Introduction
Cervical cancer was previously one of the leading
causes of cancer-related death in Japanese women and the second
most common malignancy in women worldwide (1). This disease comprises several
histologic types, of which squamous cell carcinoma is predominant
and accounts for approximately 85–90% of cases. In contrast,
adenocarcinomas/adenosquamous carcinomas are less common and
represent 10–25% of cases (2–3). The
incidence of invasive cervical adenocarcinoma and its variants has
risen dramatically among younger women over the past few decades
(4). The cause of this is not clear
but is of concern as several studies have indicated that
adenocarcinoma is associated with a worse prognosis compared to
that of squamous cell carcinoma. Compared to those with squamous
cell carcinoma, a higher proportion of lymph node involvement and
distant metastases, as well as a decline in survival across stages,
are typically found with adenocarcinoma (5).
Recently, the phosphatidylinositide 3-kinase
(PI3K)/AKT signaling pathway has been found to be a major survival
signal for cancer cells. Cell proliferation, growth, apoptosis,
autophagy, invasion, and migration are regulated by the
phosphatidylinositide 3-kinase (PI3K)/AKT serine/threonine kinase
(AKT)/mechanistic target of rapamycin kinase (mTOR) pathways, which
are putatively activated by key signals in different tumor types
(6,7). Activation is commonly conferred by
mutations in the p110α subunit of PI3K, PIK3CA, with most
mutations (>80%) occurring either in the helical domain exon 9
or the kinase domain exon 20. Until recently, molecular genetic
studies on cervical tumors have been limited. Cervical carcinomas
in U.S. populations are frequently associated with mutations in
PIK3CA, with a mutation frequency of 31% (8). Lou et al (9), analyzed 675 Latin American patients
with cervical tumors and found that 31% of squamous carcinomas and
24% of adeno and adenosquamous carcinomas harbored mutations in the
helical domain, and specifically at the E542 and E545 residues,
which were thought to result in activation of this subunit and the
PI3K/AKT pathway. In Chinese patients with cervical cancer, it was
revealed that PIK3CA mutations are more common in squamous
cell carcinomas (15.3%) than in non-squamous cell carcinomas (7.3%)
(10). However, the prevalence of
mutations in PI3KCA and how they are associated with
clinicopathological characteristics and prognosis in Japanese
patients with cervical cancer have not been studied until now.
Therefore, the objective of this work was to analyze the
relationship between amplification and somatic mutations in
PIK3CA and various clinicopathologic variables including
prognosis, in cervical cancer cell carcinomas from Japanese
patients. Furthermore, we analyzed whether mutations in
PIK3CA can predict response to PI3K/AKT/mTOR inhibition in
cervical cancer.
Materials and methods
Tissue samples
A total of 71 paraffin-embedded tumor tissue samples
were obtained from the Department of Obstetrics and Gynecology at
Shimane University Hospital; all samples were cervical squamous
cell carcinomas. In addition, 53 adenocarcinomas/adenosquamous
carcinomas were obtained from the Department of Obstetrics and
Gynecology at Seirei Hamamatsu General Hospital. Patients had
received appropriate therapy at either Shimane University Hospital
or Seirei Hamamatsu General Hospital between January 1994 and
December 2013. All specimens from cervical cancer patients were
obtained after operation and prior to any treatment. Tumor staging
was performed according to the International Federation of
Gynecology and Obstetrics (FIGO) classification (Shepherd, 2014).
The invasive squamous cell carcinomas consisted of 28 cases of
stage I disease, 11 of stage II disease, 24 of stage III disease,
and eight of stage IV disease. All tumors were classified
histologically according to the World Health Organization criteria.
The median patient age was 55 years (range 32–84 years). The
invasive adenocarcinomas/adenosquamous cell carcinomas consisted of
39 cases of stage I disease, eight of stage II disease, five of
stage III disease, and one of stage IV disease. World Health
Organization criteria were used to classify all tumors
histologically. The median patient age was 46 years (range 27–82
years). Stage I and II patients were treated with class II or class
III radical hysterectomies with pelvic lymph node dissection. Stage
I patients were treated with positive lymph node metastasis or
positive lymphovascular space invasion and all stage II patients
received concurrent chemoradiotherapy or radiotherapy as adjuvant
therapy. Stage III and IV patients were treated with concurrent
chemoradiotherapy or radiotherapy alone.
Patients with an incomplete response to radiotherapy
and patients with recurrent tumors were treated with a variety of
salvage chemotherapy agents including cisplatin, carboplatin, and
paclitaxel. The follow-up period ranged from 5 to 142 months, with
a median of 65 months. Acquisition of tissue specimens and clinical
information was approved by an institutional review board (Shimane
University and Seirei Hamamatsu General Hospital), and written
informed consent was obtained from all patients. Only patients with
follow-up data were included. The paraffin tissue blocks were
organized into tissue microarrays, each produced by removing 3-mm
diameter cores of tumors from the block. Selection of the area for
the core was made by a gynecologic oncologist (KN) and pathology
technician (KI), and was based on review of the H&E slides.
