JP and Barreau F: Complementary Roles of Nod2
in hematopoietic and nonhematopoietic cells in preventing gut
barrier dysfunction dependent on MLCK Activity. Inflamm Bowel Dis.
23:1109–1119. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang W, Wang X, Zeng B, Liu L, Tardivel
A, Wei H, Han J, MacDonald HR, Tschopp J, Tian Z and Zhou R:
Recognition of gut microbiota by NOD2 is essential for the
homeostasis of intestinal intraepithelial lymphocytes. J Exp Med.
210:2465–2476. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Allen IC, Wilson JE, Schneider M, Lich JD,
Roberts RA, Arthur JC, Woodford RM, Davis BK, Uronis JM, Herfarth
HH, et al: NLRP12 suppresses colon inflammation and tumorigenesis
through the negative regulation of noncanonical NF-κB signaling.
Immunity. 36:742–754. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zaki MH, Vogel P, Malireddi RK,
Body-Malapel M, Anand PK, Bertin J, Green DR, Lamkanfi M and
Kanneganti TD: The NOD-like receptor NLRP12 attenuates colon
inflammation and tumorigenesis. Cancer Cell. 20:649–660. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen L, Wilson JE, Koenigsknecht MJ, Chou
WC, Montgomery SA, Truax AD, Brickey WJ, Packey CD, Maharshak N,
Matsushima GK, et al: NLRP12 attenuates colon inflammation by
maintaining colonic microbial diversity and promoting protective
commensal bacterial growth. Nat Immunol. 18:541–551. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lukens JR, Gurung P, Shaw PJ, Barr MJ,
Zaki MH, Brown SA, Vogel P, Chi H and Kanneganti TD: The NLRP12
sensor negatively regulates autoinflammatory disease by modulating
interleukin-4 production in T Cells. Immunity. 42:654–664. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gevers D, Kugathasan S, Denson LA,
Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song
SJ, Yassour M, et al: The treatment-naive microbiome in new-onset
Crohn's disease. Cell Host Microbe. 15:382–392. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sartor RB and Wu GD: Roles for intestinal
bacteria, viruses, and fungi in pathogenesis of inflammatory bowel
diseases and therapeutic approaches. Gastroenterology. 152:327–339,
e324. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Frank DN, St Amand AL, Feldman RA,
Boedeker EC, Harpaz N and Pace NR: Molecular-phylogenetic
characterization of microbial community imbalances in human
inflammatory bowel diseases. Proc Natl Acad Sci USA.
104:13780–13785. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jacob V, Crawford C, Cohen-Mekelburg S,
Viladomiu M, Putzel GG, Schneider Y, Chabouni F, O'Neil S, Bosworth
B, Woo V, et al: Single delivery of high-diversity fecal microbiota
preparation by colonoscopy is safe and effective in increasing
microbial diversity in active ulcerative colitis. Inflamm Bowel
Dis. 23:903–911. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Le Chatelier E, Nielsen T, Qin J, Prifti
E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy
S, et al: Richness of human gut microbiome correlates with
metabolic markers. Nature. 500:541–546. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ridaura VK, Faith JJ, Rey FE, Cheng J,
Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et
al: Gut microbiota from twins discordant for obesity modulate
metabolism in mice. Science. 341:12412142013. View Article : Google Scholar : PubMed/NCBI
|