Introduction
Gouty arthritis (GA) is one of the most common types
of inflammatory arthritis and is caused by the deposition of
monosodium urate (MSU) crystals in and around joints. Elevated
serum urate levels are recognized as an important risk factor for
the development of GA (1). However,
only ~10% of individuals with hyperuricaemia actually develop GA.
Therefore, it has been suggested that hyperuricemia alone is not
sufficient to cause GA (2). Although
previous studies have identified several candidate risk genes
(SLC2A9, ABCG2 and URAT1) associated with raised serum urate
concentrations that increase the risk for an individual to develop
GA (3–5), the exact pathogenesis of gout remains
elusive. There may be other genes that are not associated with
urate metabolism that also contribute to susceptibility to this
disease.
Long non-coding RNAs (lncRNAs) are a group of RNA
transcripts that are >200 nt in length. Initially, they were
thought to be ‘transcriptional noise’ due to their lack of
significant open reading frames and protein-coding ability. The
human genome encoded tens of thousands of lncRNAs (6). In the past decade, an increasing number
of lncRNAs have been characterized. Certain lncRNAs have been
indicated to have important roles in diverse biological processes,
including cell development, tumorigenesis and immune response.
lncRNAs exert critical functions in the transcriptional,
post-transcriptional and epigenetic regulation of gene expression
(7). Although several studies have
identified lncRNAs involved in a series of human biological and
disease-associated processes (8,9), only
isolated examples on the regulation of rheumatic diseases through
lncRNAs have been provided and the roles of lncRNAs in the
progression of GA have remained to be fully elucidated.
In the present study, lncRNA microarrays were
performed to evaluate the global expression profile of lncRNAs in
peripheral blood mononuclear cells (PBMCs) of GA patients. In a
subsequent Bioinformatics analysis, the lncRNA functions were
annotated by Gene Ontology (GO) analysis, pathway analysis and
network analysis. Reverse transcription-quantitative (RT-q)PCR was
used to validate several random lncRNAs that were upregulated and
downregulated in the GA patients compared with those in the healthy
controls (HCs). These results provide information for future
studies on GA.
Patients and methods
Study population and ethical
statement
All participants were recruited from the Department
of Rheumatology of the Affiliated Hospital of North Sichuan Medical
College (Nanchong, China) between February 2015 and July 2016. The
study group consisted of 60 GA patients and 60 HC subjects. The
diagnoses of GA were confirmed by a clinical endocrinology
physician, according to the American College of Rheumatology
classification criteria (10). The
patients had not received any systemic anti-inflammatory treatments
or drugs to control the production and elimination of uric acid
prior to obtainment of the blood samples. According to the design
of a matched case-control study, healthy subjects with no history
of gout and without any systemic inflammatory disease were enrolled
in the present study. All the participants were of Chinese Han
descent. Blood samples from all participants were obtained in the
morning following overnight fasting for at least 12 h, and were
collected in sterile, single-use, anticoagulant-coated tubes and
immediately sent to the laboratory for genetic testing.
RNA isolation
Ficoll-Hypaque density gradient centrifugation was
performed to isolate the PBMCs from total blood samples. Total RNA
was extracted from PBMCs using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
RNA quantity and quality were measured using a NanoDrop ND-2000c
spectrophotometer (Thermo Fisher Scientific, Inc.) All of the RNA
samples were stored at −80°C until further use.
Microarray analysis
Samples from three GA patients and three HC subjects
were used for the lncRNA microarray analysis on an Agilent
SurePrint G3 Gene Expression Microarrays for Human (v3) platform
(part no. G4851C; Agilent Technologies). The RNA integrity of these
samples was assessed with an Agilent Bioanalyzer 2100 (Agilent
Technologies). In this study, RNA integrity number (RIN) and 28S to
18S rRNA ratio (28S/18S) were used as a criterion for RNA quality.
Only samples with 2,100 RIN ≥7.0 and 28S/18S ≥0.7 were used. Total
RNA was amplified and labelled with a Low Input Quick Amp Labeling
Kit, One-Color (Agilent Technologies), following the manufacturer's
protocol. Labelled cRNA (pmol Cy3/µg cRNA) was purified with an
RNeasy mini kit (Qiagen GmBH). Each slide was hybridized with 1.65
µg Cy3-labelled cRNA using a Gene Expression Hybridization Kit
(Agilent Technologies) in a hybridization oven (Agilent
Technologies) according to the manufacturer's protocol. After 17 h
of hybridization, slides were washed in staining dishes (Thermo
Fisher Scientific, Inc.) with a Gene Expression Wash Buffer Kit
(Agilent Technologies) following the manufacturer's protocol.
Slides were scanned with an Agilent microarray scanner (Agilent
Technologies) with default settings (dye channel: Green, scan
resolution=3 µm, 20 bit). Data were extracted with Feature
Extraction software 10.7 (Agilent Technologies). Raw data were
normalized with a Quantile algorithm, with Gene Spring Software
11.0 (Agilent Technologies). Differentially expressed lncRNAs and
mRNAs between the two groups were identified when fold-change
≥2.
‘Cis’ and ‘Trans’ analysis of
lncRNAs
The cis role of lncRNA refers to the lncRNA acting
on neighbouring target genes (11,12). The
protein-coding genes 10 kbp upstream or downstream of the lncRNAs
were screened as potential ‘cis’-interacting genes. The trans
predictions were made using blast to identify complementary or
similar sequences. The complementary energy between the two
sequences was calculated by using RNAplex. The genes with e ≤-30
were selected as potential ‘trans’-interacting genes (13).
Gene Ontology (GO) enrichment and
pathway analysis
GO analysis was used to identify functional terms in
different categories enriched by the differentially expressed mRNAs
(www.geneontology.org; release
2016-08-08) (14). The KEGG database
(www.genome.jp/kegg; release 79, 2016/07)
was used to determine the biological pathways that were enriched by
the differentially expressed mRNAs (15).
RT-qPCR assay
The lncRNAs selected and the sequences of the
primers that were used for qPCR are provided in Table I. GAPDH was used as the endogenous
control. In brief, total RNA was extracted as described above and
then reverse-transcribed into complementary (c)DNA using a
PrimeScript® RT Reagent kit with gDNA Eraser (Perfect
Real Time; Takara Bio Inc.) following the manufacturer's protocol.
