Introduction
Ovarian cancer is a life-threatening malignancy that
has high rates of incidence and mortality (1,2). As one
of the most effective and commonly used chemotherapeutic drugs,
cisplatin is used to treat a number of cancer types. Treatment with
cisplatin results in the formation of DNA adducts and causes DNA
damage in tumor cells through the irreversible insertion of DNA
bases (3); this prevents tumor cell
DNA replication and activates cell death (3,4). In
previous years, cisplatin has attracted extensive attention
(4,5). However, ovarian cancer cells frequently
develop cisplatin resistance during treatment, which severely
limits the efficacy of the drug and is the primary cause of
treatment failure (6). Therefore, to
enhance the sensitivity of ovarian cancer cells to cisplatin it is
first necessary to investigate the mechanism of drug
resistance.
Long non-coding RNAs (lncRNAs) are a class of
transcripts >200 nucleotides in length with no apparent
protein-coding capacity (7). The
altered expression of lncRNAs has been demonstrated to positively
and negatively regulate various biological and pathological
processes, including cellular proliferation, migration, invasion,
differentiation, apoptosis and carcinogenesis (8,9).
Numerous studies have focused on the role of lncRNAs in cancer cell
drug resistance; lncRNA colon cancer-associated transcript-1
contributed to cisplatin resistance in the non-small cell lung
cancer cell line A549/DDP by downregulating the expression of
microRNA (miR)-130a-3p (10). In
addition, knockdown of lncRNA nuclear enriched abundant transcript
1 significantly increased dexamethasone sensitivity in multiple
myeloma cell lines (11). lncRNA
LINC00152 (LINC00152) has been suggested to have an oncogenic
function in various types of cancer, and a previous study revealed
that the downregulation of LINC00152 increased the rate of
apoptosis and suppressed tumor growth in ovarian cancer by
targeting miR-125b (12). However,
the role of LINC00152 in the development of ovarian cancer remains
unclear, and few studies have evaluated the effect of LINC00152 in
chemotherapy-resistant ovarian cancer.
In the present study, the data demonstrated that
silencing LINC00152 promoted apoptosis and enhanced the cisplatin
sensitivity of ovarian cancer cells.
Materials and methods
Cell culture
The human normal ovarian cells line IOSE-80, ovarian
adenocarcinoma cell line COC1 and its cisplatin-resistant variant
COC1/DDP were obtained from the China Center for Type Culture
Collection. The cells were cultured in RPMI-1640 supplemented with
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.,) at
37°C in a humidified atmosphere (5% CO2). COC1 and
COC1/DDP cells were treated with different doze of cisplatin 24 h,
as described previously (13–15).
Then cells were then collected for further experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.,) according to the manufacturer's protocol. cDNA was generated
using the TaqMan Reverse Transcription Kit (Takara Biotechnology
Co., Ltd.) and PCR reactions were subsequently performed using the
Perfect Real Time SYBR® Premix Ex Taq™ kit (Takara
Biotechnology Co., Ltd.) with the ABI 7500 thermocycler (Thermo
Fisher Scientific, Inc.). The thermocycling conditions were as
follows: Initial denaturation at 95°C for 5 min. Amplification
cycle consists of three steps; a denaturation step at 95°C for 30
sec, an annealing step at 60°C for 45 sec and a final extension of
10 min at 72°C. U6 was used as normalization control for LINC00152
expression. GAPDH was used as an internal control for mRNAs
multi-drug resistance gene 1 (MDR1), multidrug
resistance-associated protein 1 (MRP1) and glutathione
S-transferase π (GSTπ). The results of the relative expression
levels of RNAs were analyzed using the 2−ΔΔCq method
(16). Independent experiments were
repeated 3 times. The primers used were as follows: LINC00152
forward, 5′-TGAGAATGAAGGCTGAGGTGT-3′; LINC00152 reverse,
5′-GCAGCGACCATCCAGTCATT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′; U6
reverse 5′-AACGCTTCAGGAATTTGCGT-3′; MDR1 forward,
5′-ATTGCTCACCGCCTGTCCACC-3′; MDR1 reverse,
5′-TGCTGATGCGTGCCATGCTCC-3′; MRP1 forward,
5′-CGTGTTGGTCTCTGTGTTCCTG-3′; MRP1 reverse,
5′-AGAAAGATGCTCTCTGGGTTTG-3′; GSTπ forward,
5′-TGGGCATCTGAAGCCTTTTG-3′; GSTπ reverse,
5′-GATCTGGTCACCCACGATGAA-3′; GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′; and GAPDH reverse
5′-GGCTGTTGTCATACTTCTCATGG-3′.
Transfection
Small interfering RNAs (siRNAs) targeting LINC00152
(si-1-LINC00152 and si-2-LINC00152) and an appropriate negative
control (si-NC) were transfected into COC1 and COC1/DDP cells. All
siRNAs were designed and synthesized by Guangzhou RiboBio Co., Ltd.
Cells were cultured in 6-well plates to 70–80% confluence, and the
siRNAs (100 nM) were transfected into the cells using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.,) according to the manufacturer's protocol.
Subsequent experiments were performed 48 h post-transfection.
