Introduction
Breast cancer is the most commonly diagnosed cancer
type in women, affecting approximately 10% of women during their
lifetime (1). Breast cancer is a
heterogeneous disease which is classified into several major
subtypes according to the expression of the estrogen receptor (ER),
progesterone receptor (PR) and human epidermal growth factor
receptor (HER2/neu) (2). Targeted
therapies, such as tamoxifen and trastuzumab, have improved the
outcomes of patients with ER+ or HER2+ breast
cancer (3,4). However, a number of patients with
breast cancer exhibit de novo or acquired target therapy
resistance (5,6). In addition, chemotherapy is the only
treatment option for patients with triple-negative breast cancer
(TNBC; ER−/HER2−/PR−) and targets
for the effective treatment remain to be elucidated (7). Therefore, the molecular mechanism of
breast cancer progression requires further research to fulfill
clinical needs.
Previous studies investigated the role of non-coding
RNAs in cancer progression and their application as biomarkers and
targets for the treatment of patients with cancer (8,9).
MicroRNAs (miRNAs/miRs) are small, single-stranded, non-coding RNAs
(10). Mechanistically, miRNAs bind
to complementary binding sites on the 3′-untranslated regions
(3′-UTRs) of target gene mRNAs, resulting in mRNA degradation or
inhibition of translation (11).
Dysregulation of miRNAs leads to aberrant expression of numerous
genes, which is associated with the initiation and development of
human diseases, including cancer (12). Differentially expressed miRNAs have
been shown to be associated with different molecular subtypes of
breast cancer (13). Several miRNAs,
whose expression is associated with the clinical outcomes of breast
cancer, serve as oncogenes or tumor suppressors in breast cancer
(14–16). Recently, miR-532-5p was demonstrated
to be significantly upregulated in TNBC tissues compared with
normal breast tissues (17).
However, whether and how miR-532-5p contributes to the progression
of breast cancer remains unknown.
Ras-related and estrogen-regulated growth inhibitor
(RERG) was first identified using microarray analysis in 2001
(18). Decreased expression of RERG
is observed in breast tumor tissues and is associated with poor
clinical prognosis in breast cancer (18). In breast cancer cells, overexpression
of RERG inactivated the Ras/mitogen-activated protein kinase
(MAPK)/ERK signaling, leading to the inhibition of cell
proliferation, migration and invasion (19).
In the present study, compared with the normal
tissues, miR-532-5p was significantly upregulated in breast cancer
tissues. Overexpression of miR-532-5p reduced the expression of
RERG at both mRNA and protein levels and activated MAPK/ERK
signaling in breast cancer cells. In contrast, downregulation of
miR-532-5p expression inhibited MDA-MB-231 cell proliferation and
migration, which was partially reversed by RERG knockdown. These
findings suggest an oncogenic role of miR-532-5p in breast
cancer.
Materials and methods
Patient samples
Tumor tissues and matched normal tissues were
collected from 20 female patients with breast cancer (age range,
25–65 years) between June 2014 and September 2017 at the Affiliated
Drum Tower Hospital of Nanjing University (Nanjing, China). Written
consent was obtained from all participants prior to sample
collection. Ethical approval was received from the Ethic Committee
of Nanjing University before initiation of the current study. All
experimental procedures were conducted under the supervision of the
Ethic Committee of Nanjing University. The tissues were immediately
stored in a −80°C refrigerator upon surgical removal before
subjecting to the following experiments.
Cell culture
TNBC cell lines (BT549, HS578T, MDA-MB-231) and a
normal epithelial breast cell line MCF10A were purchased from
American Type Culture Collection and used during the first six
months after purchase. MCF10A cells were cultured in Mammary
Epithelial Cell Growth Medium (Lonza Group, Ltd.) containing 100
ng/ml cholera toxin (Sigma-Aldrich; Merck KGaA). BT549, HS578T and
MDA-MB-231 cells were maintained in DMEM supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.). All cells were cultured in
a humidified incubator at 37°C with 5% CO2.