The differential expression of lncRNAs that were identified in the
microarray analysis was measured by qPCR using SYBR Green assays
(Takara Bio Inc.). The reactions were performed in a final volume
of 20 µl and included 10 µl Power SYBR Green PCR Master Mix, 0.4 µl
ROX Dye, 0.5 µl final working concentration of 10 pmol/l of each of
the forward and reverse primers, 2 µl cDNA and 6.6 µl nuclease-free
water. The reactions were performed using a QuantStudio™ 12K Flex
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The thermocycling conditions for PCR were as follows:
Pre-denaturation at 95°C for 10 min, and amplification for 40
cycles of denaturation at 95°C for 15 sec and elongation at 60°C
for 1 min. All the experiments were performed in duplicate and the
mean value was used for further analysis. Relative lncRNA
concentrations were calculated using the 2−ΔΔCq
method (16).
 | Table I.Primer sequences used for validation
of lncRNAs by reverse transcription-quantitative PCR. |
Table I.
Primer sequences used for validation
of lncRNAs by reverse transcription-quantitative PCR.
| lncRNA | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Length (bp) |
|---|
| AJ227913 |
TTTTGCCAAGGAGTGCTAAAGA |
AACCCTCTGCACCCAGTTTTC | 194 |
| AK001903 |
CCAGCGGATATTTTGGTGTTTG |
AGAAGCTATCAGGCGTTGCTG | 86 |
|
ENSG00000239182 |
TAGCACTGTTGCCTGAGCCA |
GGAAGGAGCAGCCCACAGC | 95 |
|
lnc-AP000769.1–1 |
CAAGCAGAAGCAACAGGTCA |
GAGCCAGGAAGATTGGAGAA | 144 |
|
lnc-PCYOX1L-1:1 |
GGAAAGGCAGTAATCAACTCCA |
ACTCCACAATCCCCACAGC | 171 |
|
lnc-CCDC64B-1:8 |
ACCCCCACCCCAGGTCTTC |
GCTGTGTCTCTGTCTTGGTCTCTT | 268 |
| GAPDH |
ATCATCCCTGCCTCTACTG |
AGTCAGAGGAGACCACCTG | 241 |
Statistical analysis
Statistical analysis was performed using the SPSS
22.0 software package (IBM Corp.). To analyse differences in
expression of individual lncRNAs or mRNAs in the microarray
analysis, Student's t-tests were used. Regarding the results of the
PCR analysis, the significance of differences in expression levels
of lncRNAs were determined by the Kruskal-Wallis test or
Mann-Whitney U-test. For statistical correlation, Spearman
correlation coefficient was used. Fisher's exact test was used in
the GO enrichment and KEGG pathway analyses. P<0.05 was
considered to indicate statistical significance.
Results
Overview of the lncRNA and mRNA
profiles
The expression levels of the lncRNAs and mRNAs from
three GA patients and paired control samples were analysed using
lncRNA expression microarrays, which contained a total of 63,431
lncRNAs. All lncRNAs and mRNAs that were differentially expressed
with a fold-change ≥2.0 between the GA and HC groups are provided
in Tables SI and SII. Analysis of the microarray data
revealed that 1,815 lncRNAs and 971 mRNAs were significantly
differentially expressed in GA patients compared with those in the
HC group. In total, there were 875 upregulated lncRNAs and 940
downregulated lncRNAs in GA patients compared with those in the HC
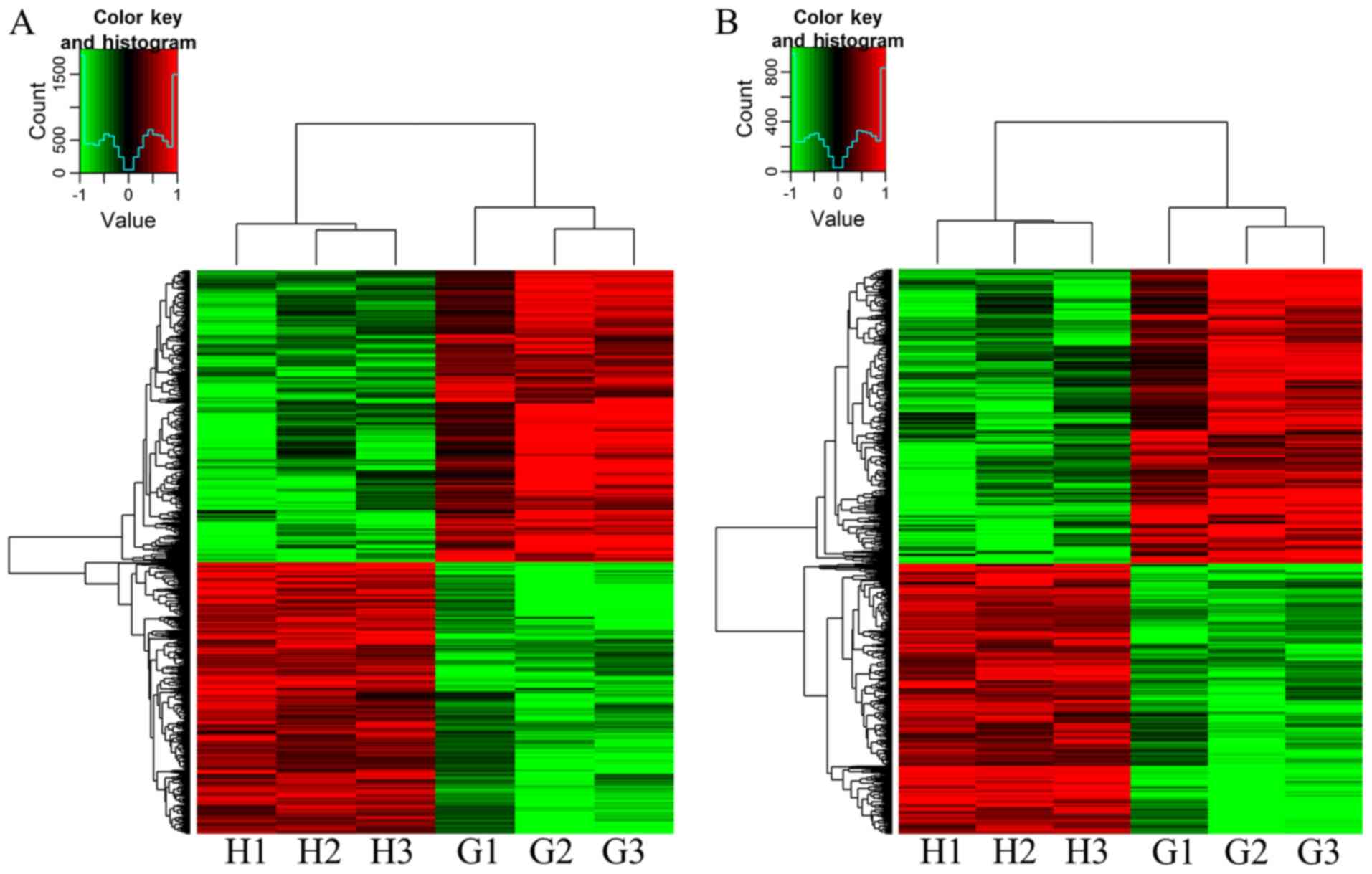
group (Fig. 1A). A total of 465
mRNAs were upregulated and 506 mRNAs were downregulated in the GA
patients compared with those in the HC group (Fig. 1B). Table
II presents the 10 most upregulated and downregulated lncRNAs
in the GA patients vs. HC group. Among these lncRNAs, AJ227913
(fold-change, 17.014) was the most upregulated lncRNA and 52094-1
(fold-change, 11.772) was the most downregulated lncRNA in the GA
patients vs. HC group. Table III
lists the 10 most upregulated and downregulated mRNAs. The most
significantly upregulated mRNA was NM_005217 (fold-change, 18.208),
and the most downregulated mRNA was NM_005353 (fold-change, 14.047)
in the GA patients vs. HC group. Studies have suggested that
several lncRNAs regulate their own transcription in ‘cis’, as well
as that of nearby genes, by recruiting remodelling factors to local
chromatin (11). Therefore, the
chromosomal co-expression was then analysed. According to this
analysis, the lncRNAs had 148 ‘cis’ genes and 145 ‘trans’ genes.
