Introduction
Helicobacter pylori (H. pylori) was
originally isolated from the gastric mucosa of patients with
chronic active gastritis in 1983 with further study revealing that
the pathogen itself caused the condition (1). H. pylori infects >50% of the
world's population (2) and it
frequently causes chronic active gastritis, gastroduodenal ulcers
(3) and gastric cancer (4). H. pylori is occasionally
associated with functional dyspepsia, unexplained iron deficiency
anemia (5) and idiopathic
thrombocytopenic purpura (6). The
standard triple therapy used to treat H. pylori is a proton
pump inhibitor (PPI) in conjunction with a 7–10 day course of two
antibiotics. This protocol eradicates H. pylori infection in
~80% of patients (7). Bismuth-based
quadruple therapies, including clarithromycin, amoxicillin and
metronidazole, are another alternative treatment (8,9).
However, some strains of H. pylori demonstrate
antibiotic-resistance, which complicates treatment. In addition,
the neurotoxicity of bismuth limits its use in the elderly and
children (10). Copious use of
antibiotics is not only accompanied by a variety of side effects,
but also increases the risk of antibiotic resistance. Therefore,
the development of safer and more effective new therapeutic agents
targeting H. pylori is an important focus of current
research. Recent studies have demonstrated that administration of
oral probiotics is greatly beneficial for the treatment of H.
pylori infection (11–13). The substances produced by probiotics
significantly inhibit VacA and flaA virulence genesin H.
pylori (12). Use of probiotics
alongside drugs often reduces the drug side effects and attenuates
the gastric mucosal inflammation (14,15). It
has been reported that Lactobacillus salivarius (L. salivarius),
Lactobacillus gasseri, Lactobacillus casei Shirota, Lactobacillus
Johnsonii La1, Lactobacillus rhamnosus GG and Saccharomyces
boulardii have either anti-H. pylori activities or
anti-inflammatory properties (16,17). The
antagonistic activities of probiotics against H. pylori are
strain-specific; however, their mechanisms of action remain unclear
(18).
Currently, mixed strain probiotics are the
most-widely studied (8). Different
strains of probiotics may possess synergetic or antagonistic
effects. In the present study, the anti-H. pylori activities
of Lactobacillus acidophilus (L. acidophilus), L. salivarius,
Clostridium butyricum (C. butyricum), Bacillus licheniformis (B.
licheniformis), Bifidobacterium infantis (B. infantis),
Bifidobacterium longum (B. longum) and Lactobacillus
bulgaricus (L. bulgaricus) were evaluated. These probiotic
strains are widely used in clinical settings to treat diarrhea and
have been reported to improve the eradication rate of H.
pylori in some clinical cases due to the secretion of
antibacterial substances including lactic acid, acetic acid and
hydrogen peroxide (19,20). The present study objective was to
evaluate the probiotic mechanisms of action against H.
pylori with the results potentially providing a new theoretical
basis for eradicating H. pylori.
Materials and methods
Bacterial strains and culture
conditions
The probiotic strains used were as follows: L.
acidophilus, L. salivarius, C. butyricum, B. licheniformis, B.
infantis, B. longum and L. bulgaricus, purchased from
Siliankang, Jinshuangqi, Changlekang and Taiwan Yaxin (Table I). The probiotics were cultured on De
Man, Rogosa and Sharpe agar plates (MRS; Oxoid; Thermo Fisher
Scientific, Inc.) at 37°C in an anaerobic humidified environment
for 24 h. Live bacteria cells were obtained by centrifuging the
cultures and washing with sterile PBS three times. The precipitate
were resuspended with RPMI 1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) and adjusted to 3×108 colony-forming
units (CFU)/ml.
 | Table I.Details of probiotic strains. |
Table I.
Details of probiotic strains.
| Species | Strain | Product |
|---|
| Lactobacillus
acidophilus | CGMCC0460.2 | Siliankang |
| Bifidobacterium
infantis | CGMCC0460.1 | Siliankang |
| Bifidobacterium
longum | NQ1501 | Jinshuangqi |
| Lactobacillus
bulgaricus | NQ2508 | Jinshuangqi |
| Clostridium
butyricum | CGMCC
N0.0313-1 | Changlekang |
| Bacillus
licheniformis | CMCC63516 |
Zhengchangsheng |
| Lactobacillus
salivarius | ATCC11741 | Taiwan Yaxin |
H. pylori ATCC43504 strain was purchased from
the American Type Culture Collection and the multidrug resistant
H. pylori strain (HP021) was isolated from the gastric
biopsy of a patient with chronic atrophic gastritis and confirmed
to have resistance to clarithromycin and levofloxacin. H.
pylori ATCC43504 strain expresses CagA and VacA proteins, which
induces nuclear factor (NF)-κB and interleukin (IL)-8 expression
(21). Both H. pylori strains
were grown on Columbia blood agar plates supplemented with 7%
defibrinated horse blood (Beijing Biotek Medical Device, Ltd.) for
at least 3 days at 37°C under microaerophilic conditions (5%
O2, 10% CO2, and 85% N2). Normal
human gastric epithelial GES-1 cells (American Type Culture
Collection) were cultured at 37°C in RPMI 1640 medium supplemented
with 10% fetal bovine serum (Biological Industries) and 100 µg/ml
penicillin/streptomycin in a humidified atmosphere with 5%
CO2. The medium was changed every other day. Before
experiments were initiated, the cells were plated in 96-well plates
at 5×103 cells/well or 2.5×105 cells/well in
6-well plates for 24 h in serum-free RPMI-1640 medium.
Cell-free supernatant (CFS)
preparation and H. pylori ATCC43504–lipopolysaccharide (LPS)
culture
Cultures of the various probiotic strains were grown
in MRS broth (Oxoid; Thermo Fisher Scientific, Inc.) in an
anaerobic humidified environment at 37°C for 96 h. CFS from the
probiotics was prepared by centrifuging (12,000 × g; 4°C; 10 min)
the respective MRS broths. CFS were filtered through a 0.2 µM
filter and stored at −20°C.
Culture broth from H. pylori ATCC43504 was
centrifuged (8,000 × g; 4°C; 10 min) and washed three times then
resuspended in PBS. H. pylori lipopolysaccharide (HP-LPS)
was obtained using an LPS extraction kit (Intron Biotechnology,
Inc.) according to the manufacturer's instructions. HP-LPS
concentrations were determined with a kinetic Limulus Amebocyte
Lysate Assay kit (Xiamen Limulus Reagent Biotechnology Co.,
Ltd.).
GES-1 cell viability
Probiotics and CFS are toxic to GES-1 cells at high
concentrations and during long incubations (17). To determine the optimal concentration
and incubation time for each probiotic and CFS, GES-1 cells were
infected with CFS and probiotics at multiplicities of infection
(MOI) of 100 and 1,000 in antibiotic-free RPMI 1640 medium at 37°C,
5% CO2 for up to 8 h. Viable GES-1 cell numbers were
determined by trypan blue staining following incubation for 2, 4, 6
and 8 h at 37°C. Non-infected cell cultures served as controls.
Cells that were not stained with trypan blue were counted as viable
cells.
GES-1 proliferation
GES-1 cells were seeded into 96-well plates at
1.5×105 cells/ml in 100 µl culture volume for 24 h. CFS
(100 µl) and probiotics were then added to the GES-1 cells at
102, 103, 104, 105,
106, 107, 108 and 109
CFU/ml. Each condition was performed in triplicate and cultured for
24 h. Cells were washed three times with sterile PBS solution then
20 µl of 4 mg/ml MTT reagents was added. The cells were cultured
for another 4 h and then centrifuged for 5 min at 800 × g. The
supernatant was discarded and 150 µl dimethyl sulfoxide was added.
Absorbance was measured at 490 nm.
Assessment of adhesion ability
Adhesion ability of probiotics on GES-1 cells was
determined as previously described by de Klerk et al
(22). In brief, probiotics were
suspended in RPMI-1640 medium to a concentration of
3×108 CFU/ml. GES-1 cells cultured on slides in 6-well
plates were infected with probiotics at a MOI of 100. Following 4 h
of incubation, each well was washed three times with sterile PBS to
remove any unbound bacteria. Cells were fixed with 4%
paraformaldehyde at room temperature for 30 min and then the
bacteria were Gram stained at room temperature for 5min. The cell
monolayer was washed with tap water and the cells were observed at
a magnification of ×1,000 under light microscope oil immersion. The
adherence of probiotics was calculated as previously described
(23,24) and was as follows: Cell adhesion
index=number of adhering probiotics/total number of cells in the
field of view ×100. Cell adhesion rate=number of cells adhered by
probiotics/total number of cells in the field of view ×100. Cell
adhesion index indicates the number of probiotics adhering to each
cell, and cell adhesion rate represents the proportion of cells
adhered by probiotics in total cells.
Inhibition of H. pylori growth
H. pylori ATCC43504 and HP021 were evenly
seeded on Columbia agar plates without antibiotics. Holes were
introduced to the agar plates with a sterile oxford cup then the
bottom of holes were sealed with 0.8% agar liquid. Live probiotics
(120 µl at 3×109 CFU/ml) were suspended in MRS broth and
CFS (120 µl) was added to the holes in the plates. The plates were
incubated under microaerophilic conditions for 72 h at 37°C, and
then the diameters of the inhibition zones were measured. PBS and
MRS medium were used as negative controls.