DNA isolation
Unstained paraffin-embedded tissues were serially
sectioned at a thickness of 10 µm and one adjacent hematoxylin and
eosin-stained section was taken for identification and selection of
tumor tissue. The carcinoma, comprising at least 85% of the total
area was marked, and using a sterile needle, gross macroscopic
dissection was performed. The dissected tissues were placed in
microcentrifuge tubes and DNA isolation was performed as described
previously (11).
Cell culture and cell lines
Hela and Hela P35 (adenocarcinoma), as well as
ME180, TCS, and CaSki (squamous cell carcinoma) human cervical cell
lines were obtained from Tohoku University (Sendai, Japan), whereas
SKGIIIa, SKGIIIb, HCS2, and BOKU (also squamous cell carcinoma)
cells were obtained from the Health Science Research Resources Bank
(Tokyo, Japan). All human cervical cancer cell lines were
maintained in Dulbecco's modified Eagle's medium (DMEM) (Life
Technologies, Gaithersburg, MD, USA) supplemented with 5% fetal
bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at
37°C in an atmosphere of 5% CO2.
Fluorescence in situ hybridization
(FISH)
FISH was performed on 5-µm paraffin sections
contained on a tissue microarray consisting of both carcinoma and
normal tissue cores. Zytolight® SPEC PIK3CA/CEN 3
dual color probes (Zytovision, Bremerhaven, Germany) were used
according to the manufacturer's protocol. Slides were denatured for
10 min at 75°C and hybridized at 37°C overnight with the probe mix.
Cell nuclei were stained with 4′,6-diamidino-2-phenylindole.
Signals were evaluated by two independent researchers using an
Olympus Bx41 fluorescence microscope (Olympus Corporation, Tokyo,
Japan). Separate narrow band-pass filters were used to detect the
ZygGreen™, ZyRed™, and DAPI signals. Approximately 100 tumor cells
were examined for each specimen at magnification ×60; a signal
ratio of experimental probe/reference probe greater than three was
considered amplification.
Mutational analysis of PIK3CA, KRAS,
and BRAF
The mutational status of KRAS and BRAF
was determined for all paraffin-embedded cervical cancer tissue
samples, whereas PIK3CA mutation status was determined for
all paraffin-embedded tissue samples as well as cell lines.
Polymerase chain reaction (PCR) was performed, followed by
nucleotide sequencing using the iCycler (Bio-Rad, Hercules, CA,
USA). Exons 9 and 20 of PIK3CA, exon 2 of KRAS, and
exon 15 of BRAF were sequenced because together, mutational
hot spots in these regions harbor nearly all published mutations
(12–15). The primers for PCR and sequencing
were manufactured by GeneLink, Inc., (Hawthorne, NY, USA), and
their sequences were described in a previous report (16). The sequences were analyzed using the
Lasergene program, DNASTAR, Inc., (Madison, WI, USA).
Cell growth assays
Cells were plated in 96-well plates at a density of
3,000 cells per well and treated with or without a potent PI3K or
mTOR inhibitor. An MTT growth assay was performed to determine the
number of cells (17). Potent
inhibitors such as temsirolimus (Selleck Chemicals, Houston, TX,
USA) and NVP-BEZ235 (Selleck Chemicals) were used to treat each of
the cell lines at doses of 50, 500, 1,000, and 3,000 nM to inhibit
PI3K/mTOR function, and cell viability was measured 98 h later.
DMSO was used at an equal amount as a control. The data were
expressed as percentage relative to the DMSO control. The mean and
SD were obtained from three experiments.
Statistical analysis
Progression-free survival was calculated as the time
between diagnosis and recurrence of disease, whereas overall
survival was calculated as the time between diagnosis of disease
and death. Kaplan-Meier curves were used to plot the survival data
and log-rank tests were performed to determine the statistical
significance of survival differences. Data were censored when
patients were lost to follow-up. The χ2 test was used
for comparisons of categorical data. Student's t-test (for
comparison of two groups) or one-way analysis of variance followed
by Tukey's post hoc test (for comparison of more than two groups)
was used to evaluate numerical data.