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay was used to assess cell
proliferation capacity. Cells were seeded into 96-well plates
(5×103 cells/well) and incubated for 24 h. COC1 cells
were treated with 0, 1, 2, 4, 8, 16 or 32 µg/ml cisplatin, and
COC1/DDP cells were treated with 0, 4, 8, 16, 32, 64 or 128 µg/ml
cisplatin for a further 24 h. CCK-8 reagent (10 µl) was then added
to each well for an additional 2 h. Absorbance was measured at 450
nm and the percentage of viable cells was calculated (normalized to
the control group). A total of 4 wells for each concentration were
examined, and the experiments were performed in triplicate.
Flow cytometric analysis of
apoptosis
Cells were treated with cisplatin for 24 h and
apoptosis was analyzed using an Annexin V-fluorescein
isothiocyanate (FITC)/propidium iodide (PI) flow cytometry assay
kit (BD Biosciences) according to the manufacturer's protocol.
Briefly, cells were collected and fixed in pre-cooled 70% ethanol
at 4°C overnight. The cells were resuspended in 300 µl cold binding
buffer, and labeled with 5 µl Annexin V-FITC (20 µg/ml) for 10 min
in the dark at room temperature. Following the addition of 5 µl PI
(50 µg/ml) and 200 µl binding buffer, the samples were incubated at
room temperature in the dark for an additional 5 min. Apoptotic
cells were quantified using the FACSCalibur™ flow cytometer (BD
Biosciences) and data were analyzed using BD Accuri™ C6 (BD
Biosciences). The experiments were independently repeated 3
times.
Western blot analysis
Following cisplatin treatment for 24 h, the cells
were harvested and total proteins were obtained using RIPA lysis
buffer (Beyotime Institute of Biotechnology). Protein concentration
was subsequently determined using the BCA method (Beyotime
Institute of Biotechnology). The proteins (30 µg/lane) were
separated using 10% SDS-PAGE gel and subsequently transferred to
PVDF membranes (EDM Millipore). Following blocking with 5% milk for
2 h at room temperature, the membranes were incubated overnight at
4°C with the following primary antibodies: Anti-Bcl-2 (1:1,000;
cat. no. 3498; Cell Signaling Technology, Inc.), anti-Bax (1:1,000;
cat. no. 5023; Cell Signaling Technology, Inc.), anti-caspase-3
(1:500; cat. no. 14220; Cell Signaling Technology, Inc.),
anti-cleaved-caspase-3 (1:500; cat. no. 9579; Cell Signaling
Technology, Inc.) and GAPDH (1:2,000; cat. no. MAB374; EMD
Millipore; Merck KGaA). The membranes were then incubated with the
corresponding horseradish peroxidase-conjugated secondary
antibodies mouse anti-rabbit (1:10,000; cat. no. sc-2357; Santa
Cruz Biotechnology, Inc.), rabbit anti-mouse (1:10,000; cat. no.
sc-358914; Santa Cruz Biotechnology, Inc.) at room temperature for
an additional 2 h, and protein expression was detected using an ECL
detection reagent (EMD Millipore; Merck KGaA). Protein levels were
normalized to that of GAPDH and the experiments were performed in
triplicate. Quantification was carried out using ImageJ software
(version 1.43; National Institutes of Health).
Statistical analysis
SPSS 17.0 software was used to conduct all
statistical analyses (SPSS, Inc). The data are presented as the
mean ± standard error of the mean and each experiment was repeated
≥3 times. The data were analyzed using the Students t-test or
one-way analysis of variance followed by Dunnett's post-hoc test or
Tukey's test. P<0.05 was considered to indicate a statistically
significant difference.