The downregulated lncRNAs had 233 ‘cis’ genes and 338 ‘trans’ genes
(detailed results are displayed in Tables SI and SII).
 | Table II.Top 10 significantly differentially
expressed lncRNAs between gouty arthritis patients and healthy
controls. |
Table II.
Top 10 significantly differentially
expressed lncRNAs between gouty arthritis patients and healthy
controls.
| A, Upregulated
lncRNAs |
|---|
|
|---|
| lncRNA name | Source
database | Fold change | P-value |
|---|
| AJ227913 | NONCODERv3 | 17.014 | 0.035 |
| AK001903 | NONCODERv3 | 10.971 | 0.016 |
| THC2686626 |
Agilent_HUMAN_G3V2 | 9.332 | 0.034 |
| TCONS_00026568 |
Agilent_HUMAN_G3V2 | 8.450 | 0.008 |
| lnc-COL20A1 | lncipedia2.1 | 7.833 | 0.038 |
|
ENST00000604411.1 | gencode_v17 | 7.741 | 0.020 |
|
lnc-AP000769.1–1 | lncipedia2.1 | 7.710 | 0.006 |
| TCONS_00026396 |
Agilent_HUMAN_G3V2 | 7.607 | 0.008 |
| TCONS_00018683 |
Agilent_HUMAN_G3V2 | 7.526 | 0.026 |
|
TCONS_l2_00003767 |
Agilent_HUMAN_G3V2 | 6.914 | 0.012 |
|
| B, Downregulated
lncRNAs |
|
| lncRNA
name | Source
database | Fold
change | P-value |
|
| 52094-1 | UCSC | 11.772 | 0.001 |
|
TCONS_l2_00000650 |
Agilent_HUMAN_G3V2 | 11.392 | 0.007 |
| CR616125 | NONCODERv3 | 9.367 | 0.012 |
| lnc-KCNT2 | lncipedia2.1 | 8.885 | 0.018 |
| BX538226 | NONCODERv3 | 8.794 | 0.001 |
|
ENST00000430519 |
Agilent_HUMAN_G3V2 | 8.410 | 0.025 |
| lnc-APTX-1 | lncipedia2.1 | 7.178 | 0.001 |
|
TCONS_l2_00007044 | UCSC | 7.000 | 0.033 |
| 37475-1 | NONCODERv3 | 6.935 | 0.017 |
| AK056081 |
Agilent_HUMAN_G3V2 | 6.894 | 0.048 |
 | Table III.Top 10 significantly differentially
expressed mRNAs between gouty arthritis patients and healthy
controls. |
Table III.
Top 10 significantly differentially
expressed mRNAs between gouty arthritis patients and healthy
controls.
| A, Upregulated
mRNAs |
|---|
|
|---|
| Primary accession
number | Gene symbol | Fold change | P-value |
|---|
| NM_005217 | DEFA3 | 18.208 | 0.050 |
| NM_014364 | GAPDHS | 14.277 | 0.003 |
| NM_173557 | RNF152 | 11.779 | 0.032 |
| NM_000584 | IL8 | 11.616 | 0.030 |
| DB514319 | NA | 11.600 | 0.028 |
| NM_002192 | INHBA | 11.220 | 0.029 |
| THC2725968 | XLOC_002791 | 10.485 | 0.017 |
|
ENST00000511330 | XLOC_003547 | 9.264 | 0.001 |
| NM_003955 | SOCS3 | 9.186 | 0.002 |
| NM_001964 | EGR1 | 8.8103 | 0.043 |
|
| B, Downregulated
mRNAs |
|
| Primary
accession number | Gene
symbol | Fold
change | P-value |
|
| NM_005353 | ITGAD | 14.047 | 0.007 |
|
ENST00000435913 | NA | 8.365 | 0.016 |
| NM_015267 | CUX2 | 8.346 | 0.042 |
| NR_026970 | LY86-AS1 | 8.068 | 0.033 |
|
ENST00000400449 | NA | 7.909 | 0.014 |
| AK095683 | PSMG4 | 7.880 | 0.037 |
| NM_021161 | KCNK10 | 7.879 | 0.022 |
|
LNCA_33_P3384628 | NA | 7.670 | 0.010 |
| NM_080747 | KRT72 | 7.656 | 0.008 |
| NR_040093 | LOC100505678 | 7.343 | 0.016 |
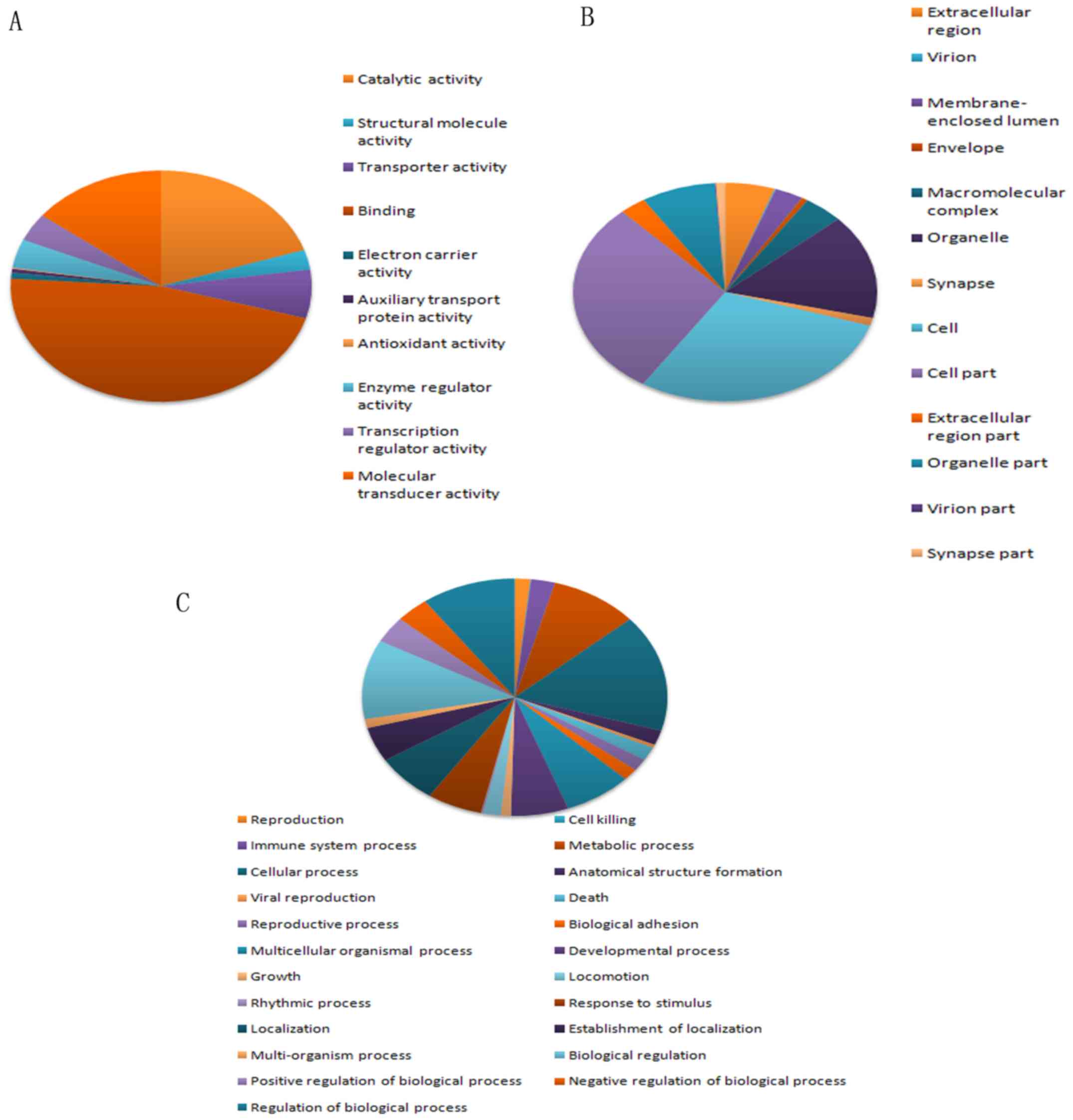