Results
Identification of PIK3CA, KRAS, and
BRAF mutations
Somatic mutations in PIK3CA were found in
four (5.6%) of 71 cervical squamous cell carcinoma samples and
three (5.7%) of 53 cervical adeno/adenosquamous cell carcinoma
samples (Table I). Interestingly,
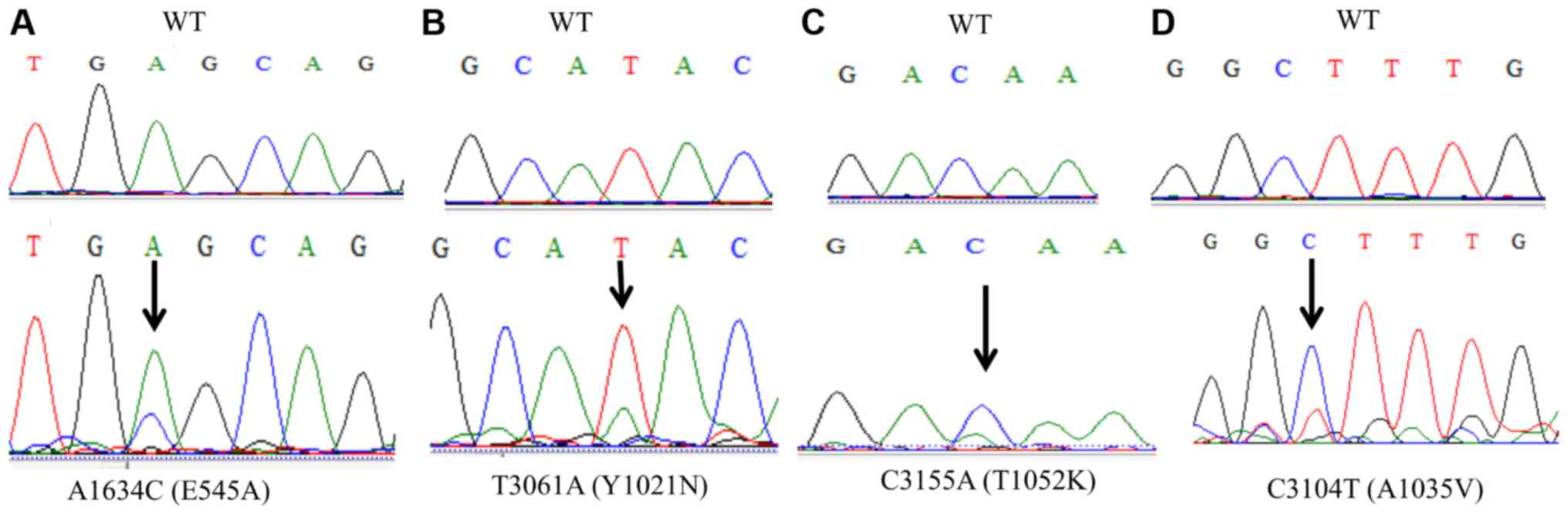
mutations in adeno/adenosquamous carcinomas only occurred within
exon 9 (E545A) (Table II; Fig. 1A) and no mutations were identified in
the catalytic domain encoded by exon 20. In contrast, mutations in
squamous cell carcinoma only occurred within exon 20 (Y1021N,
A1035V, and T1052K) and no mutations were identified in the helical
domain encoded by exon 9 (Table II;
Fig. 1B-D). Somatic mutations in
KRAS were identified in three (5.7%) of 53 cases and BRAF
mutations were identified in one (2%) of 53 cases of cervical
adeno/adenosquamous cell carcinoma (Table I). KRAS and BRAF
mutations were not present in the same samples harboring
PIK3CA mutations. No squamous cell carcinomas had detectable
oncogenic mutations in KRAS (0%, of 71) or BRAF (0%,
of 71).
 | Table I.Frequency of PIK3CA, KRAS,
BRAF and PIK3CA amplification in cervical carcinoma. |
Table I.
Frequency of PIK3CA, KRAS,
BRAF and PIK3CA amplification in cervical carcinoma.
| Histological
subtype | PIK3CA
(%) | KRAS
(%) | BRAF
(%) | PIK3CA
amplification (%) |
|---|
| SCC | 4/17 (5.6) | 0/71 (0.0) | 0/71 (0.0) | 1/71 (1.4) |
| AD/ASC | 3/53 (5.7) | 3/53 (5.7) | 1/51 (2.0) | 11/53 (20.7) |
 | Table II.Association between PIK3CA