Adhesion of H. pylori to GES-1
cells
For the infection studies, GES-1 cells were grown on
microtiter plates to form a confluent monolayer. The concentration
of each probiotic was adjusted to 1.5×107 CFU/ml. H.
pylori ATCC43504 and HP021 concentrations were then adjusted to
1.5×107 CFU/ml. GES-1 cells were pre-treated with 50 µl
of live probiotics for 2 h before infection (pre-treated group) or
following infection (post-treated group). GES-1 cells were infected
for 2 h with 50 µl of live H. pylori ATCC43504 or HP021.
Subsequently, each well was washed three times to remove any
non-adherent H. pylori. Urease activity was determined using
a modified phenol red method (18,25,26). In
brief, 200 µl of urease test solution (20% [w/v] urea and 0.012%
phenol red in phosphate buffer; pH 7.0) was added to each well of a
microtiter plate. The plate was then incubated at 37°C for 1 h. The
absorbance at 550 nm was measured with a microtiter plate
spectrophotometer (BioTek Instruments, Inc.). The adherence of
H. pylori was calculated as described by Chen et al
(18) and was as follows:
Adherence=([Optical density experimental-optical density
negative]/[optical density positive-optical density negative]
×100.
The negative control contained only GES-1 cells and
the positive control contained both GES-1 cells and H.
pylori, which were used to establish 100% adherence.
ELISA for interleukin (IL)-8
detection
LPS is a pathogenic factor of H. pylori which
can induce GES-1 cells to produce IL-8 (27). Probiotics with proven adhesive
ability were added to GES-1 cells (MOI of 100) and incubated for 2
h. Following washing with PBS to remove bacilli, HP-LPS (final
concentration of 7,000 endotoxin units [EU]/ml) was added and the
cells were incubated for another 6 h. The final culture
supernatants were centrifuged for 10 min at 12,000 × g and 4°C to
remove bacteria and cell debris. Supernatants were then aliquoted
and stored at −80°C. IL-8 concentration in the supernatant was
determined using a commercially available ELISA kit (cat. no.
VAL103; R&D Systems, Inc.). Absorbance values were measured at
450 nm using a microplate reader. Each sample was measured three
times.
Preparation of cellular lysates and
western blot analysis
GES-1 cells were pre-treated for 2 h with
probiotics, followed by HP-LPS stimulation for 60 or 120 min at a
final concentration of 7,000 EU/ml. Cytoplasmic and nuclear
extracts were then isolated using a Nuclear Extract kit (Active
Motif, Inc.). Protein concentrations were determined with an
enhanced bicinchoninic acid protein assay kit.
Each extract was mixed with ×5 loading buffer and
boiled for 5 min. The extracted proteins were then aliquoted and
stored at −80°C. A total of 30 µg of protein was loaded into each
lane, separated via 8% SDS-PAGE and then electrotransferred to a
polyvinylidene difluoride membrane. Membranes were blocked at room
temperature with 5% fat-free dried milk in Tris buffered saline
containing 0.1% Tween-20 (TBST) for 2 h. Following this, membranes
were incubated overnight at 4°C with primary antibodies
anti-toll-like receptor 4 (TLR4; cat. no. AF1478-SP; 1:1,000;
R&D Systems, Inc.), anti-NF-κB p65 (cat. no. 8242P; 1:1,000;
Cell Signaling Technology, Inc.), and anti-NFKB inhibitor-α (IκBα;
cat. no. 4814P; 1:1,000; Cell Signaling Technology, Inc.).
Anti-lamin B1 (cat. no. 66095-1-Ig; 1:5,000; ProteinTech Group,
Inc.) and anti-GAPDH (cat. no. 60004-1-Ig; 1:2,000; ProteinTech
Group, Inc.) were used to verify equal protein loading. Following
washing three times in TBST, the membranes were incubated with
horseradish peroxidase conjugated goat anti-rabbit (cat. no.
ZB-5301; 1:10,000; OriGene Technologies, Inc.) and goat anti-mouse
(cat. no. ZB-2305; 1:5,000; OriGene Technologies, Inc.) secondary
antibodies at room temperature for 1 h.
The proteins were visualized using an enhanced
chemiluminescence reagent (Beyotime Institute of Biotechnology).
The integrated optical density (IOD) of each band was analyzed
using Image-Pro Plus software v6.0 (National Institutes of
Health.). Each TLR4 and IκBα band value was normalized as the ratio
of the IOD to the GAPDH band. Each NF-κB p65 band value was
normalized as the ratio of the IOD to the lamin B1 band.
Statistical analysis
Statistical analyses were performed by SPSS v17.0
software (SPSS, Inc.). Graphs were generated using GraphPad Prism
5.2 software (GraphPad Software, Inc.). All data were presented as
the mean value ± standard deviation. Multiple comparisons were
evaluated by one-way analysis of variance followed by the
Student-Newman-Keuls and least significant difference post hoc
tests. The Dunnett method was used for comparisons with the control
group. P<0.05 was considered to indicate statistical
significance.
Results
GES-1 cell viability and growth rate
following incubation with probiotics and CFS
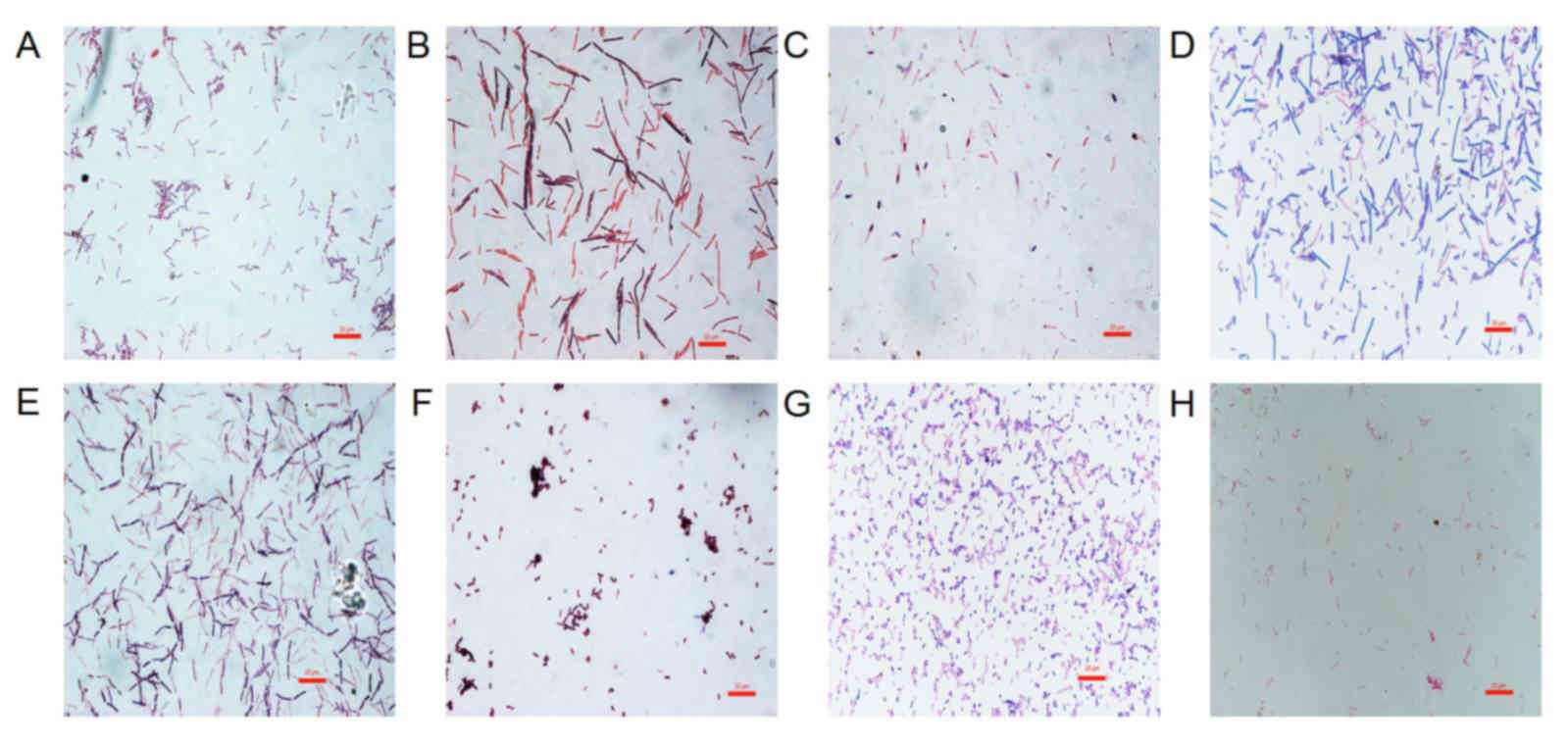
The morphological characteristics of the probiotics
and H. pylori were first evaluated following Gram staining
(Fig. 1). The probiotics in this
study were all Gram-positive bacteria. L. acidophilus was
chain rod shaped, L. bulgaricus, B. longum and B.
licheniformis were long rod shaped, B. infantis and
L. salivarius were short rod shaped. C. butyricum
contained giant spore and was enlarged at one end in a drum-hammer
shape. H. pylori was a gram-negative bacterium, which is
S-shaped or spiral. Furthermore, the viability of GES-1 cells
incubated with CFS and different titers of probiotics (MOI of 100
and 1,000) was determined by assessing the percentage of cells not
stained with trypan blue following 2, 4, 6 and 8 h of incubation.