GO analysis
GO analysis was performed to identify the
transcripts with terms that covered 3 domains (Fig. 2): Biological process, cellular
component and molecular function. Fisher's exact test was employed
to determine if there was more overlap between the differentially
expressed list and the GO annotation list than what would be
expected by chance (P<0.05 is recommended). The most highly
enriched GO terms in the category molecular function were
identified as ‘binding’, ‘catalytic activity’ and ‘molecular
transducer activity’. The most highly enriched GO terms in the
category cellular component were ‘cell’ and ‘cell part’. In the
category biological process, the most highly enriched GO term was
‘cellular process’. ‘Metabolic process’, and ‘immune system
process’ were also significantly enriched.
Pathway analysis
KEGG pathway analysis was performed for the
differentially expressed transcripts. The pathway analysis
indicated which biological pathways were significantly enriched for
the differentially expressed transcripts. A total of 19 pathways
corresponded to the differentially expressed transcripts (Table IV). The gene category ‘tumor
necrosis factor (TNF) signaling pathway’ was the most enriched
network, promoting cell survival and differentiation, as well as
immune and inflammatory responses (17). Furthermore, 8 transcripts were
significantly enriched in the pathway ‘osteoclast differentiation’,
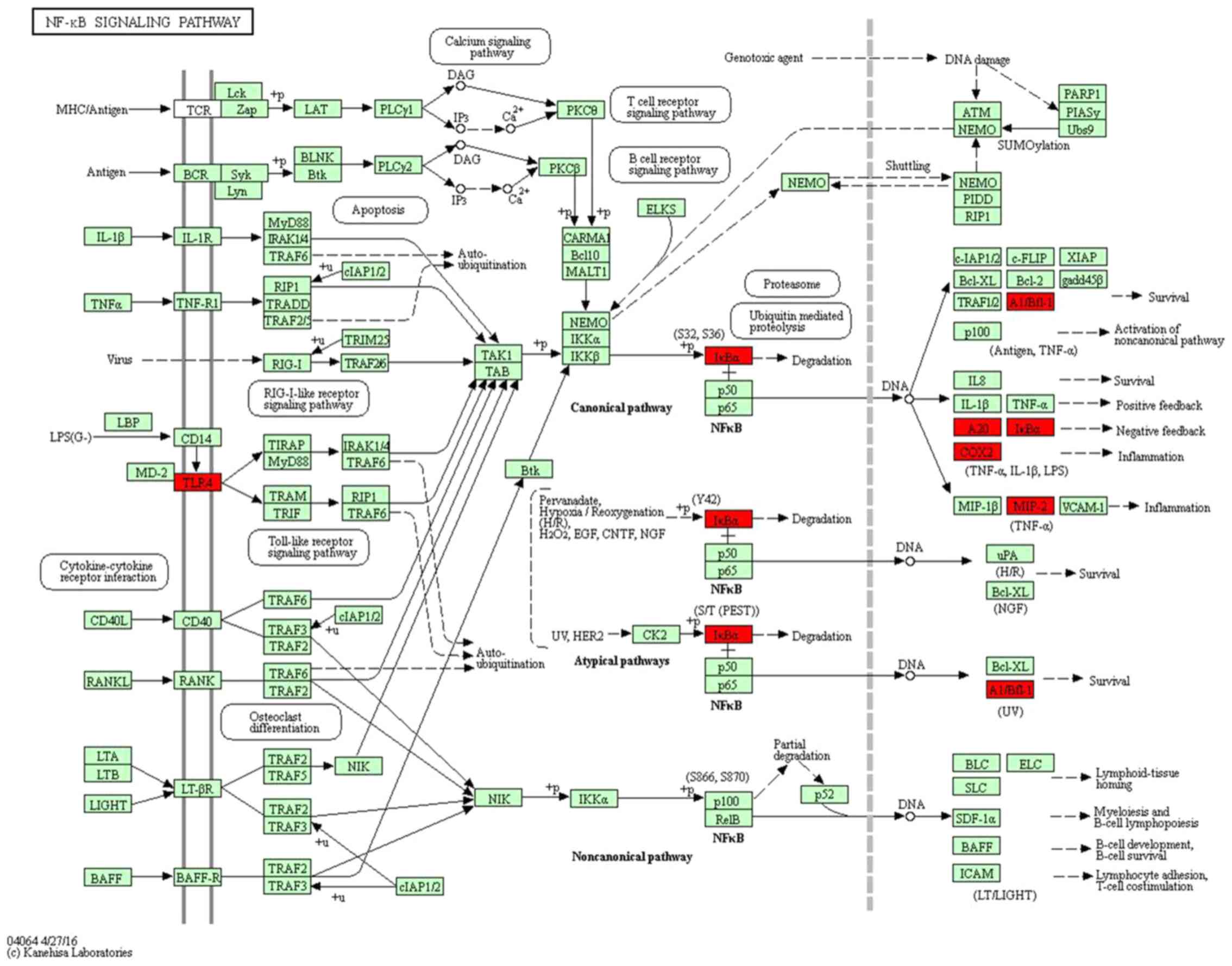
which is closely linked to bone development and repair (18). Among these pathways, the ‘NOD-like
receptor signaling pathway’ and ‘NF-κB signaling pathway’ (Fig. 3) have been previously demonstrated to
be involved in MSU-mediated inflammation through activation of
Toll-like receptors/MyD88-dependent NF-κB signaling, NLR family
pyrin domain containing 3 inflammasome and interleukin
(IL)-1β/MyD88-dependent IL-1 receptor signalling (19).
 | Table IV.KEGG pathway analysis of
differentially expressed transcripts. |
Table IV.
KEGG pathway analysis of
differentially expressed transcripts.
| Term | Description | Count | P-value | Enrichment
score |
|---|
| hsa05134 | Legionellosis | 6 | 0.001 | 5.217 |
| hsa04621 | NOD-like receptor
signaling pathway | 6 | 0.001 | 5.034 |
| hsa04610 | Complement and
coagulation cascades | 7 | 0.001 | 4.238 |
| hsa05150 | Staphylococcus
aureus infection | 5 | 0.007 | 4.195 |
| hsa04668 | TNF signaling
pathway | 11 | 0.000 | 3.913 |
| hsa05140 | Leishmaniasis | 6 | 0.004 | 3.878 |
| hsa04978 | Mineral
absorption | 4 | 0.023 | 3.679 |
| hsa05120 | Epithelial cell
signaling in Helicobacter pylori infection | 5 | 0.014 | 3.516 |
| hsa04012 | ErbB signaling
pathway | 6 | 0.010 | 3.261 |
| hsa05133 | Pertussis | 5 | 0.020 | 3.188 |
| hsa04064 | NF-κB signaling
pathway | 10 | 0.013 | 3.085 |