mutation, amplification and cervical carcinoma histological
subtype. |
Table II.
Association between PIK3CA
mutation, amplification and cervical carcinoma histological
subtype.
|
|
| PIK3CA
mutation (exon 9) | PIK3CA
mutation (exon 20) | PIK3CA
amplification |
|---|
|
|
|
|
|
|
|---|
| Histological
subtype | Patients | − | + | P-value | − | + | P-value | − | + | P-value |
|---|
| SCC | 71 | 71 | 0 | 0.0424 | 67 | 4 | 0.848 | 70 | 1 | 0.0003 |
| AD/ASC | 53 | 50 | 3 |
| 53 | 0 |
| 42 | 11 |
|
Frequency of PIK3CA gene amplification
is higher in adeno/adenosquamous cell carcinomas than in squamous
cell carcinomas
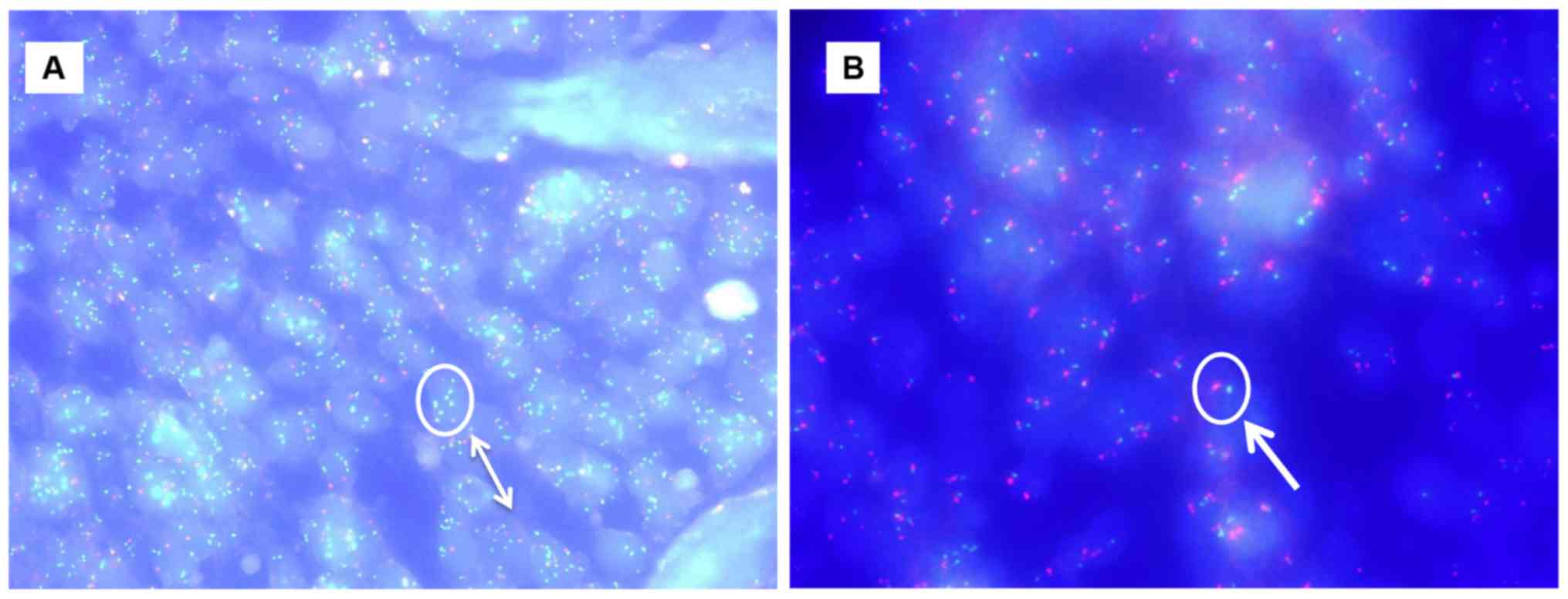
PIK3CA amplification was identified in one
(1.4%) of 71 cervical squamous cell carcinomas. In contrast,
PIK3CA amplification was identified in 11 (20.7%) of 53
adeno/adenosquamous cell carcinomas. Amplification of PIK3CA
was more frequently found of adeno/adenosquamous carcinomas than in
squamous cell carcinomas (P<0.0003, χ2-test; Table II; Fig.
2). Interestingly, amplification of PIK3CA was not found
in the same patients harboring PIK3CA mutations.
Prognostic effect of PIK3CA
amplification
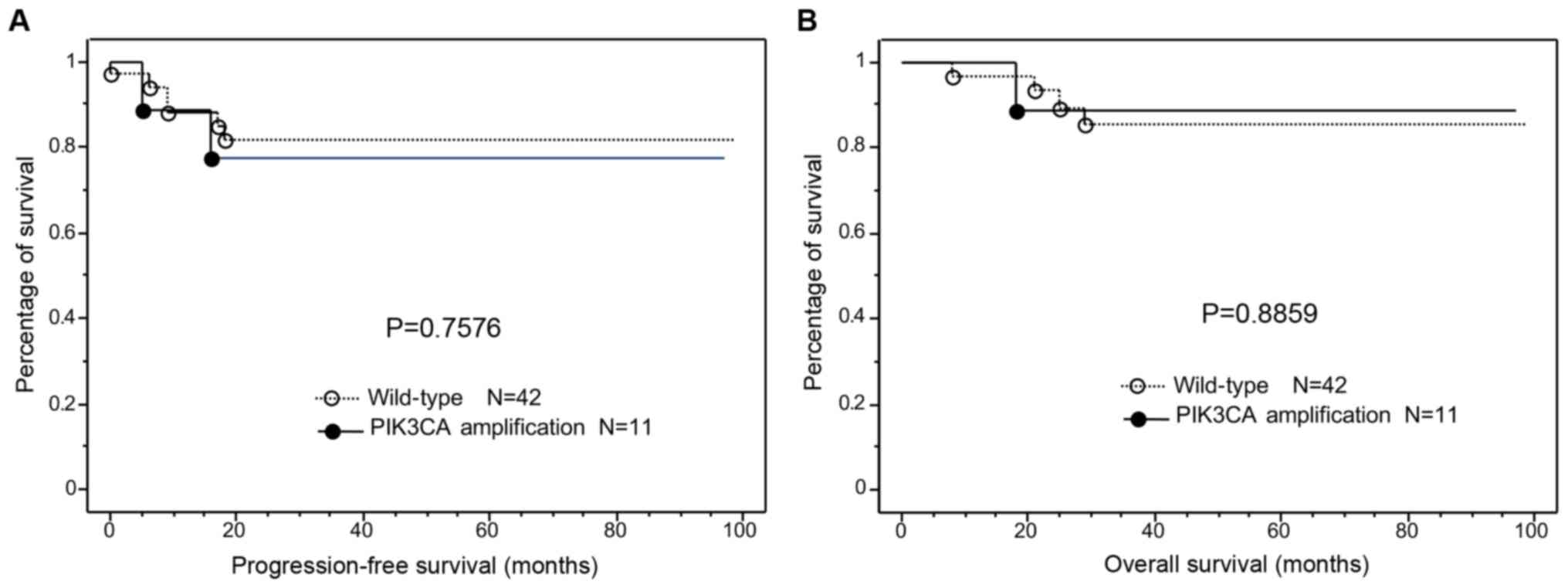
Next, we examined the prognostic ability of
PIK3CA amplification. Kaplan-Meier curves were used to plot
progression free survival and overall survival (Fig. 3), and we observed no significant
relationship between PIK3CA amplification and progression
free/overall survival (P=0.7576 and P=0.8859, respectively).