At a MOI of 100, the cytotoxicity results indicated that the CFS of
L. acidophilus, L. bulgaricus, L. salivarius, C. butyricum, B.
infantis and B. longum were toxic to GES-1 cells. The
CFS of B. licheniformis did not show any toxicity towards
GES-1 cells with an average survival rate at 8 h of 96%. L.
acidophilus, L. bulgaricus, L. salivarius, Clostridium butyricum,
Bifidobacterium infantis, Bifidobacterium longum and B.
licheniformis did not show any toxicity towards GES-1 cells
after 4 h of incubation, with an average survival rate of 97% and
no significant differences between these groups and the control
(Fig. 2A). However, GES-1 cell
viability decreased gradually after 4 h. At 6 h, the most
significant decrease in viability was observed in B.
longum-treated cells, which had a 92% survival rate. Notably,
the viability of these cells was significantly lower compared with
the cells treated with the other six bacteria (P<0.05; Fig. 2A). GES-1 cell viability following
B. licheniformis treatment decreased at the slowest rate,
with an average survival rate of 99.7%. This was significantly
higher than the survival rate of the cells treated with other
bacteria (P<0.05; Fig. 2A). There
was no significant differences in the average GES-1 survival rates
following treatment with L. acidophilus, L. bulgaricus, L.
salivarius, C. butyricum, B. longum and B. infantis at 6
h. At 8 h, the survival rate of the GES-1 cells decreased further
with cells treated with L. bulgaricus, demonstrating
significantly lower survival rate (82.3%) compared with the cells
treated with the other six probiotics (P<0.05; Fig. 2A). Furthermore, the survival rate of
cells treated with B. licheniformis was significantly
increased (96.7%) at 8 h compared with cells treated with the other
six bacteria (P<0.05; Fig.
2A).
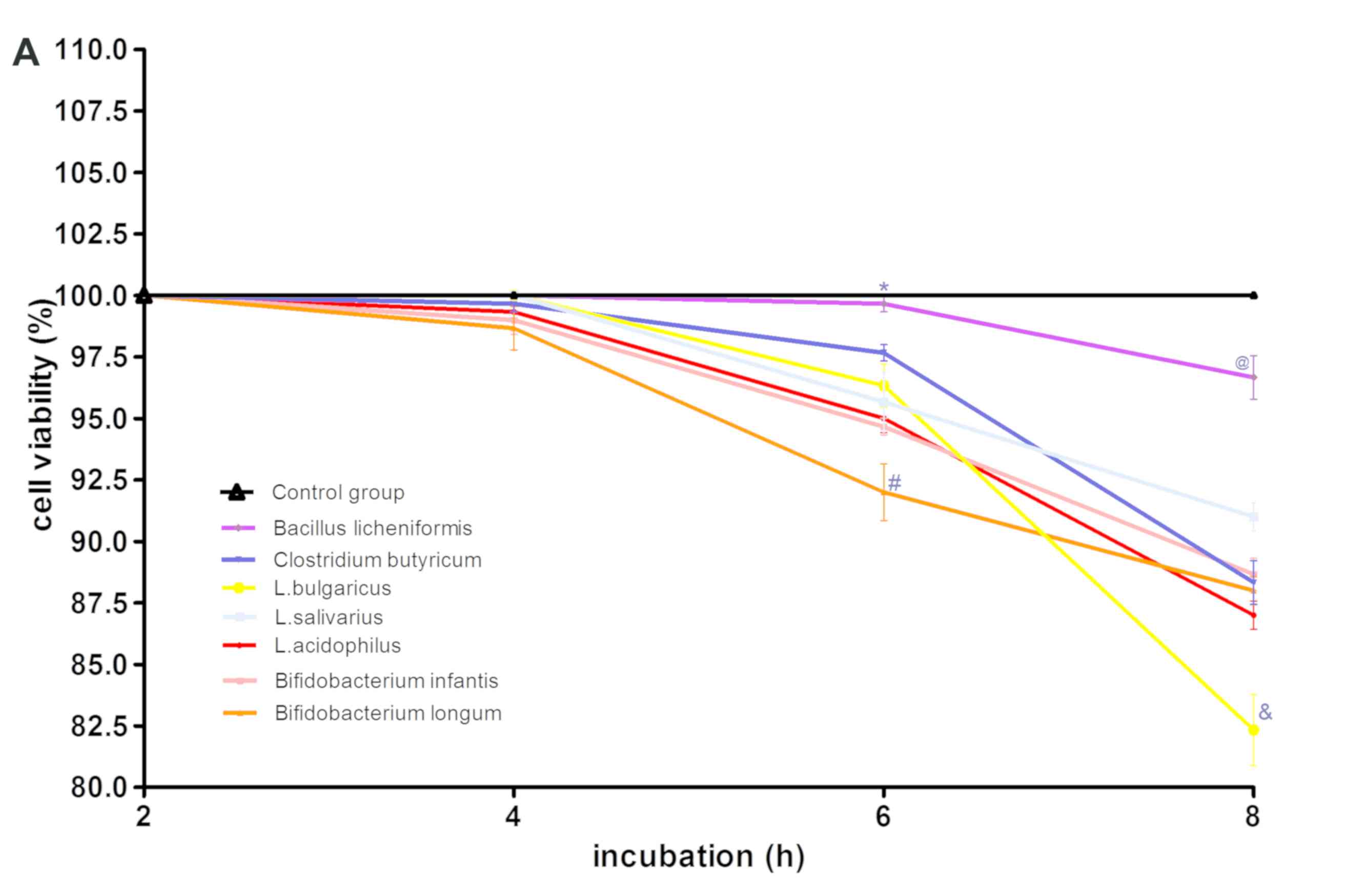 | Figure 2.Effect of various probiotics strains
on GES-1 cell viability. (A) GES-1 cell viability following culture
with L. acidophilus, L. bulgaricus, L. 0salivarius, Clostridium
butyricum, Bifidobacterium longum, Bifidobacterium infantis or
Bacillus licheniformis at a MOI of 100 and (B) MOI of 1,000.
(A) *P<0.05 vs. L. acidophilus, L. bulgaricus, L. salivarius,
Clostridium butyricum, Bifidobacterium longum, Bifidobacterium
infantis; #P<0.05 vs. L. acidophilus, L.
bulgaricus, L. salivarius, Clostridium butyricum, Bifidobacterium
infantis, Bacillus licheniformis and control group;
@P<0.05 vs. L. acidophilus, L. bulgaricus, L.
salivarius, Clostridium butyricum, Bifidobacterium infantis,
Bifidobacterium longum and control group;
&P<0.05 vs. L. acidophilus, L. salivarius,
Clostridium butyricum, Bifidobacterium infantis, Bifidobacterium
longum, Bacillus licheniformis and control group. (B)
*P<0.05 cells vs. L. acidophilus, L. bulgaricus, L.
salivarius, Clostridium butyricum, Bifidobacterium longum,
Bifidobacterium infantis; #P<0.05 vs. L.
acidophilus, L. salivarius, Clostridium butyricum, Bifidobacterium
infantis, Bifidobacterium longum, Bacillus licheniformis and
control group; @P<0.05 vs. L. acidophilus, L.
bulgaricus, L. salivarius, Clostridium butyricum, Bifidobacterium
infantis, Bifidobacterium longum and control group;
&P<0.05 cells treated with L. bulgaricus, L.
acidophilus, L. salivarius, Clostridium butyricum, Bifidobacterium
infantis, Bifidobacterium longum vs. Bacillus
licheniformis and control group. MOI, multiplicities of
infection; L, Lactobacillus. |
At a MOI of 1,000, all seven probiotic strains
exhibited no toxicity towards GES-1 cells at 2 h (Fig. 2B). However, GES-1 cell viability
decreased following 4 h of incubation, with cells treated with
L. bulgaricus (10.4% at 4 h) exhibiting significantly
reduced viability compared with cells treated with the other six
bacterial strains (P<0.05; Fig.
2B). GES-1 cell viability following B. licheniformis
treatment demonstrated the slowest decrease, with the average
survival rate of cells 100% at 4 h, which was significantly higher
when compared with cells treated with the other bacterial strains
(P<0.05; Fig. 2B). The viability
of B. licheniformis-treated GES-1 cells decreased
significantly after 6 h compared with the control group (P<0.05;
Fig. 2B). Furthermore, the viability
of these cells was 42.7% at 8 h, whereas GES-1 cells treated with
the other six bacteria did not survive past 8 h. There were no
significant differences between L. acidophilus-, L. salivarius-,
B. longum-, B. infantum- and C. butyricum-treated cells
(Fig. 2B). The probiotic half
maximal inhibitory concentration (IC50) for each group
is listed in Table II, with B.
longum demonstrating the lowest IC50 and B.
licheniformis demonstrating the highest IC50.
 | Table II.GES-1 cell growth rate in the
presence of various probiotic strains. |
Table II.