| hsa05131 | Shigellosis | 4 | 0.047 | 2.943 |
| hsa04380 | Osteoclast
differentiation | 8 | 0.006 | 2.898 |
| hsa05132 | Salmonella
infection | 5 | 0.034 | 2.780 |
| hsa05142 | Chagas disease
(American trypanosomiasis) | 6 | 0.021 | 2.759 |
| hsa05161 | Hepatitis B | 7 | 0.033 | 2.293 |
| hsa04024 | cAMP signaling
pathway | 9 | 0.023 | 2.163 |
| hsa05205 | Proteoglycans in
cancer | 9 | 0.028 | 2.100 |
| hsa05166 | HTLV–I
infection | 8 | 0.020 | 2.031 |
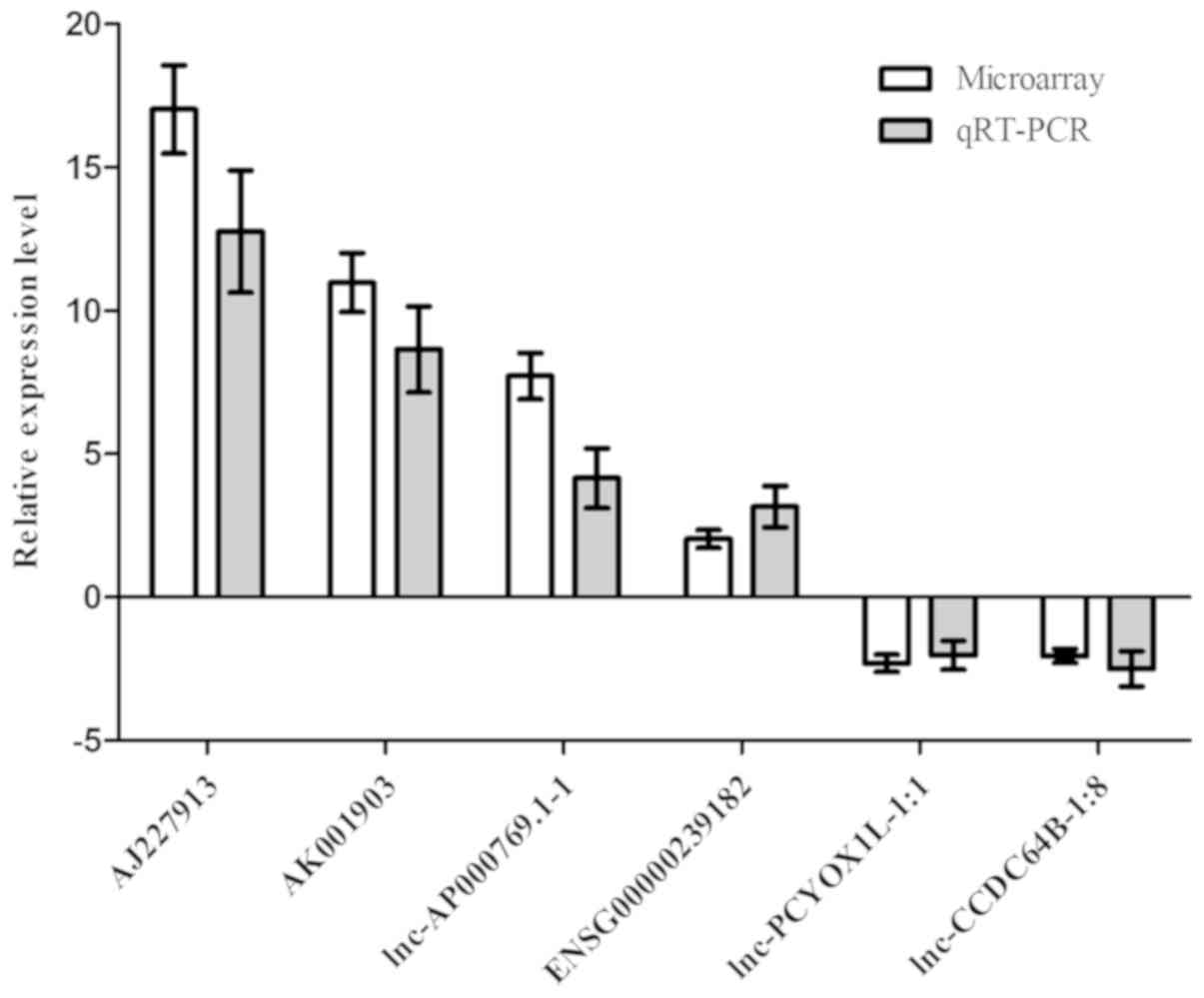
RT-qPCR validation
To validate the results of the microarray, four of
the upregulated lncRNAs and two of the downregulated lncRNAs were
selected for the RT-qPCR analysis of 60 GA patients and 60 HC
subjects from a Han Chinese population. The results confirmed that
the expression of AJ227913 (P<0.05), AK001903 (P<0.05),
ENSG00000239182 (P<0.05) and lnc-AP000769.1–1 (P<0.05) was
significantly increased in gouty arthritis patients compared with
that in healthy subjects, whereas the expression of lnc-PCYOX1L-1:1
(P<0.05) and lnc-CCDC64B-1:8 (P<0.05) was significantly
decreased in GA patients compared with that in healthy subjects.
The fold changes of the normalized levels for the six lncRNAs were
17.02, 10.97, 7.71, 2.03, 2.31 and 2.05, respectively, in the gene
chip analysis comparing the GA to the HC groups. Regarding the
expression levels in the RT-qPCR analysis, these fold changes were
10.76, 7.64, 4.15, 3.16, 2.03 and 2.50, respectively, in the GA
group compared to those in the control group. Thus, the results of
the RT-qPCR and the microarray data were consistent (Fig. 4). Among the lncRNAs analysed,
AJ227913 had the largest difference in the expression between the
two groups. Furthermore, Spearman correlation analysis suggested
that the expression levels of AJ227913 in GA patients were
correlated with urea (r=0.618, P<0.05), creatinine (r=0.382,
P<0.05) and cystatin C (r=0.482, P<0.05; Table SIII).
Discussion
Protein-coding genes account for only ~2% of the
human genome, while a large proportion of the genome is transcribed
to generate non-coding RNAs that have been estimated to comprise up
to 98–99% of the genome (20). It
has become apparent that mutations and abnormal regulation of
certain lncRNAs have important roles in the occurrence and
development of various rheumatic diseases (21). Luo et al (22) indicated that 8,868 lncRNAs were
highly differentially expressed in systemic lupus erythematosus
(SLE) samples compared with those in a healthy group, and overall,
aberrant expression profiles of lncRNAs may provide important
insight into the pathogenesis of SLE (23). Downregulation of lnc-dendritic cell
lncRNA (lnc-DC) in the plasma of patients with SLE may regulate
type 17 T-helper cell differentiation by regulating the expression
of signal transducer and activator of transcription 3; thus, the