Results
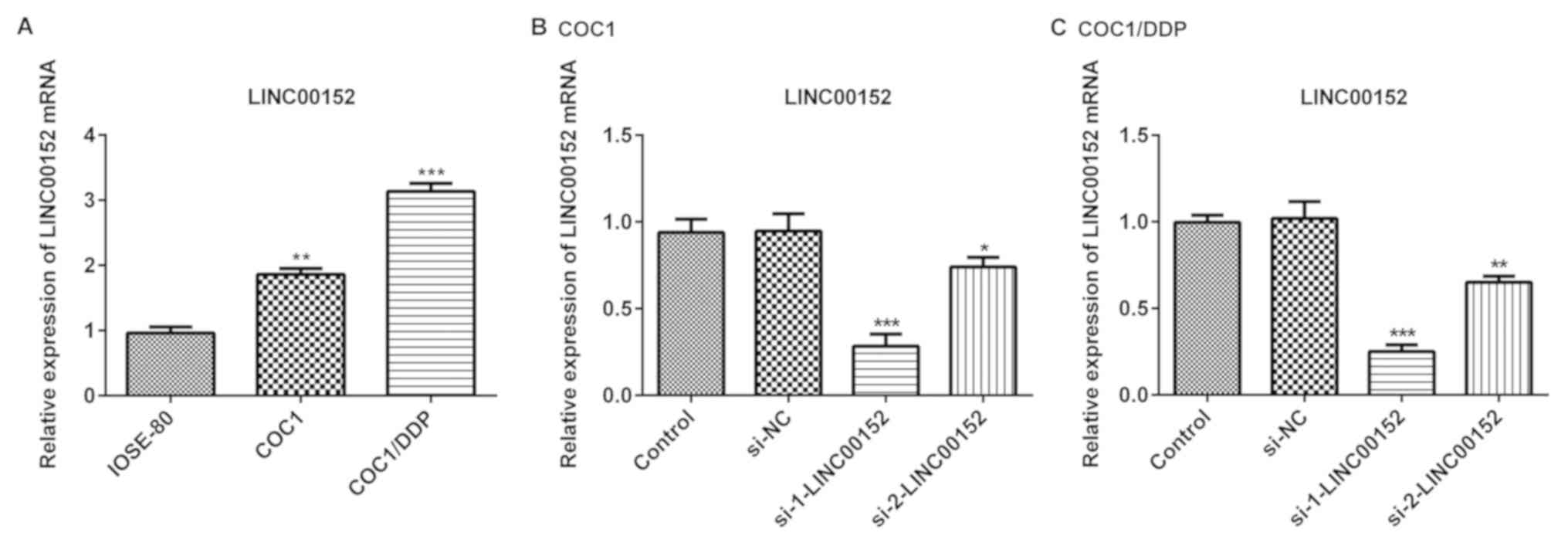
Expression level of LINC00152 is
upregulated in epithelial ovarian cancer cells
In the present study, the level of LINC00152
expression was detected in COC1 and COC1/DDP ovarian cancer cell
lines, and in normal ovarian cells (IOSE-80). As demonstrated in
Fig. 1A, LINC00152 expression level
was significantly increased in the ovarian cancer cells compared
with the normal ovarian cells. Furthermore, LINC00152 expression
levels in COC1/DDP cisplatin-resistant cells were markedly
increased compared with the cisplatin-sensitive COC1 cells
(Fig. 1A). To further investigate
the role of LINC00152 in ovarian cancer cells, siRNAs
(si-1-LINC00152 and si-2-LINC00152) and the negative control
(si-NC) were transfected into COC1 and COC1/DDP cells. Transfection
efficiency was detected using RT-qPCR 48 h post-transfection. The
results demonstrated that the inhibitory effect of si-1-LINC00152
on LINC00152 expression level was improved compared with that of
si-2-LINC00152 in the COC1 (Fig. 1B)
and COC1/DDP cells (Fig. 1C).
LINC00152 knockdown enhances the
sensitivity of ovarian cancer cells to cisplatin
To additionally investigate the effects of LINC00152
knockdown on cisplatin resistance, LINC00152 knockdown cells were
treated with a concentration gradient of cisplatin, and the levels
of proliferation were assessed using the CCK-8 assay 24 h after
treatment. As indicated in the Fig.
2, the viability of si-1-LINC00152-transfected COC1 cells was
significantly decreased compared with that of the si-NC-transfected