GES-1 cell growth rate in the
presence of various probiotic strains.
| Probiotics | Half maximal
inhibitory concentration (colony-forming units) |
|---|
| Bifidobacterium
longum |
6.56×107 |
| Bifidobacterium
infantis |
6.76×107 |
| Lactobacillus
acidophilus |
8.57×107 |
| Lactobacillus
salivarius |
1.55×108 |
| Lactobacillus
bulgaricus |
2.09×108 |
| Clostridium
butyricum |
3.0×108 |
| Bacillus
licheniformis |
3.33×109 |
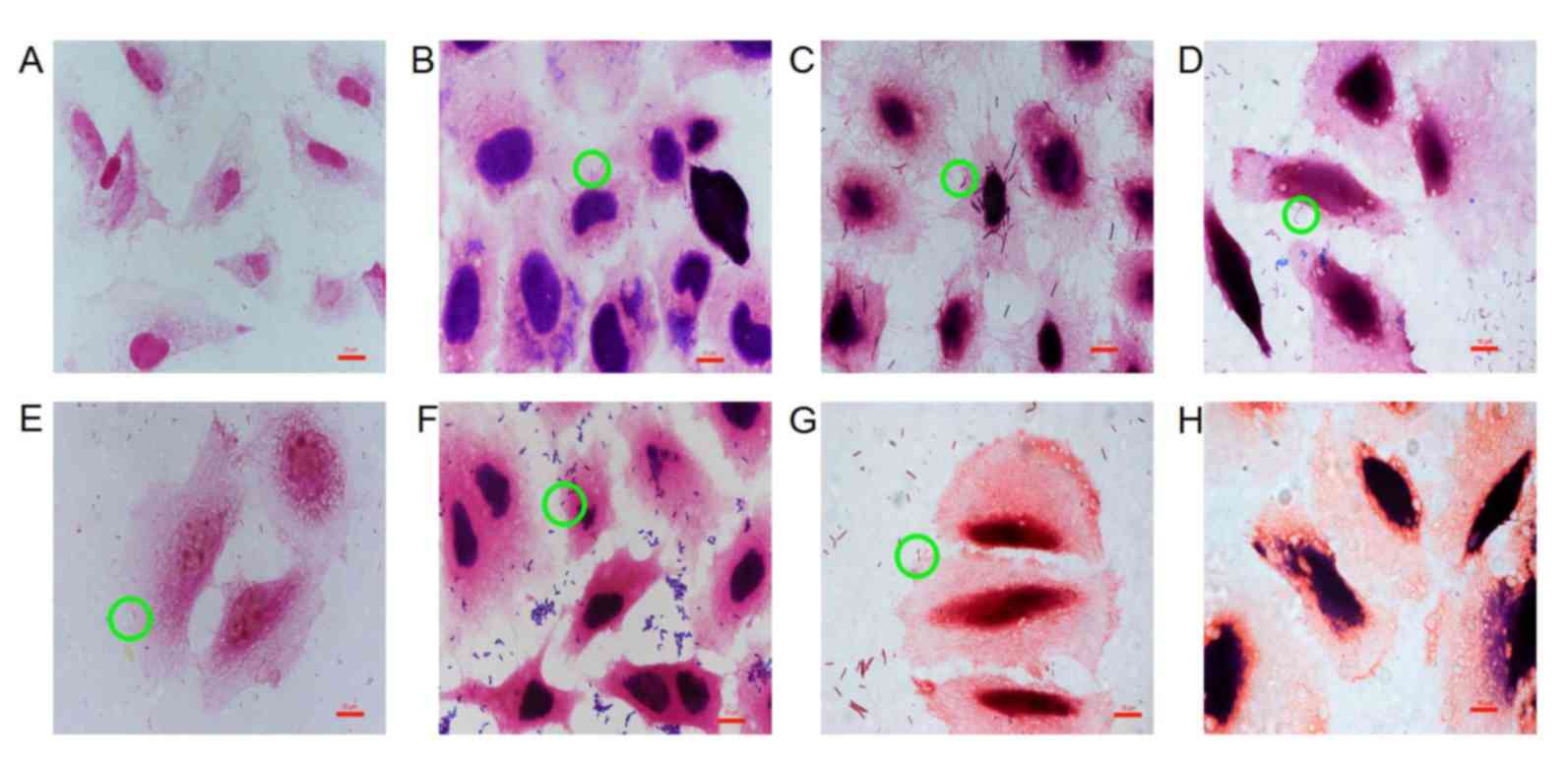
Adhesion of probiotics to GES-1
cells
Probiotics adhered to GES-1 cells with different
cell adhesion indexes and cell adhesion rates (Table III; Fig.
3). Staining of GES-1 cells for each probiotic group varied
because metabolites produced by different probiotics were able to
change the pH values of the culture medium, thereby affecting
viability and staining (28). The pH
values of the culture medium were pH 4 (L. acidophilus), pH
5 (L. bulgaricus), pH 3.8 (B. infantis), pH 4.2
(B. longum), pH 4.8 (L. salivarius), pH 5.5 (C.
butyricum) and pH 6.8 (B. licheniformis).
 | Table III.Cell adhesion index and adhesion rate
at the same concentration onto GES-1 cells. |
Table III.
Cell adhesion index and adhesion rate
at the same concentration onto GES-1 cells.
| Probiotic
strain | Adhesion index
(%) | Adhesion rate
(%) |
|---|
| Lactobacillus
acidophilus | 891.2±24.35 | 56.6±8.35 |
| Bifidobacterium
infantis | 394.4±31.00 | 37.8±7.12 |
| Bifidobacterium
longum | 328.2±23.04 | 21.2±5.12 |
| Clostridium
butyricum | 24.4±12.78 | 11.2±0.84 |
| Bacillus
licheniformis | 38.00±7.97 | 17.8±4.60 |
| Lactobacillus
bulgaricus | 499.2±27.83 | 50.0±8.12 |
| Lactobacillus
salivarius | 229.8±22.19 | 20.6±5.41 |
As indicated in Fig.
4, the cell adhesion index was consistent with the cell
adhesion rate, with the highest to lowest results as follows: L.
acidophilus>L. bulgaricus> B.
infantis>B. longum>L. salivarius> B.
licheniformis> C. butyricum. There was no significant
difference in the cell adhesion index between C. butyricum
and B. licheniformis (Fig.
4A); however, there were significant differences between the
other five bacterial groups when compared with each other
(P<0.05; Fig. 4A). There was no
significant difference in the cell adhesion rate between L.
bulgaricus and L. acidophilus. In addition, no
significant differences in the cell adhesion rate were observed
between B. longum, C. butyricum, B. licheniformis and L.
salivarius (Fig. 4B).
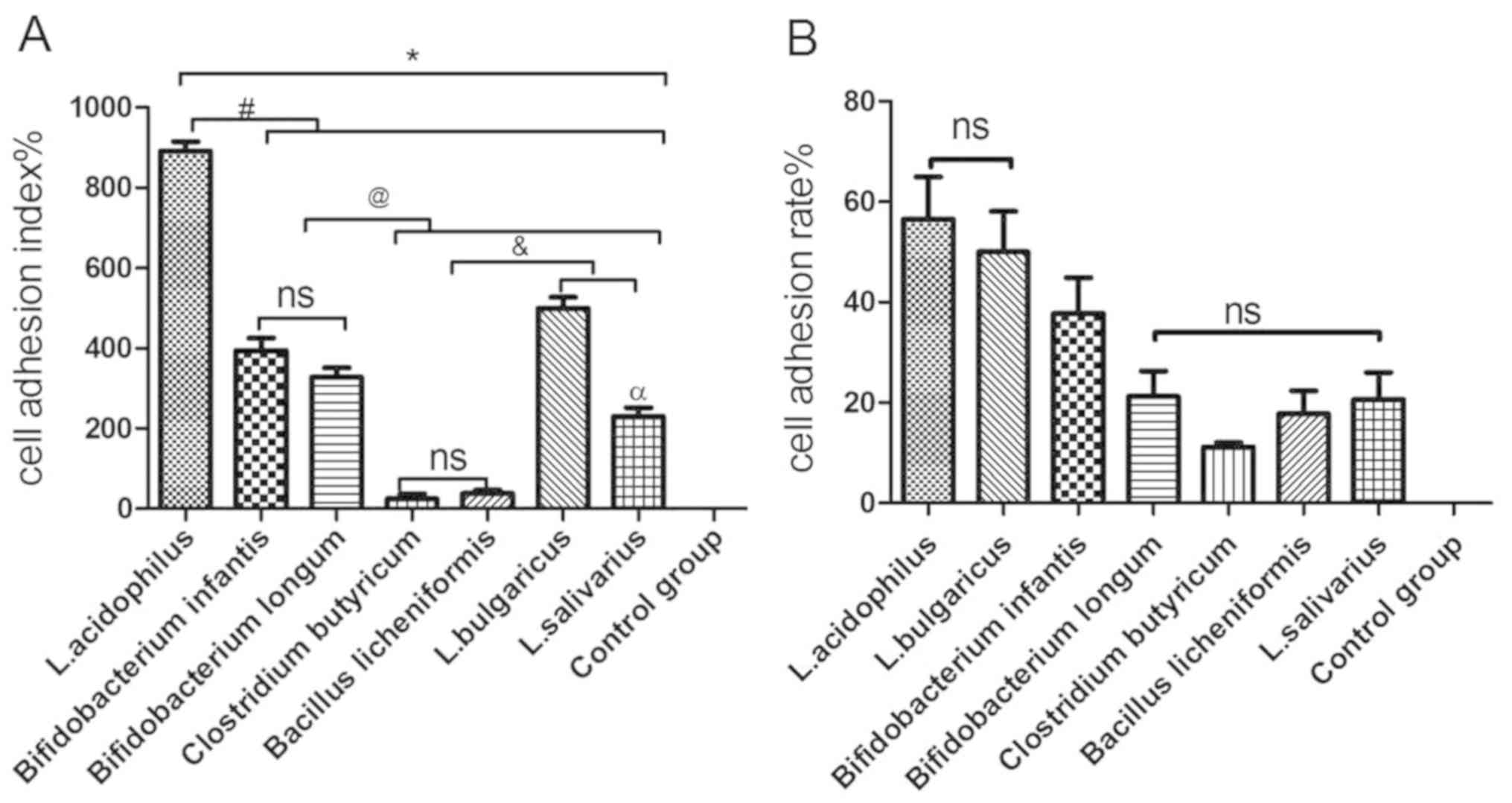 | Figure 4.Adhesion of various probiotic strains
to GES-1 cells. (A) Cell adhesion index of L. acidophilus, L.
bulgaricus, L. salivarius, Clostridium butyricum, Bifidobacterium
longum, Bifidobacterium infantis and Bacillus
licheniformis to GES-1 cells. (B) Cell adhesion rate of L.
acidophilus, L. bulgaricus, L. salivarius, Clostridium butyricum,
Bifidobacterium longum, Bifidobacterium infantis and
Bacillus licheniformis to GES-1 cells. *P<0.05 vs.
control group; #P<0.05 vs. L. acidophilus;
@P<0.05 vs. Bifidobacterium infantis and
Bifidobacterium longum; &P<0.05 vs.