plasma levels of lnc-DC may be a potential biomarker for SLE
(24,25). In rheumatoid arthritis (RA) patients,
MALAT1 increases the expression of caspase-3 and caspase-9 and
promotes cell growth and apoptosis of RA fibroblast-like
synoviocyte cells. Methotrexate treatment increases the expression
of lincRNA-p21 and inhibits NF-κB activity compared with those in
untreated RA patients (26,27). Patients with osteoarthritis (OA) were
indicated to have high levels of lncRNA-CIR. Treatment of
chondrocytes in vitro with IL-1β and TNF-α significantly
increased the expression of lncRNA-CIR compared with that in the
controls. Furthermore, silencing of lncRNA-CIR by small interfering
RNA reduced the expression of matrix metalloproteinase (MMP)13 and
ADAM metallopeptidase with thrombospondin type 1 motif 5 compared
with that in the controls. Thus, lncRNA-CIR may act as a potential
target in OA therapy (28,29). To examine the functional implications
of lncRNAs in GA, the expression profiles of lncRNAs in GA patients
and in HCs were determined using microarrays.
In the present study, the lncRNA expression profiles
in 3 GA patients and 3 HCs comprising 63,431 lncRNAs were assessed
by microarray analysis. A total of 875 lncRNAs were significantly
upregulated and 940 lncRNAs were significantly downregulated, which
was by >2-fold. A total of 6 lncRNAs, which were not previously
reported by studies on other diseases, were then randomly selected
for validation in 60 other GA patients and in paired HCs by
RT-qPCR. The results of the RT-qPCR analysis were consistent with
those of the microarray analysis. The differential expression of
lncRNAs in GA implied that lncRNAs may have a crucial role in the
onset and development of GA. However, further validations and
functional studies are warranted in the future. In the present
study, a significant increase in the expression level of AJ227913
in GA patients compared with that in HCs was identified based on
the microarray analysis. The AJ227913 expression levels in GA
patients had a close correlation with urea (r=0.618, P<0.05),
creatinine (r=0.382, P<0.05) and cystatin C (r=0.482, P<0.05)
levels based on the Spearman correlation analysis. Most circulating
uric acid is freely filtered in the kidney. Urea, creatinine,
cystatin C and uric acid are crude indicators of renal filtration
and excretory function. However, uric acid is excreted in urine in
healthy humans. Uric acid excretion may be impaired by kidney
disease, leading to chronic hyperuricaemia. High uric acid levels
in the body may lead to glomerular filtration disorders and a
decrease in renal tubular secretion. It may increase the risk of
gout or gouty nephropathy (30,31).
These results suggest that AJ227913 may be involved in the dynamic
balance of the production and elimination of uric acid in GA
patients.