Overexpression and inhibition of
miR-532-5p
miR-negative control (NC) inhibitor, miR-532-5p
inhibitor, miR-NC mimic and miR-532-5p mimic were purchased from
Suzhou GenePharma Co., Ltd. miR-NC inhibitor and miR-NC mimic
served as negative controls for miR-532-5p inhibitor and miR-532-5p
mimic, respectively. For the manipulation of miR-532-5p, 50 nM
miR-532-5p mimic, miR-532-5p inhibitor, miR-NC mimic or miR-NC
inhibitor was transfected into MDA-MB-231 cells using Lipofectamine
3000® (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. After 48 h, the RNA and
protein were extracted from cells and subjected to the subsequent
experiments. The following sequences were used: miR-NC inhibitor,
5′-UUCUCCGAACGUGUCACGU-3′; miR-532-5p inhibitor,
5′-ACGGUCCUACACUCAAGGCAUG-3′; miR-NC mimic,
5′-AUUGGAACGAUACAGAGAAGA−3′; and miR-532-5p mimic,
5′-CAUGCCUUGAGUGUAGGACCGU-3′.
Knockdown of RERG
Control siRNA and RERG siRNA were purchased from
Suzhou GenePharma Co., Ltd. For the knockdown of RERG, 50 nM RERG
siRNA or control siRNA was transfected into MDA-MB-231 cells using
Lipofectamine RNAiMax (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. After 48 h, the RNA and
protein were extracted from cells and subjected to the subsequent
experiments. The following sequences were used: Control siRNA,
5′-UAAGGCUAUGAAGAGAUAC-3′; RERG siRNA,
5′-CAUCUAGGAUGUUCUUAAGUGGC-3′.
Cell proliferation assay
Cell proliferation was evaluated using a Cell
Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.)
following the manufacturer's protocol. In brief, 1,000 MDA-MB-231
cells/well were seeded into 96-well plates. On the next day, the
aforementioned miR-NC inhibitor or miR-532-5p inhibitor (50 nM) and
control siRNA or RERG siRNA (50 nM) were co-transfected into
MDA-MB-231 cells using Lipofectamine® RNAiMAX (Thermo
Fisher Scientific, Inc.). After 8 h, the medium containing
transfection reagents was replaced with fresh DMEM supplemented
with 10% FBS. At 0, 24, 48, 72 and 96 h, 10 µl CCK-8 solution was
added into each well and incubated for 2 h. The medium containing
the CCK-8 solution was then transferred into another 96-well plate,
where absorbance was measured at a wavelength of 450 nm to reflect
the cell number in each group for the calculation of cell
proliferation.
Cell migration assay
The cell migration ability was determined using a
wound healing assay. Briefly, 1×106 MDA-MB-231 cells
were seeded in each well of six-well plates and cultured until
reaching 90% confluence. A wound was created at the central area of
the cell monolayer by scratching the plate with a 10 µl pipette
tip. The medium was discarded and the cells were rinsed with PBS
twice. Subsequently, DMEM with no serum was added into each well.
The images of the migrated area were captured using an inverted
light microscope at 0 and 30 h. The percentage migrated area was
calculated with Image Pro Plus (Media Cybernetics, Inc.).
RNA isolation and RT-qPCR
Total RNA was isolated from cells and tissues using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The RNA
concentration was determined using a NanoDrop™ 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). For mRNA
quantification, RNA was reverse transcribed into cDNA with a
PrimeScript RT Master Mix kit (Takara Bio, Inc.) following the
manufacturer's instructions. qPCR was performed using a SYBR Green
qPCR Master Mix kit (Takara Bio, Inc.). The stem-loop method was
used for detection of miRNA expression (20). For miRNA quantification, RNA was
reverse-transcribed using a Mir-X miRNA First Strand Synthesis kit
(Takara Bio, Inc.) followed by RT-qPCR with Mir-X™ miRNA qRT-PCR
SYBR kit (Takara Bio, Inc.). The thermocycling conditions for the
qPCR was as follows: Initial denaturation at 95°C for 30 sec,
followed by 35 cycles of 95°C for 10 sec and 60°C for 30 sec. GAPDH
and U6 served as internal controls for mRNA and miRNA,
respectively. The relative expression of mRNA and miRNA was
calculated using the 2−ΔΔCq method (21). The following primer sequences were
used: GAPDH forward, 5′-CCTGCACCACCAACTGCTTA-3′ and reverse,
5′-GGCCATCCACAGTCTTCTGAG-3′; RERG forward,
5′-GAATCAACCTACCGACACCAAG-3′ and reverse,
5′-CTCCCTCTGAATGGTATCTTCCT-3′; U6 forward, 5′-GCGCGTCGTGAAGCGTTC-3′
and reverse 5′-GGTCGGCTTTCAGTCGGATGTT-3′; miR-532-5p forward
5′-TGGGTCCTTGCCTTGAGTGTAG-3′ and reverse
5′-GGTCGGCTTTCAGTCGGATGTT-3′; stem-loop-miR-532-5p,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACGGTC-3′; and
stem-loop-U6,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAATATG−3′.