Clostridium butyricum and Bacillus licheniformis;
αP<0.05 vs. L. bulgaricus. L, Lactobacillus;
ns, not significant. |
Live probiotics and CFS inhibit H.
pylori growth
CFS from probiotic culture and live probiotics were
screened for inhibitory effects against HP021 by measuring the
diameters of inhibitory growth zones on agar plates (Table IV). Fresh MRS medium, used as a
negative control, also exhibited significant activity against HP021
and formed inhibition zones of ~27 mm, which was larger than all
the tested probiotics (P<0.05; Table
IV). This suggested that there were likely unknown anti-HP
substances in MRS medium. Probiotics consumed some of the anti-HP
substances of MRS, and likely secreted some anti-HP substances.
Live L. bulgaricus demonstrated significantly greater
anti-H. pylori activity compared with the other live
probiotics, with inhibition zones of ~25 mm (P<0.05; Table IV). Live B. licheniformis did
not display any anti-H. pylori activity. By contrast, there
were no significant differences in inhibition of H. pylori
growth by live L. acidophilus, B. infantis, B. longum, L.
salivarius and C. butyricum. The CFS from all the
probiotics also inhibited H. pylori activities; however,
there were no significant differences between the probiotic groups
(Table IV). Similar results were
observed when H. pylori ATCC43504 was used (data not
shown).
 | Table IV.HP021 growth in the presence of live
probiotics and cell-free supernatant. |
Table IV.
HP021 growth in the presence of live
probiotics and cell-free supernatant.
|
| Average zone of
inhibition (mm) |
|---|
|
|
|
|---|
| Probiotic
strain | Live probiotic | Cell-free
supernatant |
|---|
| Lactobacillus
acidophilus | 20.33±1.53 | 21.00±1.00 |
| Bifidobacterium
infantis | 20.00±1.00 | 20.67±1.53 |
| Bifidobacterium
longum | 20.67±0.58 | 21.91±0.58 |
| Clostridium
butyricum | 22.33±0.58 | 21.33±0.58 |
| Lactobacillus
bulgaricus |
25.33±0.58b | 22.33±0.58 |
| Bacillus
licheniformis | 0 | 22.67±0.58 |
| Lactobacillus
salivarius | 21.00±1.00 | 22.24±0.58 |
| Man, Rogosa and
Sharpe broth |
27.00±1.00a |
27.00±1.00a |
Probiotics and CFS suppress adhesion
of H. pylori to GES-1 cells
The urease activity of HP021 and ATCC43504 were
examined to evaluate the effect of CFS and probiotics on HP021 and
H. pylori ATCC43504 adherence to GES-1 cells. The adherence
rate of H. pylori was 100% without probiotic or CFS
treatment. Results indicated that pre- or post-treatment with B.
licheniformis CFS did not reduce the adhesion of H.
pylori to GES-1 cells compared with HP group (Fig. 5A). However, urease activity was
significantly reduced following probiotic treatment, with the
exception of C. butyricum and B. licheniformis
treatment compared with the HP group (Fig. 5A). No significant differences between
the two H. pylori strains were found (the data not shown).
The adherence rate of H. pylori dropped to ~50% with L.
acidophilus treatment. Compared with non-treated cells, the
adherence rate of HP021 did not decrease with C. butyricum
or B. licheniformis treatment (Fig. 5B). These results demonstrated that
pre-treatment with L. acidophilus, L. bulgaricus, L. salivarius,
B. infantis and B. longum inhibited the adherence of
H. pylori to GES-1 cells.
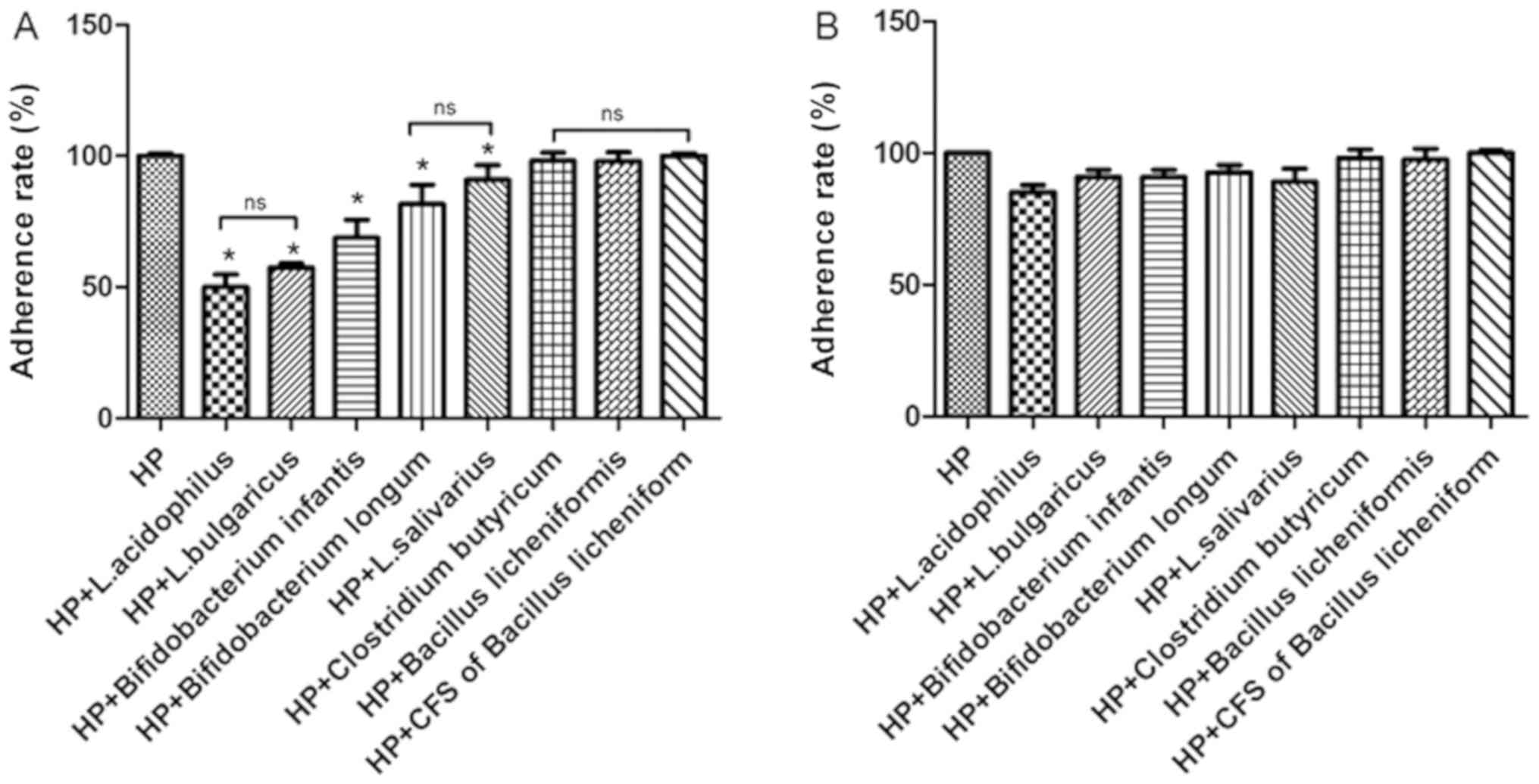 | Figure 5.Adherence rate of HP to GES-1 cells
in the presence of different probiotics at a MOI of 100. (A)
Adherence rate of HP to GES-1 cells pretreated with L.
acidophilus, L. bulgaricus, L. salivarius, Clostridium butyricum,
Bifidobacterium infantis, Bifidobacterium longum, Bacillus
licheniformis and CFS of Bacillus licheniformis. (B)
Adherence rate of HP to GES-1 cells treated with L. acidophilus,
L. bulgaricus, L. salivarius, Clostridium butyricum,
Bifidobacterium infantis, Bifidobacterium longum, Bacillus
licheniformis and CFS of Bacillus licheniformis
following infection (post-treated). *P<0.05 vs. HP group. HP,
Helicobacter pylori; L, Lactobacillus; CFS, cell-free
supernatant; ns, not significant. |
Certain probiotics significantly
affect IL-8 production in GES-1 cells
Under normal physiological conditions, GES-1 cells
secrete a small amount of IL-8 (29). The addition of HP-LPS to GES-1 cells
for 6 h significantly increased the IL-8 levels compared with
non-treated cells (P<0.05; Fig.
6). No decrease in IL-8 secretion following treatment with the
CFS of B. licheniformis was observed compared with HP-LPS
group. However, following pre-treatment with L. bulgaricus
and L. salivarius, there was a significant decrease in IL-8
secretion induced by HP-LPS compared with HP-LPS group (P<0.05;
Fig. 6). Notably, IL-8 levels
following HP-LPS plus L. acidophilus, B. infantis or B.
longum treatment were higher compared with HP-LPS only
treatment.