GO and pathway analyses were performed to predict
the biological functions of the differentially expressed lncRNAs
and potential mechanisms in GA progression. According to this
analysis, the mostly highly enriched GO terms by the differentially
expressed transcripts were binding, catalytic activity, molecular
transducer activity, cellular process, cell and cell part. The GO
project is a framework for modelling biology that may be used to
describe the functional classification of differentially expressed
transcripts. The pathway analysis suggested that the differentially
expressed transcripts were associated with 19 pathways. The gene
category ‘TNF signaling pathway’ promotes cell survival and
differentiation, as well as immune and inflammatory responses
(17). The ‘osteoclast
differentiation’ process is known to be important for bone
development and repair (18). In
addition, the ‘NOD-like receptor signaling pathway’ and ‘NF-κB
signaling pathway’ have been reported to participate in
MSU-mediated signaling cascades, which induce
inflammasome-dependent gouty inflammation (19). These results have prompted us to
investigate the molecular mechanisms of the pathogenesis of gout,
which is important for further studies.
Gene expression is a highly complex, regulated
process with numerous levels of regulation (epigenetic,
transcriptional, post-transcriptional, translational and
posttranslational). lncRNAs act as essential regulators of gene
expression at the epigenetic level. The results of the ‘cis’
analysis performed in the present study suggested that AJ227913 may
regulate IL-8 expression. IL-8 is a member of the chemokine family
and is a key inflammatory mediator, which has an important role in
regulating inflammation and immunity in various rheumatic diseases.
Significantly higher concentrations of IL-8 were detected in the
synovial fluid of RA patients than in those of HCs. IL-8 may
stimulate neutrophils to produce cartilage-degrading enzymes, which
may lead to joint tissue damage in RA. Inhibiting IL-8 production
has been reported to reduce joint damage, which may be a novel
target for the treatment of RA (32–34).
Previous studies have indicated a marked increase in IL-8 in the
exudate from patients with psoriatic arthritis (PsA) compared with
that in the controls. An IL-8 monoclonal antibody was approved as a
topical treatment for PsA in China. The therapeutic effect of
anti-IL-8 is thought to be associated with a decrease in the
accumulation of neutrophils and inflammatory cells, leading to the
reduction of inflammation in patients with PsA (35–37).
These results suggest a pathogenic role for IL-8 in rheumatic
diseases. GA is a type of inflammatory arthritis that is caused by
the deposition of MSU crystals. Several lines of evidence indicate
that IL-8 is an essential mediator of neutrophil-mediated acute
inflammation. The levels of IL-8 are increased during the acute and
intercritical phases of GA. MSU may stimulate the secretion of IL-8
by human neutrophils. In rabbits with MSU crystal-induced
arthritis, intra-articular injection of anti-IL-8 significantly
attenuated crystal-induced joint swelling (38–40).
Therefore, it was hypothesized that AJ227913 may regulate IL-8
expression in GA patients, and is thus involved in the pathogenesis
of GA. Further research focusing on the roles of AJ227913 may
provide a novel strategy for the treatment of GA.
In summary, the present study determined the
expression profiles of patients with GA vs. HCs using microarray
and identified a collection of differentially expressed lncRNAs. To
the best of our knowledge, the present study was the first to
examine the lncRNA profiles in GA. The present results may indicate
that the differential expression of various lncRNAs has an
important role in the development and progression of GA. Further
studies will focus on the biological function of lncRNAs involved
in GA.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The authors acknowledge the financial support from
the Applied Basic Research Program of Sichuan Province (grant no.
2017JY0151), the Science and Technology Support Program of Nanchong
(grant nos. NSMC20170436 and 18SXHZ0513) and the Development of
Scientific Research Plan of Doctoral Scientific Research Foundation
of North Sichuan Medical College (grant no. CBY14-QD-07).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ conducted the majority of the experiments and
wrote the manuscript. XG and JZ conceived and designed the study.
YP and HL performed microarray analysis. CY and JL performed PCR
assays. QY, YH and YQ collected blood samples and analyzed the
clinical data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Affiliated Hospital of the North Sichuan Medical
College (Nantong, China; IRB: 2015-EA-016). All of the participants
who participated in the study provided written informed consent at
the time of enrolment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Richette P and Bardin T: Gout. Lancet.
375:318–328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kuo CF, Grainge MJ, Zhang W and Doherty M:
Global epidemiology of gout: Prevalence, incidence and risk
factors. Nat Rev Rheumatol. 11:649–662. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang X, Yang X, Wang M, Li X, Xia Q, Xu
S, Xu J, Cai G, Wang L, Xin L, et al: Association between SLC2A9
(GLUT9) gene polymorphisms and gout susceptibility: An updated
meta-analysis. Rheumatol Int. 36:1157–1165. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Matsuo H, Takada T, Ichida K, Nakamura T,
Nakayama A, Ikebuchi Y, Ito K, Kusanagi Y, Chiba T, Tadokoro S, et
al: Common defects of ABCG2, a high-capacity urate exporter, cause
gout: A function-based genetic analysis in a Japanese population.
Sci Transl Med. 1:5ra112009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tan PK, Farrar JE, Gaucher EA and Miner
JN: Coevolution of URAT1 and uricase during primate evolution:
Implications for serum urate homeostasis and gout. Mol Biol Evol.
33:2193–2200. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pennisi E: Cell biology. Lengthy RNAs earn
respect as cellular players. Science. 344:10722014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang C, Wang L, Ding Y, Lu X, Zhang G,
Yang J, Zheng H, Wang H, Jiang Y and Xu L: LncRNA structural
characteristics in epigenetic regulation. Int J Mol Sci.
18:E26592017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Saha P, Verma S, Pathak RU and Mishra RK:
Long Noncoding RNAs in mammalian development and diseases. Adv Exp
Med Biol. 1008:155–198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kwok ZH and Tay Y: Long noncoding RNAs:
lincs between human health and disease. Biochem Soc Trans.
45:805–812. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Khanna D, Khanna PP, Fitzgerald JD, Singh
MK, Bae S, Neogi T, Pillinger MH, Merill J, Lee S, Prakash S, et
al: 2012 American college of rheumatology guidelines for management
of gout. Part 2: Therapy and anti-inflammatory prophylaxis of acute
gouty arthritis. Arthritis Care Res (Hoboken). 64:1447–1461. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gomez JA, Wapinski OL, Yang YW, Bureau JF,
Gopinath S, Monack DM, Chang HY, Brahic M and Kirkegaard K: The
NeST Long ncRNA controls microbial susceptibility and epigenetic
activation of the interferon-gamma locus. Cell. 152:743–754. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lai F, Orom UA, Cesaroni M, Beringer M,
Taatjes DJ, Blobel GA and Shiekhattar R: Activating RNAs associate
with Mediator to enhance chromatin architecture and transcription.