Association between PIK3CA mutational
status and growth inhibition
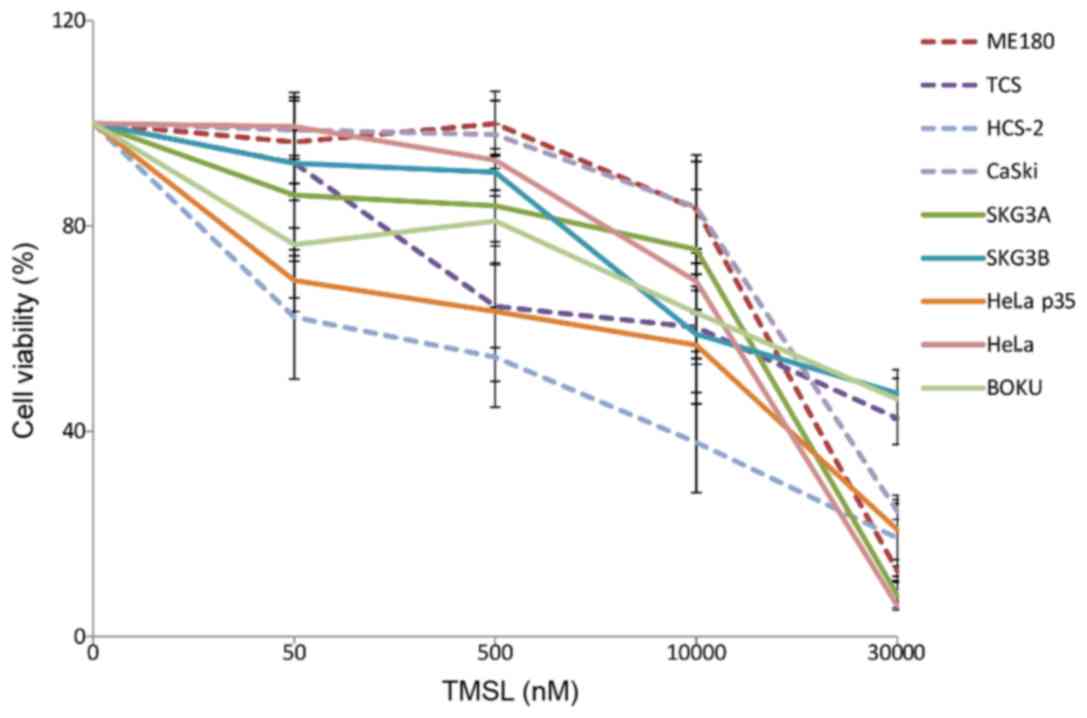
Squamous cell carcinoma and adeno/adenosquamous cell
lines were first analyzed for PIK3CA mutational status. Of
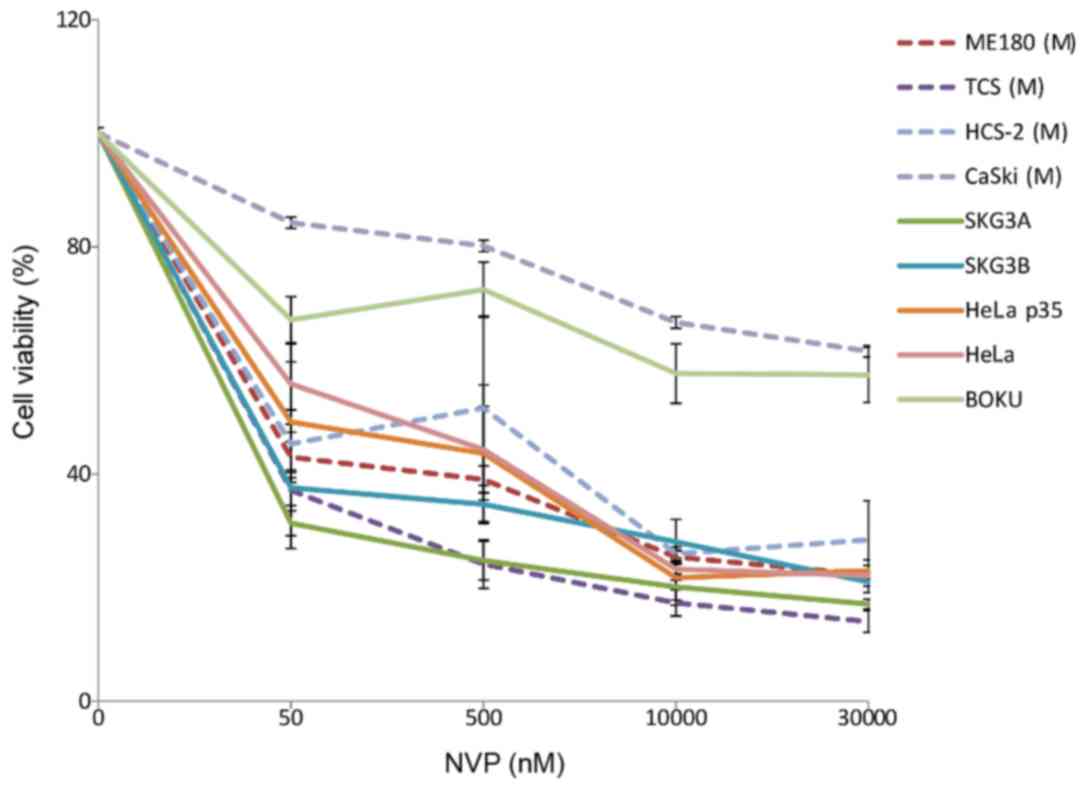
nine cervical cancer cell lines, PIK3CA mutations were
observed in four (ME180, TCS, HCS-2, and CASKI), but only in Exon
9. Next, we used the potent inhibitor temsirolimus (formerly known
as CCI-799) and NVP-BEZ235 to examine the relationship between
mutational status and growth inhibition. Growth of the
PIK3CA mutation-containing cell lines was not inhibited by
any of the inhibitors (Figs. 4 and
5). Treatment with PI3K/AKT/mTOR
inhibitors failed to inhibit proliferation in all four cell lines
harboring PIK3CA mutations.
Discussion
PIK3CA mutations were found in only four of
71 (5.6%) cervical squamous cell carcinoma samples, and were
identified in exon 20 (P=0.848). Similar mutations were present in
three of 53 (5.7%) cervical adeno/adenosquamous cell carcinoma
samples, and these were identified in exon 9 (P=0.042). No tumors
harbored mutations in both the helical and kinase domain. These
results indicated that mutations are more common in exon 9 for
adeno/adenosquamous cell carcinoma and in exon 20 for cervical
squamous carcinomas in Japanese cervical cancer patients. Our
findings suggest that adeno/adenosquamous carcinomas could be
distinguished from squamous cell carcinomas based on genetic
alterations. Lou et al (9),
found that PIK3CA mutations in the helical domain are
significantly more prevalent in squamous cell carcinomas than in
adenocarcinomas (P=0.017) in Latin American patients with cervical
cancer, which is not consistent with the results of our study.
Xiang et al (10), identified
105 cases (13.6%) with PIK3CA mutations from a cohort of
Chinese patients with cervical carcinoma. The most common mutations
were found in exon 9, at residues 545 and 542, in 59 and 32
samples, respectively. The H1047R mutation has been reported in
four cases, and several rare non synonymous base substitutions were
also identified in Chinese patients, some of which have been
reported in the Catalog of Somatic Mutation in Cancer (COSMIC)