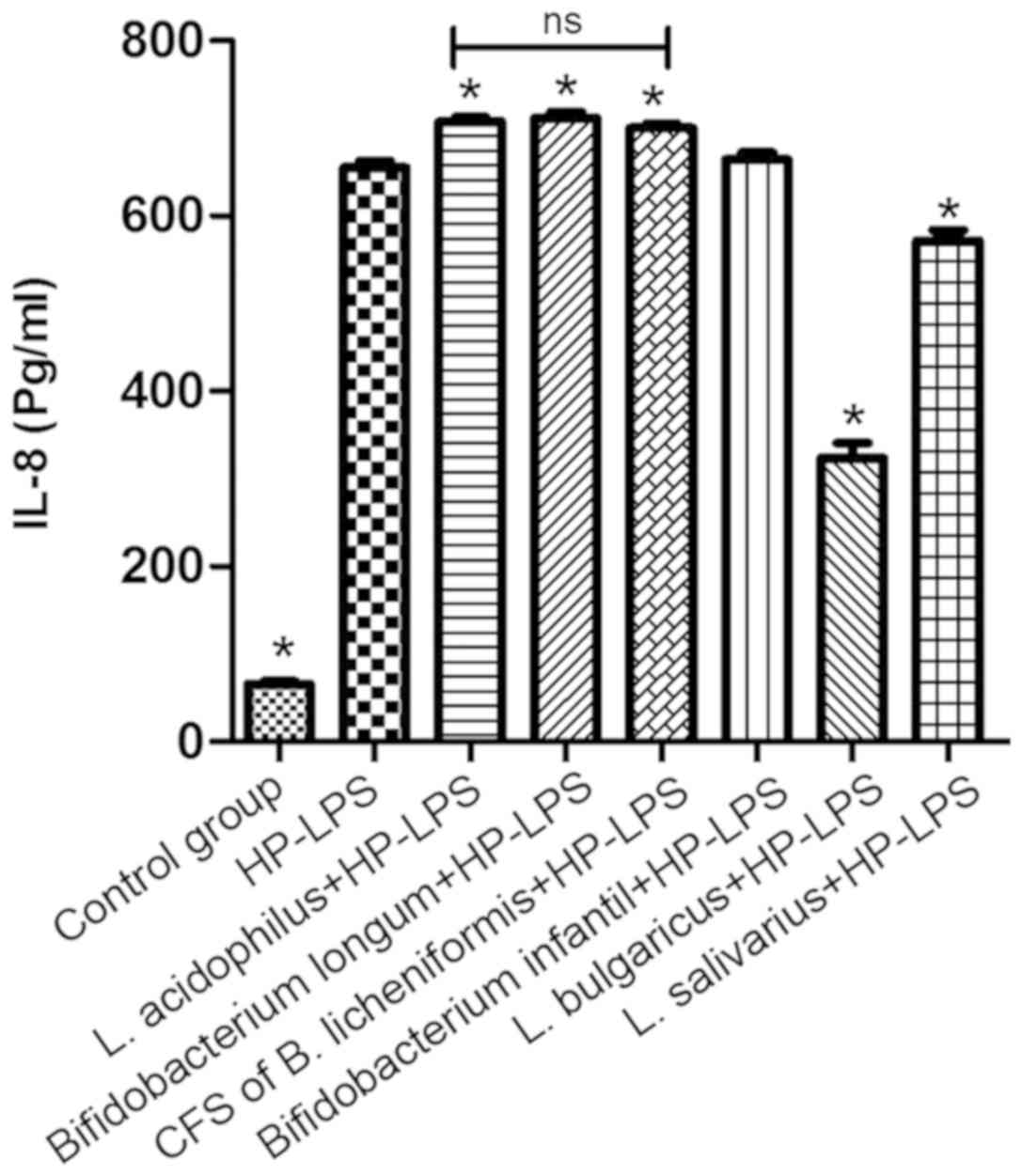 | Figure 6.Effect of L. acidophilus, L.
bulgaricus, L. salivarius, Bifidobacterium infantis,
Bifidobacterium longum CFS of Bacillus licheniformis on
IL-8 production following HP-LPS treatment. *P<0.05 vs. HP-LPS.
HP, Helicobacter pylori; LPS, lipopolysaccharide; IL,
interleukin; NS, not significant, L, Lactobacillus. |
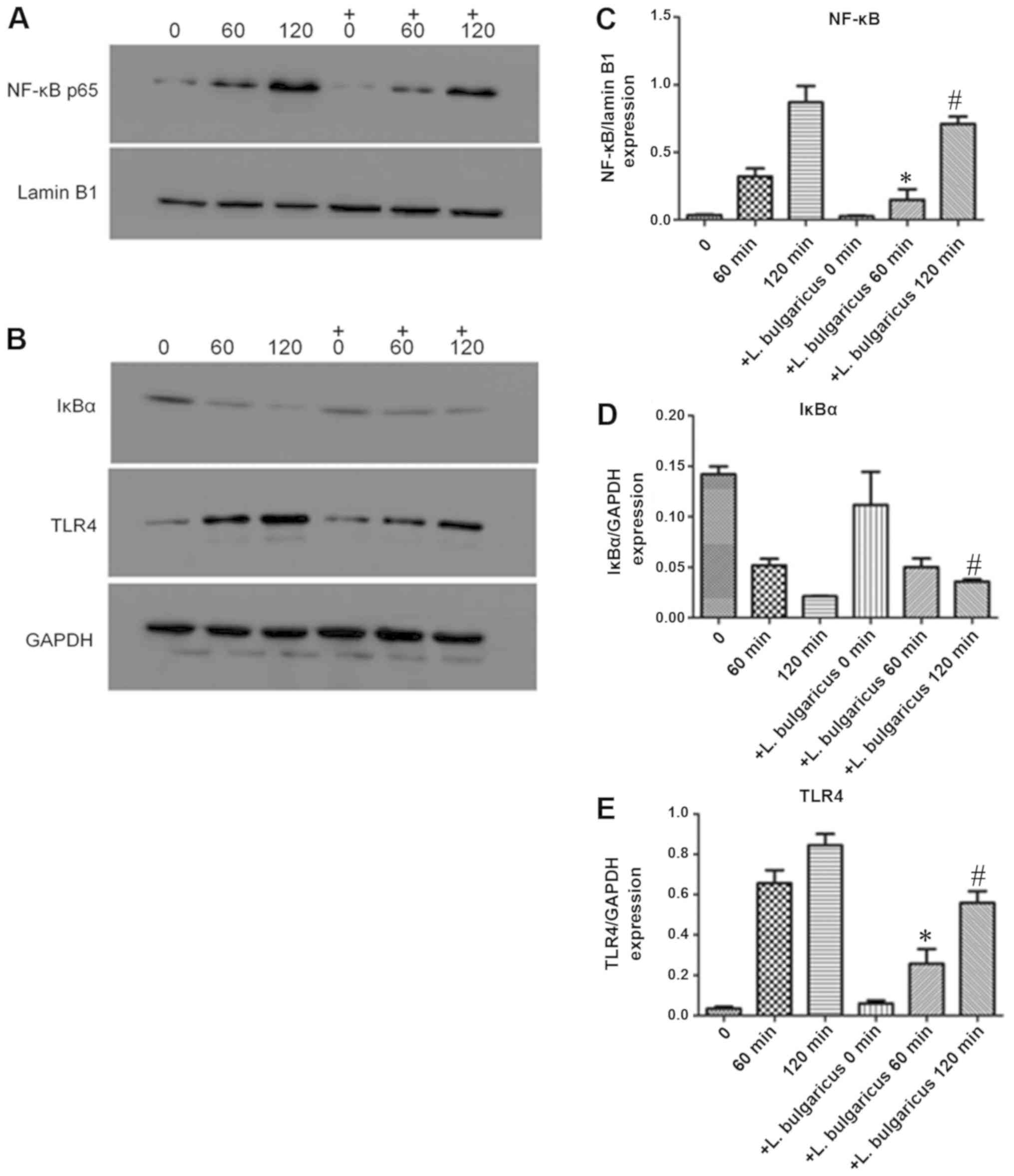
Probiotics attenuate the TLR4-NF-κB
p65 signaling pathway following HP-LPS activation
To explore the molecular mechanism of H.
pylori on GES-1 growth, expression levels of markers associated
with inflammation, TLR-4, IκBα and NF-κB, p65 were detected. H.
pylori ATCC43504 contains CagA protein that induces activation
of NF-κB and IL-8 whereas HP021 does not contain CagA protein.
Therefore, H. pylori ATCC43504 was selected for the NF-κB
activation and IL-8 production experiment. L. bulgaricus
reduced IL-8 production most significantly. Therefore, L.
bulgaricus was selected for the NF-κB activation experiment.
Treatment of GES-1 cells with 7,000 EU/ml of HP-LPS upregulated the
expression of TLR-4, increased the degradation of IκBα and induced
the translocation of NF-κB p65 into nucleus in a time-dependent
manner (Fig. 7A and B).
Pre-treatment for 2 h with viable L. bulgaricus (MOI of 100)
significantly inhibited the effects of HP-LPS on the TLR4/NFκB
pathway by increasing cytoplasmic IκBa and decreasing nuclear NF-κB
p65 levels (Fig. 7C-E). The levels
of NFκB and TLR-4 in L. bulgaricus pre-treatment 60 and 120
min groups were significantly lower than those of HP-LPS 60 and 120
mins groups, respectively (P<0.05; Fig. 7C and E). The level of IκBα in L.
bulgaricus pre-treatment 120 min group was significantly higher
than HP-LPS 120 min group (P<0.05; Fig. 7D). These results indicated that the
TLR4/NF-κB signaling pathway may be a critical target for
probiotics to alter GES-1 cell biological functions.