Nature. 494:497–501. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
The Gene Ontology Consortium: Expansion of
the Gene Ontology knowledgebase and resources. Nucleic Acids Res.
45(D1): D331–D338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kalliolias GD and Ivashkiv LB: TNF
biology, pathogenic mechanisms and emerging therapeutic strategies.
Nat Rev Rheumatol. 12:49–62. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shapiro F: Bone development and its
relation to fracture repair. The role of mesenchymal osteoblasts
and surface osteoblasts. Eur Cell Mater. 15:53–76. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qing YF, Zhang QB and Zhou JG: Innate
immunity functional gene polymorphisms and gout susceptibility.
Gene. 524:412–414. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mattick JS: Non-coding RNAs: The
architects of eukaryotic complexity. EMBO Rep. 2:986–991. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stachurska A, Zorro MM, van der Sijde MR
and Withoff S: Small and long regulatory RNAs in the immune system
and immune diseases. Front Immunol. 5:5132014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luo Q, Li X, Xu C, Zeng L, Ye J, Guo Y,
Huang Z and Li J: Integrative analysis of long non-coding RNAs and
messenger RNA expression profiles in systemic lupus erythematosus.
Mol Med Rep. 17:3489–3496. 2018.PubMed/NCBI
|
|
23
|
Li LJ, Zhao W, Tao SS, Li J, Xu SZ, Wang
JB, Leng RX, Fan YG, Pan HF and Ye DQ: Comprehensive long
non-coding RNA expression profiling reveals their potential roles
in systemic lupus erythematosus. Cell Immunol. 319:17–27. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S,
Jiang Z, Xu J, Liu Q and Cao X: The STAT3-binding long noncoding
RNA lnc-DC controls human dendritic cell differentiation. Science.
344:310–313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu GC, Li J, Leng RX, Li XP, Li XM, Wang
DG, Pan HF and Ye DQ: Identification of long non-coding RNAs GAS5,
linc0597 and lnc-DC in plasma as novel biomarkers for systemic
lupus erythematosus. Oncotarget. 8:23650–23663. 2017.PubMed/NCBI
|
|
26
|
Pan F, Zhu L, Lv H and Pei C: Quercetin
promotes the apoptosis of fibroblast-like synoviocytes in
rheumatoid arthritis by upregulating lncRNA MALAT1. Int J Mol Med.
38:1507–1514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Spurlock CF III, Tossberg JT, Matlock BK,
Olsen NJ and Aune TM: Methotrexate inhibits NF-κB activity via long
intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol.
66:2947–2957. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Q, Zhang X, Dai L, Hu X, Zhu J, Li L,
Zhou C and Ao Y: Long noncoding RNA related to cartilage injury
promotes chondrocyte extracellular matrix degradation in
osteoarthritis. Arthritis Rheumatol. 66:969–978. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang CL, Peng JP and Chen XD: LncRNA-CIR
promotes articular cartilage degeneration in osteoarthritis by
regulating autophagy. Biochem Biophys Res Commun. 505:692–698.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mount DB: The kidney in hyperuricemia and
gout. Curr Opin Nephrol Hypertens. 22:216–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Prasad Sah OS and Qing YX: Associations
between hyperuricemia and chronic kidney disease: A Review.
Nephrourol Mon. 7:e272332015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Georganas C, Liu H, Perlman H, Hoffmann A,
Thimmapaya B and Pope RM: Regulation of IL-6 and IL-8 expression in
rheumatoid arthritis synovial fibroblasts: The dominant role for
NF-kappa B but not C/EBP beta or c-Jun. J Immunol. 165:7199–7206.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De Benedetti F, Pignatti P, Bernasconi S,
Gerloni V, Matsushima K, Caporali R, Montecucco CM, Sozzani S,
Fantini F and Martini A: Interleukin 8 and monocyte chemoattractant
protein-1 in patients with juvenile rheumatoid arthritis. Relation
to onset types, disease activity, and synovial fluid leukocytes. J
Rheumatol. 26:425–431. 1999.PubMed/NCBI
|
|
34
|
Lun SW, Wong CK, Tam LS, Li EK and Lam CW:
Decreased ex vivo produxtion of TNF-alpha and IL-8 by peripheral
blood cells of patients with rheumatoid arthritis after infliximab
therapy. Int Immuno pharmacol. 7:1668–1677. 2007. View Article : Google Scholar
|
|
35
|
König A, Krenn V, Gillitzer R, Glöckner J,
Janssen E, Gohlke F, Eulert J and Müller-Hermelink HK: Inflammatory
infiltrate and interleukin-8 expression in the synovium of
psoriatic arthritis-an immunohistochemical and mRNA analysis.
Rheumatol Int. 17:159–168. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Biasi D, Carletto A, Caramaschi P,
Bellavite P, Maleknia T, Scambi C, Favalli N and Bambara LM:
Neutrophil functions and IL-8 in psoriatic arthritis and in
cutaneous psoriasis. Inflammation. 22:533–543. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tsai YC and Tsai TF: Anti-interleukin and
interleukin therapies for psoriasis: Current evidence and clinical
usefulness. Ther Adv Musculoskelet Dis. 9:277–294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kienhorst LB, van Lochem E, Kievit W,
Dalbeth N, Merriman ME, Phipps-Green A, Loof A, van Heerde W,
Vermeulen S, Stamp LK, et al: Gout is a chronic inflammatory
disease in which high levels of interleukin-8 (CXCL8),
myeloid-related protein 8/myeloid-related protein 14 complex, and
an altered proteome are associated with diabetes mellitus and
cardiovascular disease. Arthritis Rheumatol. 67:3303–3313. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hachicha M, Naccache PH and McColl SR:
Inflammatory microcrystals differentially regulate the secretion of
macrophage inflammatory protein 1 and interleukin 8 by human
neutrophils: A possible mechanism of neutrophil recruitment to
sites of inflammation in synovitis. J Exp Med. 182:2019–2025. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nishimura A, Akahoshi T, Takahashi M,
Takagishi K, Itoman M, Kondo H, Takahashi Y, Yokoi K, Mukaida N and
Matsushima K: Attenuation of monosodium urate crystal-induced
arthritis in rabbits by a neutralizing antibody against
interleukin-8. J Leukoc Biol. 62:444–449. 1997. View Article : Google Scholar : PubMed/NCBI
|