database. Two mutations were identified in Chinese patients that
were not previously reported. In addition, mutations in
PIK3CA were found in 93 (15.3%) of the 606 patients with
squamous cell carcinomas, nine (8.9%) of the 101 patients with
adenocarcinomas, and one (2.3%) of the 44 patients with
adenosquamous carcinoma. Therefore, PIK3CA mutations also
occurred more frequently in squamous cell carcinomas than in
non-squamous cell carcinomas in Chinese patients; moreover,
PIK3CA mutations were mostly activating helical domain
mutations, specifically E542K and E545K (10). The cancer Genome Atlas Research
Network described the genomic and molecular characterization of
cervical cancer and observed that most PIK3CA mutations
occurred at the helical domain of exon 9 (E542K and E545K) and that
PIK3CA (P=0.01) were differentially expressed between
keratin-low and keratin-high squamous cluster gene family members
(18). Comparing the results of the
current study with those of previous studies, we hypothesized that
the difference in prevalence is due to a difference in genetic
background between the Japanese population and other ethnic
groups.
PIK3CA gene amplification was also detected
in this study and our results suggested that 11 of 53 (20.7%)
samples were associated with PIK3CA gene amplification in
adeno/adenosquamous cell carcinoma. In contrast, one of 71 (1.4%)
cervical squamous cell carcinomas exhibited PIK3CA gene
amplification. This event occurred at a significantly higher
prevalence (P=0.0003) in adeno/adenosquamous cell carcinoma than in
cervical squamous cell carcinomas. Our data suggest that cervical
squamous cell carcinoma and adenocarcinoma have distinct gene
expression profiles that might arise from different pathways
involved in carcinogenesis. We previously found that Notch3 is
significantly overexpressed in cervical squamous cell carcinomas
compared to expression in cervical cancer adenocarcinomas (19). Wright et al (5), identified KRAS mutations only in
adenocarcinomas (17.5% (AC) vs. 0% (SCC); P=0.01), which is similar
to our results; in addition, a novel EGFR mutation was
previously detected only in squamous cell carcinomas (0% (AC) vs.