Protein isolation and western
blotting
Proteins were isolated from MDA-MB-231 cells with a
RIPA lysis buffer (Sigma-Aldrich; Merck KGaA). The concentration of
protein lysates was determined using a BCA Protein Assay kit
(Thermo Fisher Scientific, Inc.). Anti-RERG (cat. no. SAB2107423;
1:2,000) and GAPDH (cat. no. G8795; 1: 5,000) antibodies were
purchased from Sigma-Aldrich (Merck KGaA). Anti-dual specificity
mitogen-activated protein kinase kinase (MEK; cat. no. 4694;
1:1,000), ERK1/2 (cat. no. 4695; 1:1,000), phosphorylated (p)-MEK
(cat. no. 2338; 1:2,000) and p-ERK1/2 (cat. no. 9101; 1:2,000)
antibodies were purchased from Cell Signaling Technology, Inc.
Horseradish peroxidase-conjugated secondary antibodies against
mouse (cat. no. KC-MM-035, 1:10,000) and rabbit (cat. no.
KC-RB-035, 1:10,000) were purchased from Aksomics, Inc. For western
blotting, 10 µg protein/lane was separated using SDS-PAGE (8% gel),
transferred onto a PVDF membrane and blocked in 5% nonfat milk for
1 h at room temperature. The PVDF membrane was incubated with the
aforementioned primary antibodies overnight at 4°C. Subsequently,
the PVDF membrane was washed and incubated with secondary
antibodies for 1 h at room temperature. The bands were visualized
using a Pierce™ ECL Western Blotting Substrate (Thermo Fisher
Scientific, Inc.). Densitometric analysis was performed using the
Image J software (version 1.51; National Institute of Health).
Dual-luciferase reporter assay
The 3′-UTR sequence of RERG mRNA was amplified from
cDNA of MCF10A cells using Takara Ex Taq® Hot Start
polymerase (Takara Bio, Inc.) and ligated into the pGL3-basic
plasmid (Promega Corporation). Two point mutations were introduced
into the pGL3-RERG 3′-UTR-wild-type (WT) construct using
QuickChange Site-Directed Mutagenesis kit (Agilent Technologies,
Inc.) to generate pGL3-RERG 3′UTR-mutant (Mut). For the dual
luciferase reporter assay, 2×106 MDA-MB-231 cells were
transfected with pGL3-RERG 3′-UTR-WT or pGL3-RERG 3′-UTR-Mut and
miR-532-5p mimic or miR-NC mimic using Lipofectamine®
RNAiMAX (Thermo Fisher Scientific, Inc.). Relative luciferase
activity was calculated by normalizing to that of Renilla
luciferase. After 48 h, the relative luciferase activity of each
well was detected using the Dual Luciferase Reporter Assay System
(Promega Corporation).
Statistical analysis
Data were analyzed using GraphPad Prism 7.0
(GraphPad Software, Inc.) and are presented as the mean ± SD. The
correlation between miR-532-5p and RERG was analyzed using
Pearson's correlation analysis. Differences between normal breast
tissues and breast cancer tissues were analyzed by paired Student's
t-test, while the differences between two independent groups were
analyzed using Student's t-test. Differences between multiple
groups were analyzed with one-way ANOVA followed by Newman-Keuls
test. P<0.05 was considered to indicate a statistically
significant difference. All experiments were repeated three
times.