Discussion
The present study assessed the effects of live
probiotics and their CFS on H. pylori growth, adherence to
gastric epithelial cells and H. pylori-induced inflammation.
The findings demonstrated that L. acidophilus and L.
bulgaricus significantly inhibited the adherence of H.
pylori to GES-1 cells and also decreased IL-8 production by
GES-1 cells following stimulation with HP-LPS. In addition, L.
bulgaricus inhibited the TLR4/IκBα/NFκB signaling pathway in a
time-dependent manner.
H. pylori causes gastritis, as well as
gastric and duodenal ulcers (2,30). In
the past few decades, H. pylori treatment has improved due
to the administration of PPI plus antibiotics. However, the
emergence of drug-resistant H. pylori strains has
complicated the eradication of the bacteria. Therefore, research is
focusing on developing new strategies for treating H.
pylori. Recent clinical trials have demonstrated that the
administration of certain exogenous probiotics may improve the
eradication of H. pylori (31). In addition, some probiotic strains
can attenuate the mucosal inflammation induced by H. pylori
(32). However, the molecular
mechanism of these biological effects remains unclear.
The present study identified that the anti-H.
pylori effects of live probiotics or their CFS were highly
strain dependent. Seven probiotic strains commonly used in clinical
settings were chosen. The H. pylori standard strain
(ATCC43504), containing CagA and VacA virulence factors, and the
clarithromycin and levofloxacin-resistant HP021 strain, originally
isolated from the gastric mucosa of a patient, were also
investigated. Results revealed that all seven probiotics
significantly affected GES-1 cell viability following longer
incubation times and higher MOIs. The CFS of Lactobacillus
culture is a 96-h fermentation product containing high
concentrations of acetic acid and other bactericidal substances.
The present study determined that Lactobacillus CFS has a pH
of 2, which was toxic to gastric epithelial cells in vitro.
GES-1 cell death occurred within 2 h when treated with L.
acidophilus, B. infantis, B. longum, L. salivarius, L.
bulgaricus and C. butyricum CFS. Only B.
licheniformis CFS had a pH of 7 therefore, was not considered
toxic to GES-1 cells. Hence, only B. licheniformis CFS was
selected for investigation into the adhesion of H. pylori to
GES-1 cells and IL-8 production.
It is well established that adhesion to mucosal
surfaces is a key step in the pathogenesis of H. pylori
(33). The inhibition of H.
pylori colonization by probiotics is strain specific. Chen
et al (18) reported that
CFS, live and dead lactobacilli inhibits H. pylori
adhesion to SGC7901 cells. Hsieh et al (16) identified that L. johnsonii
MH-68 and salicinius AP-32 effectively suppress H. pylori
viability and reduce H. pylori colonization in the gastric
mucosa of mice. Aiba et al (34) proved that L. salivarius is
capable of producing a high amount of lactic acid and inhibiting
the growth of H. pylori. In the present study, five
probiotic strains adhered to GES-1 cells, with L.
acidophilus displaying the strongest adhesive ability, followed
by L. bulgaricus. However, C. butyricum and B.
licheniformis did not demonstrate any notable adhesive ability.
Further study is required to identify the mechanism of adhesion to
the cell surface of L. acidophilus and L. bulgaricus.
Pre-treatment with L. acidophilus, L. bulgaricus, L. salivarius,
B. infantis and B. longum reduced the ability of H.
pylori to adhere to GES-1 cells to differing extents. The
inhibitory effects of L. acidophilus and L.
bulgaricus were greater than L. salivarius, B. infantis
and B. longum at the same concentration. Notably, the CFS of
B. licheniformis did not reduce the adhesion of H.
pylori to GES-1 cells compared with the HP group. The
inhibitory effect of probiotic pre-treatment against H.
pylori adherence is likely mediated via increasing production
of mucin (35) or competition to
bind H. pylori adhesion sites by probiotics (36). The adhesion of H. pylori to
GES-1 cells was not inhibited when cells were treated with
probiotics post-infection. Therefore, it appears that probiotics
cannot reduce the adhesion rate of H. pylori when it is
already adhered to gastric epithelial cells. Therefore, probiotics
may be more effective in a preventive rather than therapeutic
role.
Certain probiotic strains produce bactericidal
substances that are either secreted into the culture supernatant or
expressed on the cell surface, which can significantly inhibit
H. pylori (37). The agar
plate diffusion assays performed in the present study determined
that live bacteria and their CFS from L. acidophilus, B.
infantis, B. longum, L. bulgaricus, C. butyricum and L.
salivarius inhibited the growth of H. pylori ATCC43504
and HP021 strains. Live B. licheniformis did not appear to
inhibit the growth of H. pylori ATCC43504 or HP021, but the
CFS of B. licheniformis did demonstrate anti-H.
pylori properties. A possible mechanism of action is the
secretion of lactic acid by the probiotics, as the metabolic end
products of lactic acid fermentation and organic acids are capable
of interfering with the growth of pathogens. Notably, El-Adawi
et al (37) demonstrated that
lactic acid may be a potent antimicrobial. However, B.
licheniformis is not a lactic acid-producing bacterium, and the
nature of the antimicrobial substance contained within its CFS
requires further characterization.
Human immunity plays an important role in the
development of clinical diseases. Pro-inflammatory cytokine
production occurs during H. pylori infection, with the
inflammatory reactions potentially leading to chronic inflammation
rather than eliminating H. pylori (38,39). The
transcription factor NF-κB can be activated by IL-1β, LPS,
peptidoglycan and tumor necrosis factor-α during H. pylori
infection (40). NF-κB is critical
modulator of cytokine expression (41). TLRs are cell transmembrane and
pathogen-associated molecular pattern receptors that have a central
role in the recognition of microbial pathogens and may be a first
line of immunity against H. pylori (42). HP-LPS-induced inflammation in gastric
mucosa demonstrates similar pathological characteristics to the
mucosal inflammation initiated by H. pylori infection
(43). The present study determined
that H. pylori infection induced TLR4 and IL-8
pro-inflammatory cytokine expression in vitro. The findings
demonstrated that pre-treatment with viable L. bulgaricus
for 2 h prevented TLR4 signaling and IL-8 production stimulated by
HP-LPS. This strongly supports the hypothesis that certain soluble
proteins secreted by L. bulgaricus and/or cell-bound
components of L. bulgaricus exert inhibitory effects on the
TLR4 signaling pathways in GES-1 cells. Other cytokines such as
IL-1β, IL-10, IL-6 and Smad family member 7 are also involved in
the response of H. pylori to epithelial cells (17,44). The
present study suggested that suppression of the TLR4/NF-κB
signaling pathway occurred in a time-dependent manner and was
mediated through the stabilization of IκBα.
In conclusion, the present study identified that
two probiotic strains, L. acidophilus and L.
bulgaricus, were effective in reducing the H. pylori
load. One possible mechanism of L. bulgaricus on H.
pylori activity was implied to be via modulation of the
TLR4/IκBα/NF-κB signaling pathway. Considering the safety and
health function of probiotics, food containing L.
acidophilus and L. bulgaricus may have potential as an
adjuvant therapy for gastric diseases caused by H. pylori,
and displays promise as a preventive measure against H.
pylori infection.
Acknowledgements
The authors would like to thank Professor Jianzhong
Zhang from the China Center of Disease Prevention Center for the
H. pylori culture technical support.
Funding
No funding was received.
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
SHY, ZL and GLH performed the experiments and
analyzed data. LY designed the experiment, analyzed data and wrote
the manuscript. LDY analyzed data and was involved in discussions
about the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The clinical study was approved by the Ethics
Committee of Shengjing Hospital and the patient provided written
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Warren JR and Marshall B: Unidentified
curved bacilli on gastric epithelium in active chronic gastritis.
Lancet. 1:1273–1275. 1983.PubMed/NCBI
|
|
2
|
Kusters JG, van Vliet AH and Kuipers EJ:
Pathogenesis of Helicobacter pylori infection. Clin
Microbiol Rev. 19:449–490. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Salama NR, Hartung ML and Müller A: Life
in the human stomach: Persistence strategies of the bacterial
pathogen Helicobacter pylori. Nat Rev Microbiol. 11:385–399.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fock KM, Graham DY and Malfertheiner P:
Helicobacter pylori research: Historical insights and future
directions. Nat Rev Gastroenterol Hepatol. 10:495–500. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Todo K, Ohmae T, Osamura T, Imamura T and
Imashuku S: Severe Helicobacter pylori gastritis-related
thrombocytopenia and iron deficiency anemia in an adolescent
female. Ann Hematol. 95:835–836. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Franchini M, Vescovi PP, Garofano M and
Veneri D: Helicobacter pylori-associated idiopathic
thrombocytopenic purpura: A narrative review. Semin Thromb Hemost.
38:463–468. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gao W, Cheng H, Hu F, Li J, Wang L, Yang
G, Xu L and Zheng X: The evolution of Helicobacter pylori
antibiotics resistance over 10 years in Beijing, China.