7.5% (SCC); P=0.24). Taken together, our results and those of
previous studies suggest that cervical squamous cell carcinoma and
adenocarcinoma have distinct molecular profiles.
Activating mutations and amplification of
PIK3CA, the gene that encodes the catalytic subunit of
phosphatidylinositol 3-kinase (PI3K), have also been reported in
23–36% of cervical cancer specimens (20–22).
Based on observational studies, activation of the PI3K pathway has
been associated with higher rates of local recurrence after
radiotherapy and decreased survival (22,23).
Next, we examined the relationship between PIK3CA
amplification and prognosis and found that PIK3CA
amplification did not correlate with progression free survival and
overall survival. Possible reasons for the different results in our
study compared to those of others, including PIK3CA mutation
rate in cervical cancer and the prognostic effect of PIK3CA
amplification, might be different patient cohorts or sample
sizes.
Mutations in PIK3CA are becoming a promising
target for newly discovered anticancer drugs. Janku et al
(24), observed that in patients
with advanced breast, ovarian, endometrial, and cervical cancers,
an association between PIK3CA mutations and positive
response to PI3K/AKT/mTOR inhibitors was apparent. McIntyre et
al (22), recently identified
that among cervical cancer patients with PIK3CA mutations
treated with radical chemoradiotherapy, there was a strong
association with overall survival in FIGO stage IB/II but not stage
III/IVA. In the present study, PI3K/AKT/mTOR inhibitors such as
temsirolimus and NVP-BEZ235 were tested in cervical cancer cell
lines; it was found that mutational status does not correlate with
growth inhibition with any of the tested inhibitors. These
observations suggest that PIK3CA inhibition might not
represent a potential therapeutic option for the treatment of
cervical cancers with associated mutations.
In conclusion, our data demonstrate that a
proportion of Japanese cervical cancer patients harbor mutations in
PIK3CA. Helical domain-encoding PIK3CA mutations are
more frequent in adeno/adenosquamous cell carcinomas, whereas
kinase domain-encoding PIK3CA mutations are more frequent in
cervical squamous cell carcinomas. Our study also suggests that
PIK3CA amplification occurs at a significantly higher rate
(P=0.0003) in adeno/adenosquamous cell carcinoma than in cervical
squamous cell carcinomas, and that PIK3CA amplification does
not correlate with progression free survival or overall survival.
The mutation status of PIK3CA also did not predict
sensitivity to PI3K/AKT/mTOR inhibitors in cervical carcinoma cells
in vitro. This result suggests that PIK3CA mutations
might not be an important parameter for predicting treatment
response in Japanese cervical cancer patients.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Ministry of Education, Culture, Sports, Science and Technology in
Japan (grant no. 15K10717).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on responsible
request.
Authors' contributions
SR and KeN drafted the manuscript. KoN, TI, MI, and
TM performed data collection, analysis and interpretation of data.
SR and KI conducted the experimental trials. YO, SN and NI
performed pathological diagnosis. KeN participated in the design of
the study. SK conceived the study, participated in its design and
coordination, and drafted the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from the Ethics
Committee of Shimane Medical University. All patients provided
written informed consent for the procedure and study
participation.
Patient consent for publication
All patients approved their data for
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yeasmin S, Nakayama K, Rahman MT, Rahman
M, Ishikawa M, Katagiri A, Iida K, Nakayama N, Otuski Y, Kobayashi
H, et al: Biological and clinical significance of NAC1 expression
in cervical carcinomas: A comparative study between squamous cell
carcinomas and adenocarcinomas/adenosquamous carcinomas. Hum
Pathol. 43:506–519. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith HO, Tiffany MF, Qualls CR and Key
CR: The rising incidence of adenocarcinoma relative to squamous
cell carcinoma of the uterine cervix in the United States-a 24-year
population-based study. Gynecol Oncol. 78:97–105. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chan PG, Sung HY and Sawaya GF: Changes in
cervical cancer incidence after three decades of screening US women
less than 30 years old. Obstet Gynecol. 102:765–773. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu S, Semenciw R and Mao Y: Cervical
cancer: The increasing incidence of adenocarcinoma and
adenosquamous carcinoma in younger women. CMAJ. 164:1151–1152.