COC1 cells. A similar effect was observed in COC1/DDP cells. At a
cisplatin concentration of 4 or 32 µg/ml, the viability of COC1 and
COC1/DDP cells was close to 50%, respectively (Fig. 2), and these 2 concentrations were
subsequently selected for subsequent investigation.
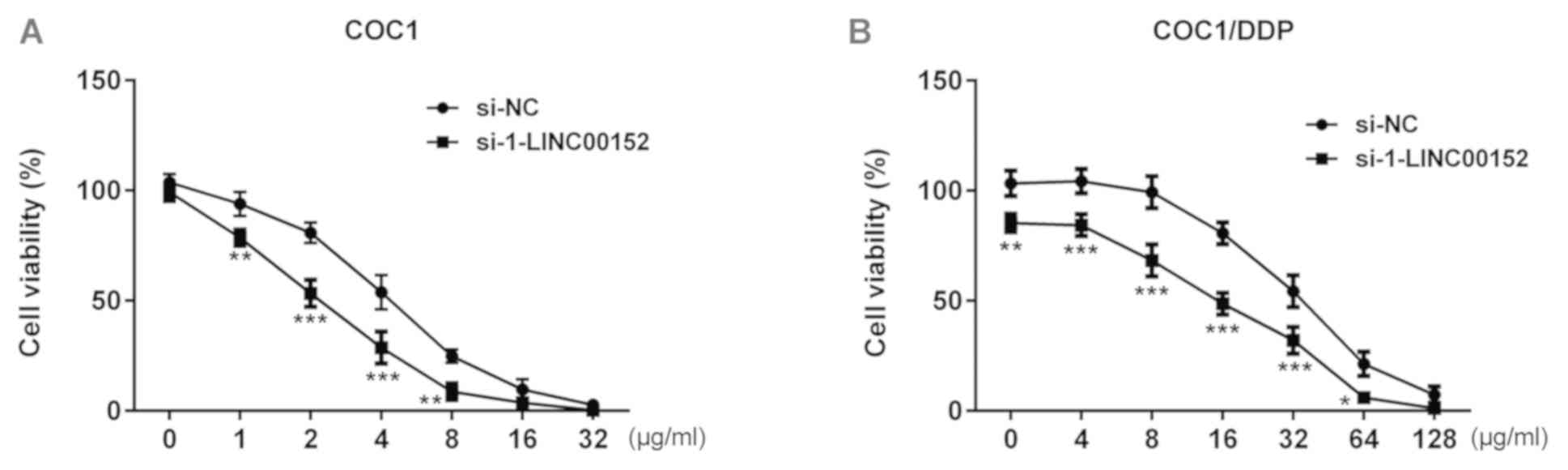 | Figure 2.Effect of LINC00152 knockdown on
cisplatin sensitivity. (A) At 24 h post-transfection with
si-1-LINC00152 or si-NC, COC1 cells were treated with 0, 1, 2, 4,
8, 16 or 32 µg/ml cisplatin for a further 24 h, and cell viability
was assessed using the CCK-8 assay. (B) At 24 h post-transfection
with si-1-LINC00152 or si-NC, COC1/DDP cells were treated with 0,
4, 8, 16, 32, 64 or 128 µg/ml cisplatin for a further 24 h, and
viability was assessed using the CCK-8 assay. *P<0.05,
**P<0.01 and ***P<0.001 vs. si-NC. CCK-8, Cell Counting
Kit-8; si, small interfering RNA; NC, negative control. |
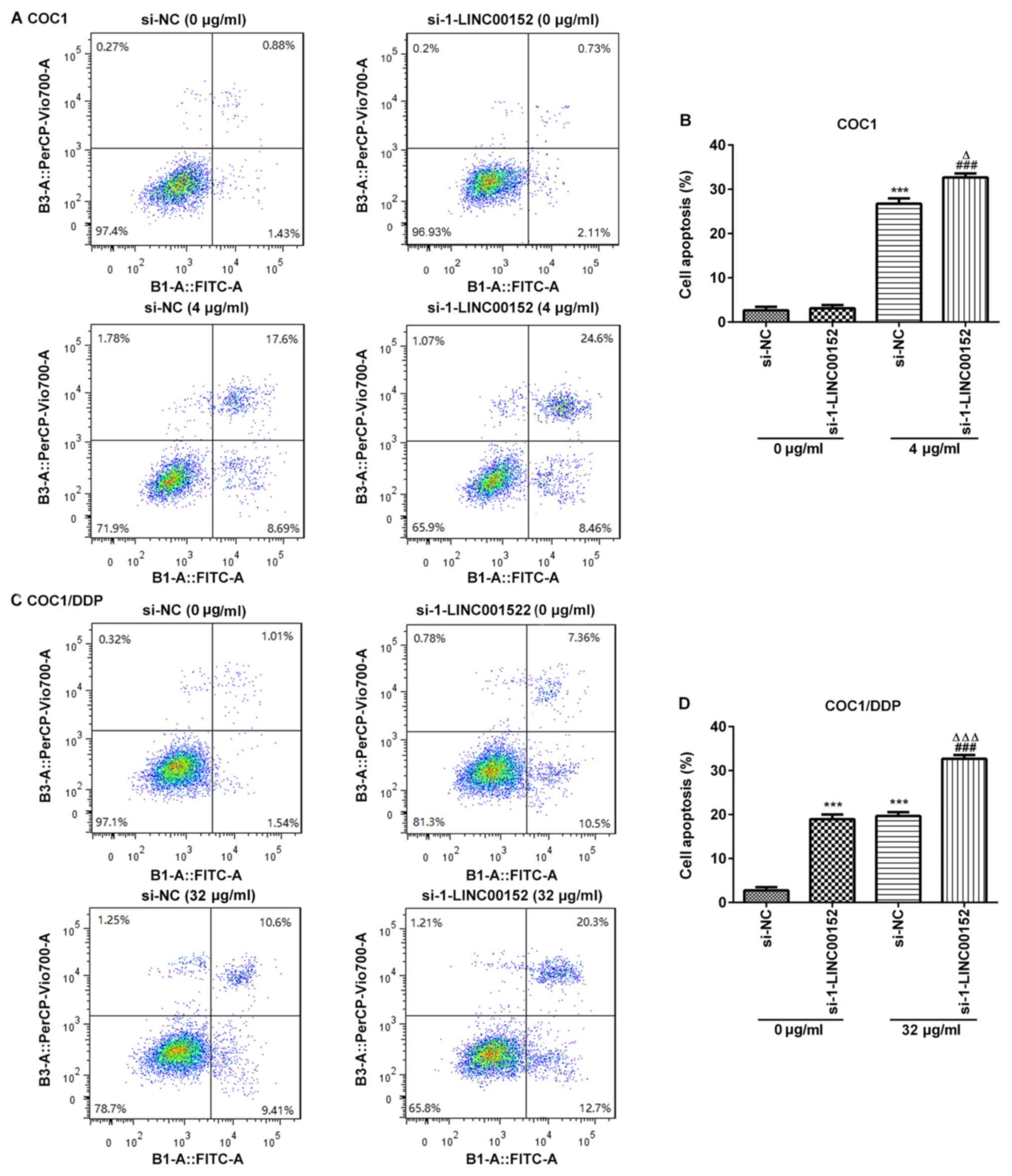
LINC00152-silencing promotes apoptosis
in cisplatin-treated COC1 and COC1/DDP cells
To investigate the underlying mechanism of the
LINC00152 knockdown-associated increase in cisplatin sensitivity,
flow cytometric analyses were conducted to assess the rate of
apoptosis in ovarian cancer cells. The results demonstrated that
the apoptotic rate was significantly increased in the
cisplatin-treated COC1 and COC1/DDP cells following LINC00152
knockdown (Fig. 3). Notably,
si-1-LINC00152 transfection alone had no significant effect on the
level of apoptosis in COC1 cells (Fig.