Results
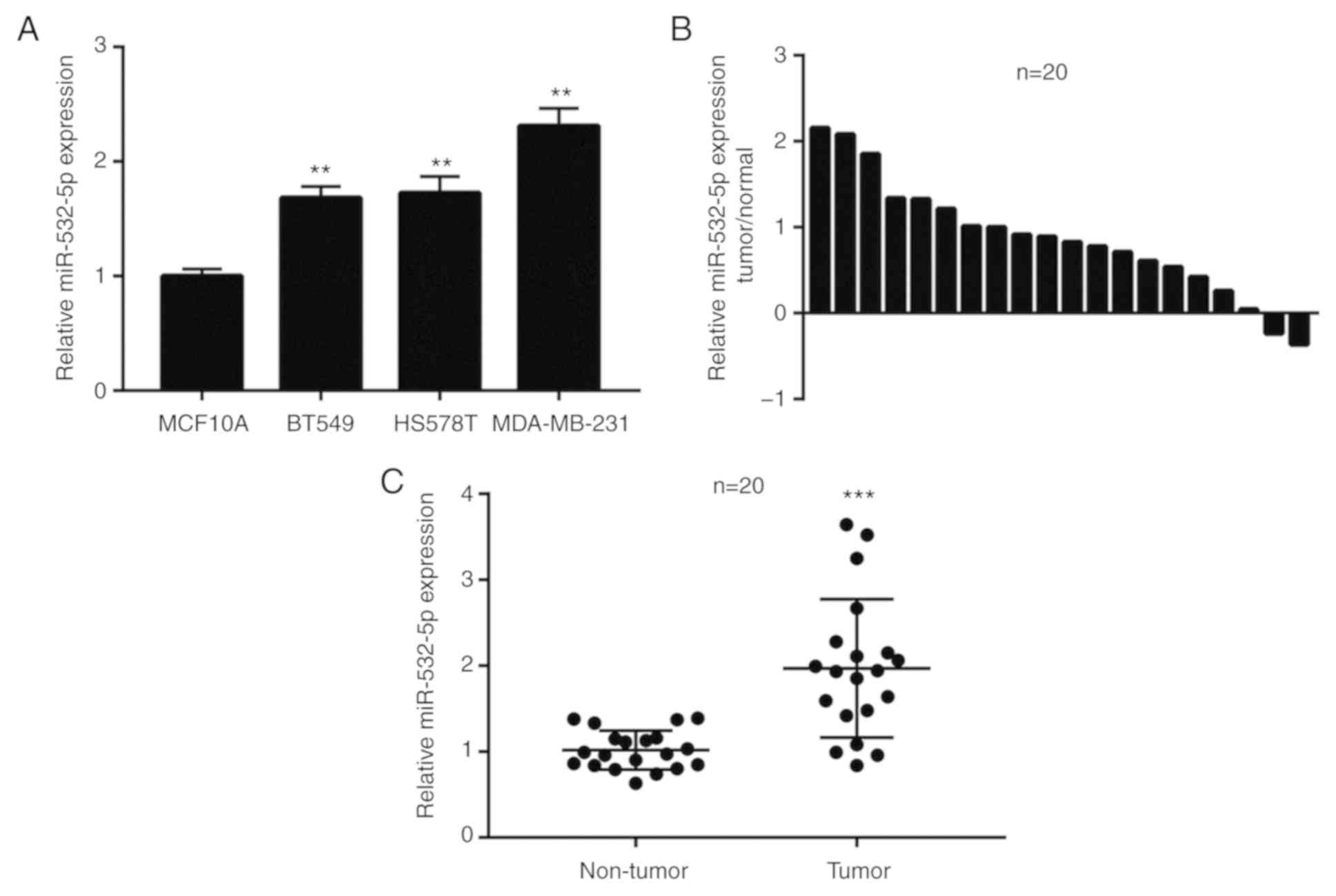
miR-532-5p expression levels is
increased in breast cancer cells and tumor tissues
To investigate the role of miR-532-5p in breast
cancer, RT-qPCR was performed to detect the difference in
miR-532-5p expression levels between the immortalized
non-tumorigenic breast epithelial cell line MCF10A and TNBC cell
lines, including BT549, HS578T and MDA-MB-231. Compared with
MCF10A, the expression of miR-532-5p was significantly upregulated
in three TNBC cell lines (Fig. 1A).
Additionally, in 20 pairs of normal breast tissues and breast tumor
tissues, miR-532-5p expression was increased in 90% (18/20) of
breast cancer tissues (Fig. 1B).
Compared with normal breast tissues, the expression of miR-532-5p
was significantly elevated in breast cancer tissues (Fig. 1C). These data indicated that
miR-532-5p may be associated with breast cancer.