Helicobacter. 15:460–466. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chey WD, Leontiadis GI, Howden CW and Moss
SF: ACG Clinical Guideline: Treatment of Helicobacter pylori
Infection. Am J Gastroenterol. 112:212–239. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Graham DY, Fagoonee S and Pellicano R:
Increasing role for modified bismuth-containing quadruple therapies
for Helicobacter pylori eradication. Minerva Gastroenterol
Dietol. 63:77–79. 2017.PubMed/NCBI
|
|
10
|
Masannat Y and Nazer E: Pepto bismuth
associated neurotoxicity: A rare side effect of a commonly used
medication. W V Med J. 109:32–34. 2013.PubMed/NCBI
|
|
11
|
Kafshdooz T, Akbarzadeh A, Majdi
Seghinsara A, Pourhassan M, Nasrabadi HT and Milani M: Role of
probiotics in managing of Helicobacter pylori infection: A
review. Drug Res (Stuttg). 67:88–93. 2017.PubMed/NCBI
|
|
12
|
Urrutia-Baca VH, Escamilla-Garcia E, de la
Garza-Ramos MA, Tamez-Guerra P, Gomez-Flores R and Urbina-Rios CS:
In vitro antimicrobial activity and downregulation of Virulence
gene expression on Helicobacter pylori by reuterin.
Probiotics Antimicrob Proteins. 10:168–175. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Garcia-Castillo V, Zelaya H, Ilabaca A,
Espinoza-Monje M, Komatsu R, Albarracín L, Kitazawa H,
Garcia-Cancino A and Villena J: Lactobacillus fermentum
UCO-979C beneficially modulates the innate immune response
triggered by Helicobacter pylori infection in vitro. Benef
Microbes. 9:829–841. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Çekin AH, Şahintürk Y, Akbay Harmandar F,
Uyar S, Yolcular BO and Çekin Y: Use of probiotics as an adjuvant
to sequential H. pylori eradication therapy: Impact on
eradication rates, treatment resistance, treatment-related side
effects, and patient compliance. Turk J Gastroenterol. 28:3–11.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Marcial G, Villena J, Faller G, Hensel A
and de Valdez GF: Exopolysaccharide-producing Streptococcus
thermophilus CRL1190 reduces the inflammatory response caused
by Helicobacter pylori. Benef Microbes. 8:451–461. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hsieh PS, Tsai YC, Chen YC, Teh SF, Ou CM
and King VA: Eradication of Helicobacter pylori infection by
the probiotic strains Lactobacillus johnsonii MH-68 and
L. salivarius ssp. salicinius AP-32. Helicobacter.
17:466–477. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang YJ, Chuang CC, Yang HB, Lu CC and
Sheu BS: Lactobacillus acidophilus ameliorates H.
pylori-induced gastric inflammation by inactivating the Smad7 and
NFκB pathways. BMC Microbiol. 12:382012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen X, Liu XM, Tian F, Zhang Q, Zhang HP,
Zhang H and Chen W: Antagonistic activities of lactobacilli against
Helicobacter pylori growth and infection in human gastric
epithelial cells. J Food Sci. 77:M9–M14. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hassan ST and Šudomová M: Probiotics as
dietary supplements for eradication of Helicobacter pylori
infection in children: A role beyond infection. Children (Basel).
3:E272016.PubMed/NCBI
|
|
20
|
Lee CY, Shih HC, Yu MC, Lee MY, Chang YL,
Lai YY, Lee YC, Kuan YH and Lin CC: Evaluation of the potential
inhibitory activity of a combination of L. acidophilus, L.
rhamnosus and L. sporogenes on Helicobacter pylori: A
randomized double-blind placebo-controlled clinical trial. Chin J
Integr Med. 23:176–182. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Peng YC, Ho SP, Shyu CL, Chang CS and
Huang LR: Clarithromycin modulates Helicobacter
pylori-induced activation of nuclear factor-κB through
classical and alternative pathways in gastric epithelial cells.
Clin Exp Med. 14:53–59. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
de Klerk N, Maudsdotter L, Gebreegziabher
H, Saroj SD, Eriksson B, Eriksson OS, Roos S, Lindén S, Sjölinder H
and Jonsson AB: Lactobacilli reduce Helicobacter pylori
attachment to host gastric epithelial cells by inhibiting adhesion
gene expression. Infect Immun. 84:1526–1535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deng X, Ma H, Yu Q, et al: In vitro
investigate the role of the Lactobacillus from human and
Lactobacillus Acidophilus on the growth of the
Helicobacter pylori Sydney Strainl (SS1) and the effects of
the adhesive ability to gastric epithelial cell. Sichuan Med J.
36:1256–1260. 2015.
|
|
24
|
Wang B, Wei H, Yuan J, Li Q, Li Y, Li N
and Li J: Identification of a surface protein from
Lactobacillus reuteri JCM1081 that adheres to porcine
gastric mucin and human enterocyte-like HT-29 cells. Curr
Microbiol. 57:33–38. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Amin M, Anwar F, Naz F, Mehmood T and
Saari N: Anti- Helicobacter pylori and urease inhibition
activities of some traditional medicinal plants. Molecules.
18:2135–2149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sgouras D, Maragkoudakis P, Petraki K,
Martinez-Gonzalez B, Eriotou E, Michopoulos S, Kalantzopoulos G,
Tsakalidou E and Mentis A: In vitro and in vivo inhibition of
Helicobacter pylori by Lactobacillus casei strain Shirota.
Appl Environ Microbiol. 70:518–526. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou C, Ma FZ, Deng XJ, Yuan H and Ma HS:
Lactobacilli inhibit interleukin-8 production induced by
Helicobacter pylori lipopolysaccharide-activated Toll-like
receptor 4. World J Gastroenterol. 14:5090–5095. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bettinger C and Zimmermann HW: New
investigations on hematoxylin, hematein, and hematein-aluminium
complexes. II. Hematein-aluminium complexes and hemalum staining.
Histochemistry. 96:215–228. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu C, Wan Y, Guo T and Chen X: Effect of
hydrogen sulfide on the expression of CSE, NF-κB, and IL-8 mRNA in
GES-1 cells with Helicobacter pylori infection. Zhong Nan Da
Xue Xue Bao Yi Xue Ban. 38:977–983. 2013.(In Chinese). PubMed/NCBI
|
|
30
|
Amieva M and Peek RM Jr: Pathobiology of
Helicobacter pylori -induced gastric cancer.
Gastroenterology. 150:64–78. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oh B, Kim BS, Kim JW, Kim JS, Koh SJ, Kim
BG, Lee KL and Chun J: The effect of probiotics on gut microbiota
during the Helicobacter pylori eradication: Randomized
controlled trial. Helicobacter. 21:165–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Panpetch W, Spinler JK, Versalovic J and
Tumwasorn S: Characterization of Lactobacillus salivarius
strains B37 and B60 capable of inhibiting IL-8 production in
Helicobacter pylori -stimulated gastric epithelial cells.
BMC Microbiol. 16:2422016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De Falco M, Lucariello A, Iaquinto S,
Esposito V, Guerra G and De Luca A: Molecular mechanisms of
Helicobacter pylori pathogenesis. J Cell Physiol.
230:1702–1707. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Aiba Y, Suzuki N, Kabir AM, Takagi A and
Koga Y: Lactic acid-mediated suppression of Helicobacter
pylori by the oral administration of Lactobacillus
salivarius as a probiotic in a gnotobiotic murine model. Am J
Gastroenterol. 93:2097–2101. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mack DR, Michail S, Wei S, McDougall L and
Hollingsworth MA: Probiotics inhibit enteropathogenic E. coli
adherence in vitro by inducing intestinal mucin gene expression. Am
J Physiol. 276:G941–G950. 1999.PubMed/NCBI
|
|
36
|
Valeur N, Engel P, Carbajal N, Connolly E
and Ladefoged K: Colonization and immunomodulation by Lactobacillus
reuteri ATCC 55730 in the human gastrointestinal tract. Appl
Environ Microbiol. 70:1176–1181. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
El-Adawi H, El-Sheekh M, Khalil M, El-Deeb
N and Hussein M: Lactic acid bacterial extracts as anti-
Helicobacter pylori: A molecular approach. Ir J Med Sci.
182:439–452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jang S, Kim J and Cha JH: Cot kinase plays
a critical role in Helicobacter pylori -induced IL-8
expression. J Microbiol. 55:311–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dixon BR, Radin JN, Piazuelo MB, Contreras
DC and Algood HM: IL-17a and IL-22 induce expression of
antimicrobials in gastrointestinal epithelial cells and may
contribute to epithelial cell defense against Helicobacter
pylori. PLoS One. 11:e01485142016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee MH, Yang JY, Cho Y, Woo HJ, Kwon HJ,
Kim DH, Park M, Moon C, Yeon MJ, Kim HW, et al: Inhibitory effects
of Menadione on Helicobacter pylori growth and
Helicobacter pylori -induced inflammation via NF-βB
inhibition. Int J Mol Sci. 20:E11692019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cha B, Lim JW and Kim H: Jak1/Stat3 is an
upstream signaling of NF-κB activation in Helicobacter
pylori -induced IL-8 production in gastric epithelial AGS
cells. Yonsei Med J. 56:862–866. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Villena J and Kitazawa H: Modulation of
intestinal TLR4-inflammatory signaling pathways by probiotic
microorganisms: Lessons learned from Lactobacillus jensenii TL2937.
Front Immunol. 4:5122014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Stein SC, Faber E, Bats SH, Murillo T,
Speidel Y, Coombs N and Josenhans C: Helicobacter pylori
modulates host cell responses by CagT4SS-dependent translocation of
an intermediate metabolite of LPS inner core heptose biosynthesis.
PLoS Pathog. 13:e10065142017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yu HJ, Liu W, Chang Z, Shen H, He LJ, Wang
SS, Liu L, Jiang YY, Xu GT, An MM and Zhang JD: Probiotic BIFICO
cocktail ameliorates Helicobacter pylori induced gastritis.
World J Gastroenterol. 21:6561–6571. 2015. View Article : Google Scholar : PubMed/NCBI
|