3A and B); however, apoptosis was markedly promoted in the
COC1/DDP cells (Fig. 3C and D).
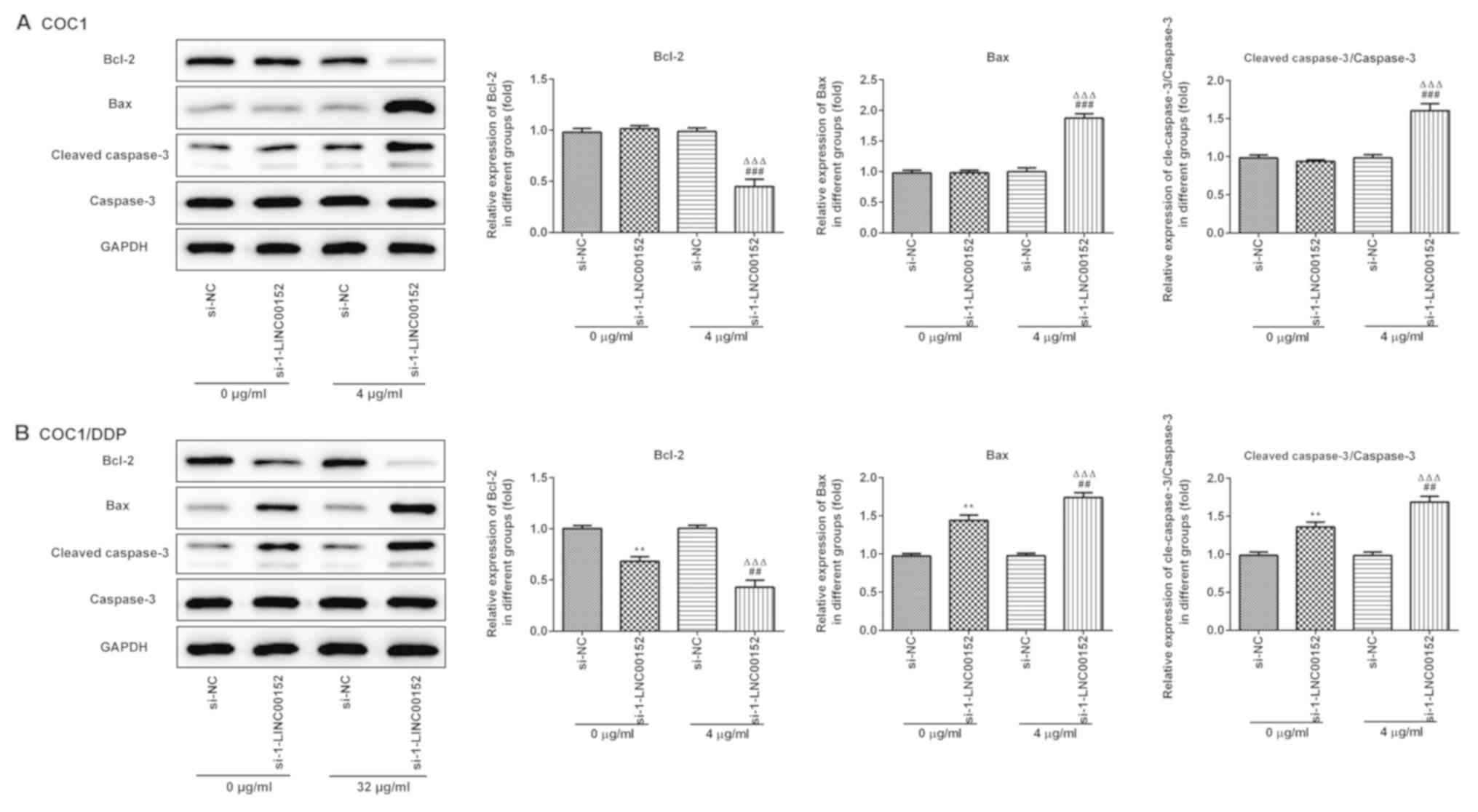
Furthermore, the western blot analysis results indicated that
LINC00152 knockdown significantly decreased Bcl-2, and increased
Bax and Cleaved caspase-3 expression levels in the
cisplatin-treated (Fig. 4A) COC1 and
(Fig. 4B) COC1/DDP cells. Again,
si-1-LINC00152 transfection alone had no significant effect on the
expression levels of these proteins in the COC1 cells (Fig. 4A), which was consistent with the
results of the apoptosis assay.