2001.PubMed/NCBI
|
|
5
|
Wright AA, Howitt BE, Myers AP, Dahlberg
SE, Palescandolo E, Van Hummelen P, MacConaill LE, Shoni M, Wagle
N, Jones RT, et al: Oncogenic mutations in cervical cancer: Genomic
differences between adenocarcinomas and squamous cell carcinomas of
the cervix. Cancer. 119:3776–3783. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vivanco I and Sawyer CL: The
phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chang L, Graham PH, Hao J, Ni J, Bucci J,
Cozzi PJ, Kearsley JH and Li Y: PI3K/Akt/mTOR pathway inhibitors
enhance radiosensitivity in radioresistant prostate cancer cells
through inducing apoptosis, reducing autophagy, suppressing NHEJ
and HR repair pathways. Cell Death Dis. 5:e14372014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wright AA, Howitt BE, Myers AP, Dahlberg
SE, Palescandolo E, Van Hummelen P, MacConaill LE, Shoni M, Wagle
N, Jones RT, et al: Oncogenic mutations in cervical cancer: Genomic
differences between adenocarcinomas and squamous cell carcinomas of
the cervix. Cancer. 119:3776–3783. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lou H, Villagran G, Boland JF, Im KM, Polo
S, Zhou W, Odey U, Juárez-Torres E, Medina-Martínez I,
Roman-Basaure E, et al: Genome Analysis of Latin American Cervical
Cancer: Frequent activation of the PIK3CA pathway. Clin
Cancer Res. 21:5360–5370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiang L, Jiang W, Li J, Shen X, Yang W,
Yang G, Wu X and Yang H: PIK3CA mutation analysis in Chinese
patients with surgically resected cervical cancer. Sci Rep.
11:140352015. View Article : Google Scholar
|
|
11
|
Nakayama K, Takebayashi Y, Namiki T,
Tamahashi N, Nakayama S, Uchida T, Miyazaki K and Fukumoto M:
Comprehensive allelotype study of ovarian tumors of low malignant
potential: Potential differences in pathways between tumors with
and without genetic predisposition to invasive carcinoma. Int J
Cancer. 94:605–609. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jones S, Wang TL, Shih leM, Mao TL,
Nakayama K, Roden R, Glas R, Slamon D, Diaz LA Jr, Vogelstein B, et
al: Frequent mutations of chromatin remodeling gene ARID1A in
ovarian clear cell carcinoma. Science. 330:228–231. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singer G, Kurman RJ, Chang HW, Cho SK and
Shih IeM: Diverse tumorigenic pathways in ovarian serous carcinoma.
Am J Pathol. 160:1223–1228. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singer G, Oldt R III, Cohen Y, Wang BG,
Sidransky D, Kurman RJ and Shih IeM: Mutations in BRAF and KRAS
characterize the development of low-grade ovarian serous carcinoma.
J Natl Cancer Inst. 95:484–486. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakayama K, Nakayama N, Kurman RJ, Cope L,
Pohl G, Samuels Y, Velculescu VE, Wang TL and Shih IeM: Sequence
mutations and amplification of PIK3CA and AKT2 genes in
purified ovarian serous neoplasms. Cancer Biol Ther. 5:779–785.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nakayama K, Miyazaki K, Kanzaki A,
Fukumoto M and Takebayashi Y: Expression and cisplatin sensitivity
of copper-transporting P-type adenosine triphosphatase (ATP7B) in
human solid carcinoma cell lines. Oncol Rep. 8:1285–1287. 2011.
|
|
18
|
Cancer Genome Atlas Research Network;
Albert Einstein College of Medicine; Analytical Biological
Services; Barretos Cancer Hospital; Baylor College of Medicine;
Beckman Research Institute of City of Hope; Buck Institute for
Research on Aging; Canada's Michael Smith Genome Sciences Centre;
Harvard Medical School; Helen F. Graham Cancer Center &Research
Institute at Christiana Care Health Services et al, . Integrated
genomic and molecular characterization of cervical cancer. Nature.
543:378–384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yeasmin S, Nakayama K, Rahman MT, Rahman
M, Ishikawa M, Iida K, Otsuki Y, Kobayashi H, Nakayama S and
Miyazaki K: Expression of nuclear Notch3 in cervical squamous cell
carcinomas and its association with adverse clinical outcomes.
Gynecol Oncol. 117:409–416. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Janku F, Lee JJ, Tsimberidou AM, Hong DS,
Naing A, Falchook GS, Fu S, Luthra R, Garrido-Laguna I and Kurzrock
R: PIK3CA mutations frequently coexist with RAS and BRAF
mutations in patients with advanced cancers. PLoS One.
6:e227692011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bertelsen BI, Steine SJ, Sandvei R, Molven
A and Laerum OD: Molecular analysis of the PI3K-AKT pathway in
uterine cervical neoplasia: Frequent PIK3CA amplification
and AKT phosphorylation. Int J Cancer. 118:1877–1883. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McIntyre JB, Wu JS, Craighead PS, Phan T,
Köbel M, Lees-Miller SP, Ghatage P, Magliocco AM and Doll CM:
PIK3CA mutational status and overall survival in patients
with cervical cancer treated with radical chemoradiotherapy.
Gynecol Oncol. 128:409–414. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim TJ, Lee JW, Song SY, Choi JJ, Choi CH,
Kim BG, Lee JH and Bae DS: Increased expression of pAKT is
associated with radiation resistance in cervical cancer. Br J
Cancer. 94:1678–1682. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Janku F, Wheler JJ, Westin SN, Moulder SL,
Naing A, Tsimberidou AM, Fu S, Falchook GS, Hong DS, Garrido-Laguna
I, et al: PI3K/AKT/mTOR inhibitors in patients with breast and
gynecologic malignancies harboring PIK3CA mutations. J Clin
Oncol. 30:777–782. 2012. View Article : Google Scholar : PubMed/NCBI
|