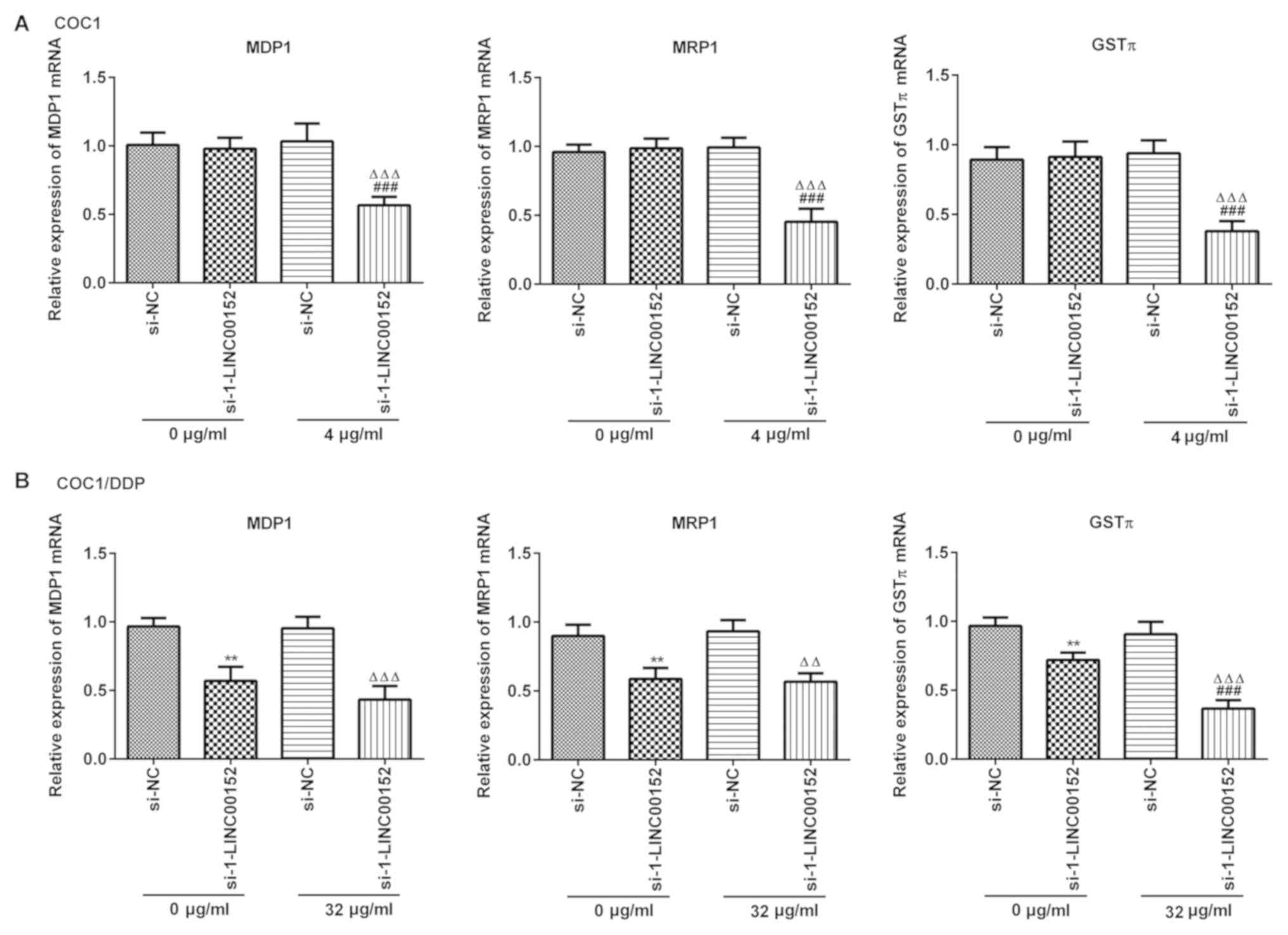
LINC00152 silencing downregulates the
expression level of MDR1, MRP1 and GSTπ in ovarian cancer
cells
The mRNA expression levels of MDR1, MRP1 and GSTπ
were detected using RT-qPCR. LINC00152 silencing markedly decreased
the level of MDR1, MRP1 and GSTπ expression in COC1 (Fig. 5A) and COC1/DDP (Fig. 5B) cells following exposure to
cisplatin. However, knockdown of LINC00152 alone did not affect
mRNA expression levels in COC1 cells (Fig. 5A).
Discussion
At present, the most common treatments for ovarian
cancer are surgery and chemotherapy. However, the level of drug
resistance in cancer cells has a clear effect on patient prognosis
(6,17,18).
Accumulating evidence has suggested that numerous lncRNAs are
involved in the occurrence and development of ovarian cancer
(19–21), and that they may considerably affect
drug resistance (22). Xu et
al (23) indicated that lncRNA
EIBC was highly expressed in ovarian cancer tissues, and that the
upregulation of EIBC promoted tumor growth and improved cisplatin
resistance via inhibiting the Wnt/β-catenin signaling pathway. Wang
et al (24) demonstrated that
the downregulation of Hox transcript antisense intergenic RNA
significantly decreased the levels of cell proliferation and
invasion, and reversed cisplatin resistance in SKOV-3CDDP/R cells.
In addition, Zhang et al (25) indicated that Linc00312 promoted
apoptosis and enhanced cisplatin sensitivity through the
Bcl-2/Caspase-3 pathway in SKOV3/DDP cells. These studies indicated
that lncRNAs have great potential in the treatment of cisplatin
resistance. Another previous study suggested that LINC00152 confers
oxaliplatin resistance by regulating the expression of miR-193a-3p,
and that it may be an independent prognostic factor for colon
cancer (26). Notably, a recent
study by Chen et al (12)
demonstrated that LINC00152 was upregulated in ovarian cancer
tissues and cell lines, and that LINC00152 knockdown promoted
apoptosis and inhibited tumor growth, suggesting a pro-cancer
effect of LINC00152. To the best of our knowledge, in the present
study, the sensitization effect of LINC00152 on cisplatin was
evaluated for the first time in ovarian cancer cells. The results
indicated that the expression level of LINC00152 was markedly
increased in the COC1/DDP cisplatin-resistant cells compared with
the cisplatin-sensitive COC1 and normal ovarian cells, concluding
that LINC00152 knockdown enhanced cisplatin sensitivity in both
COC1 and COC1/DDP cells.
Previous studies have revealed that the expression
of multidrug resistance genes is closely associated with lncRNAs in
tumors (22,27); silencing of lncRNA
metastasis-associated lung adenocarcinoma transcript decreased
MDR1, MRP5 and prolow-density lipoprotein receptor-related protein
1 (LRP1) expression levels and reversed temozolomide resistance in
glioblastoma cells (28).
Additionally, downregulation of lncRNA X-inactive specific
transcript inhibited GSTπ expression, promoted apoptosis and
increased the sensitivity of colorectal cancer cells to doxorubicin
(29). The present study suggested
that LINC00152 silencing promoted apoptosis and downregulated the
levels of MDR1, MRP1 and GSTπ expression in cisplatin-treated COC1
and COC1/DDP cells.
Notably, it was also identified that LINC00152
knockdown alone had no significant effect on apoptosis and the
expression of drug resistance-associated genes (MDR1, MRP1 and
GSTπ) in COC1 cells. Contrastingly, the data from the study by Chen
et al (12) demonstrated that
LINC00152 silencing significantly promoted apoptosis in the ovarian
cancer SKOV3 and A2780 cell lines. This result is inconsistent with
ours, potentially due to the selection of different cell lines.
However, subsequent results from the present study indicated that
LINC00152 knockdown markedly enhanced apoptosis in COC1/DDP cells
with or without cisplatin treatment. Therefore, it was hypothesized
that LINC00152 may serve a prominent role in the formation of
cisplatin resistance in COC1 cells, and the acquisition of
additional functions involved in this process.
There were certain limitations to the present study.
Firstly, only the effect of LINC00152 silencing on the cisplatin
resistance of COC1 and COC1/DDP cells was investigated; whether the
overexpression of LINC00152 has an effect on ovarian cancer was not
explored. In addition, only COC1 and COC1/DDP cells were used.
Whether LINC00152 serves a role in other ovarian cancer cell lines
is unknown. Future studies involving other cell lines and an animal
model are required to confirm the results.
In conclusion, the present study revealed that the
altered expression of LINC00152 may be associated with the
cisplatin-resistance mechanisms of ovarian cancer cells. Knockdown
of LINC00152 improved the chemosensitivity of epithelial ovarian
cancer cells to cisplatin by increasing apoptosis and decreasing
the expression levels of MDR1, MRP1 and GSTπ. Although future
experiments are required for confirmation, the upregulation of
LINC00152 may be involved in the formation of cisplatin resistance
in COC1/DDP cells.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ and HL participated in data analysis and
manuscript preparation. HZ performed the experiments and HL revised
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mezzanzanica D: Ovarian cancer: A
molecularly insidious disease. Chin J Cancer. 34:1–3. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Galluzzi L, Senovilla L, Vitale I, Michels
J, Martins I, Kepp O, Castedo M and Kroemer G: Molecular mechanisms
of cisplatin resistance. Oncogene. 31:1869–1883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rancoule C, Guy JB, Vallard A, Ben Mrad M,
Rehailia A and Magné N: 50th anniversary of cisplatin. Bull Cancer.
104:167–176. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Armstrong DK, Bundy B, Wenzel L, Huang HQ,
Baergen R, Lele S, Copeland LJ, Walker JL and Burger RA;
Gynecologic Oncology Group, : Intraperitoneal cisplatin and
paclitaxel in ovarian cancer. N Engl J Med. 354:34–43. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tomao F, Marchetti C, Romito A, Di Pinto
A, Di Donato V, Capri O, Palaia I, Monti M, Muzii L and Benedetti
Panici P: Overcoming platinum resistance in ovarian cancer
treatment: From clinical practice to emerging chemical therapies.
Expert Opin Pharmacother. 18:1443–1455. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wieczorek E and Reszka E: mRNA, microRNA
and lncRNA as novel bladder tumor markers. Clin Chim Acta.
477:141–153. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Marchese FP, Raimondi I and Huarte M: The
multidimensional mechanisms of long noncoding RNA function. Genome
Biol. 18:2062017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu B, Zhang H, Wang Z, Zhang F, Wei H and
Li L: LncRNA CCAT1/miR-130a-3p axis increases cisplatin resistance
in non-small-cell lung cancer cell line by targeting SOX4. Cancer
Biol Ther. 18:974–983. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu Y and Wang H: LncRNA NEAT1 promotes
dexamethasone resistance in multiple myeloma by targeting
miR-193a/MCL1 pathway. J Biochem Mol Toxicol. 32:e220082018.
View Article : Google Scholar
|
|
12
|
Chen P, Fang X, Xia B, Zhao Y, Li Q and Wu
X: Long noncoding RNA LINC00152 promotes cell proliferation through
competitively binding endogenous miR-125b with MCL-1 by regulating
mitochondrial apoptosis pathways in ovarian cancer. Cancer Med.
7:4530–4541. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma J, Yang J, Wang C, Zhang N, Dong Y,
Wang C, Wang Y and Lin X: Emodin augments cisplatin cytotoxicity in
platinum-resistant ovarian cancer cells via ROS-dependent MRP1
downregulation. Biomed Res Int. 2014:1076712014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang T, Guan M, Jin HY and Lu Y: Reversal
of multidrug resistance by small interfering double-stranded RNAs
in ovarian cancer cells. Gynecol Oncol. 97:501–507. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang F, Zhu Y, Fang S, Li S and Liu S:
Effect of lanthanum chloride on tumor growth and apoptosis in human
ovarian cancer cells and xenograft animal models. Exp Ther Med.
16:1143–1148. 2018.PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pogge von Strandmann E, Reinartz S, Wager
U and Müller R: Tumor-Host cell interactions in ovarian cancer:
Pathways to therapy failure. Trends Cancer. 3:137–148. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kujawa KA and Lisowska KM: Ovarian
cancer-from biology to clinic. Postepy Hig Med Dosw (Online).
69:1275–1290. 2015.(In Polish). View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mitra R, Chen X, Greenawalt EJ, Maulik U,
Jiang W, Zhao Z and Eischen CM: Decoding critical long non-coding
RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat
Commun. 8:16042017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Worku T, Bhattarai D, Ayers D, Wang K,
Wang C, Rehman ZU, Talpur HS and Yang L: Long non-coding RNAs: The
new horizon of gene regulation in ovarian cancer. Cell Physiol
Biochem. 44:948–966. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martini P, Paracchini L, Caratti G,
Mello-Grand M, Fruscio R, Beltrame L, Calura E, Sales G, Ravaggi A,
Bignotti E, et al: lncRNAs as novel indicators of patients'
prognosis in stage I epithelial ovarian cancer: A retrospective and
multicentric study. Clin Cancer Res. 23:2356–2366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu J, Wu J, Fu C, Teng F, Liu S, Dai C,
Shen R and Jia X: Multidrug resistant lncRNA profile in
chemotherapeutic sensitive and resistant ovarian cancer cells. J
Cell Physiol. 233:5034–5043. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu QF, Tang YX and Wang X: LncRNA EBIC
promoted proliferation, metastasis and cisplatin resistance of
ovarian cancer cells and predicted poor survival in ovarian cancer
patients. Eur Rev Med Pharmacol Sci. 22:4440–4447. 2018.PubMed/NCBI
|
|
24
|
Wang Y, Wang H, Song T, Zou Y, Jiang J,
Fang L and Li P: HOTAIR is a potential target for the treatment of
cisplatin-resistant ovarian cancer. Mol Med Rep. 12:2211–2216.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang C, Wang M, Shi C, Shi F and Pei C:
Long non-coding RNA Linc00312 modulates the sensitivity of ovarian
cancer to cisplatin via the Bcl-2/Caspase-3 signaling pathway.
Biosci Trends. 12:309–316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yue B, Cai D, Liu C, Fang C and Yan D:
Linc00152 Functions as a competing endogenous RNA to confer
oxaliplatin resistance and holds prognostic values in colon cancer.
Mol Ther. 24:2064–2077. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo F, Cao Z, Guo H and Li S: The action
mechanism of lncRNA-HOTAIR on the drug resistance of non-small cell
lung cancer by regulating Wnt signaling pathway. Exp Ther Med.
15:4885–4889. 2018.PubMed/NCBI
|
|
28
|
Li H, Yuan X, Yan D, Li D, Guan F, Dong Y,
Wang H, Liu X and Yang B: Long non-coding RNA MALAT1 decreases the
sensitivity of resistant glioblastoma cell lines to temozolomide.
Cell Physiol Biochem. 42:1192–1201. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu J, Zhang R, Yang D, Li J, Yan X, Jin
K, Li W, Liu X, Zhao J, Shang W and Yu T: Knockdown of long
non-coding RNA XIST inhibited doxorubicin resistance in colorectal
cancer by upregulation of miR-124 and downregulation of SGK1. Cell
Physiol Biochem. 51:113–128. 2018. View Article : Google Scholar : PubMed/NCBI
|