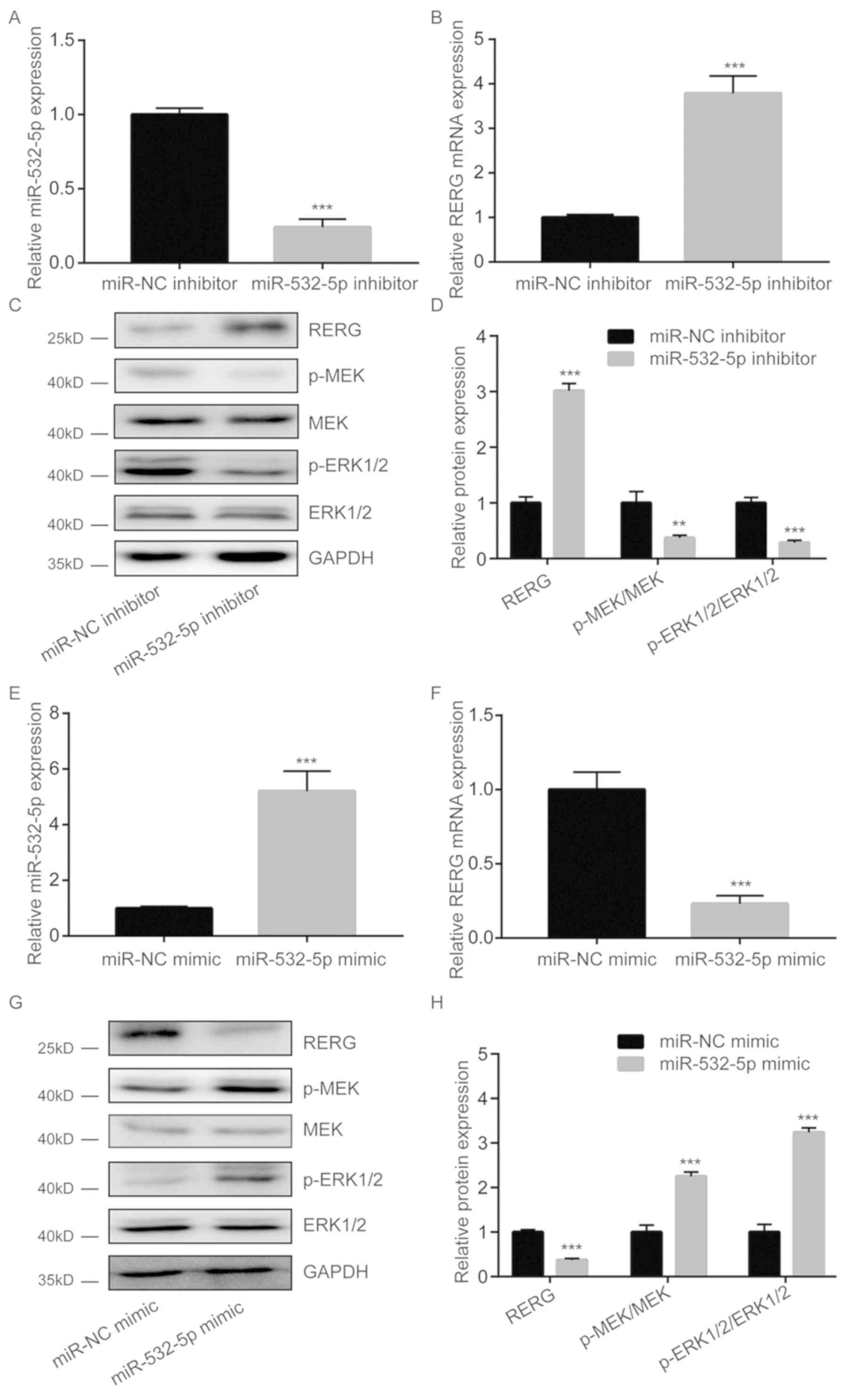
miR-532-5p inhibits RERG expression
and activates the MAPK/ERK pathway in breast cancer cells
Following downregulation of miR-532-5p expression by
miR-532-5p inhibitor transfection, RERG mRNA levels were
significantly increased (Fig. 2A and
B). Furthermore, western blotting results showed that RERG
protein levels were also increased upon miR-532-5p downregulation
in MDA-MB-231 cells (Fig. 2C and D).
In the present study, miR-532-5p downregulation decreased the
protein levels of p-MEK and p-ERK1/2 but had no effect on total MEK
and ERK1/2 expression in MDA-MB-231 cells (Fig. 2C and D), suggesting inactivation of
the MAPK/ERK pathway. However, transfection with miR-532-5p mimic
increased miR-532-5p expression levels (Fig. 2E), reduced RERG expression (Fig. 2F) and increased p-MEK/MEK and
p-ERK1/2/ERK1/2 ratios in MDA-MB-231 cells (Fig. 2G and H). These findings suggest that
miR-532-5p could activate the MAPK/ERK signaling via repression of
RERG expression in breast cancer cells.
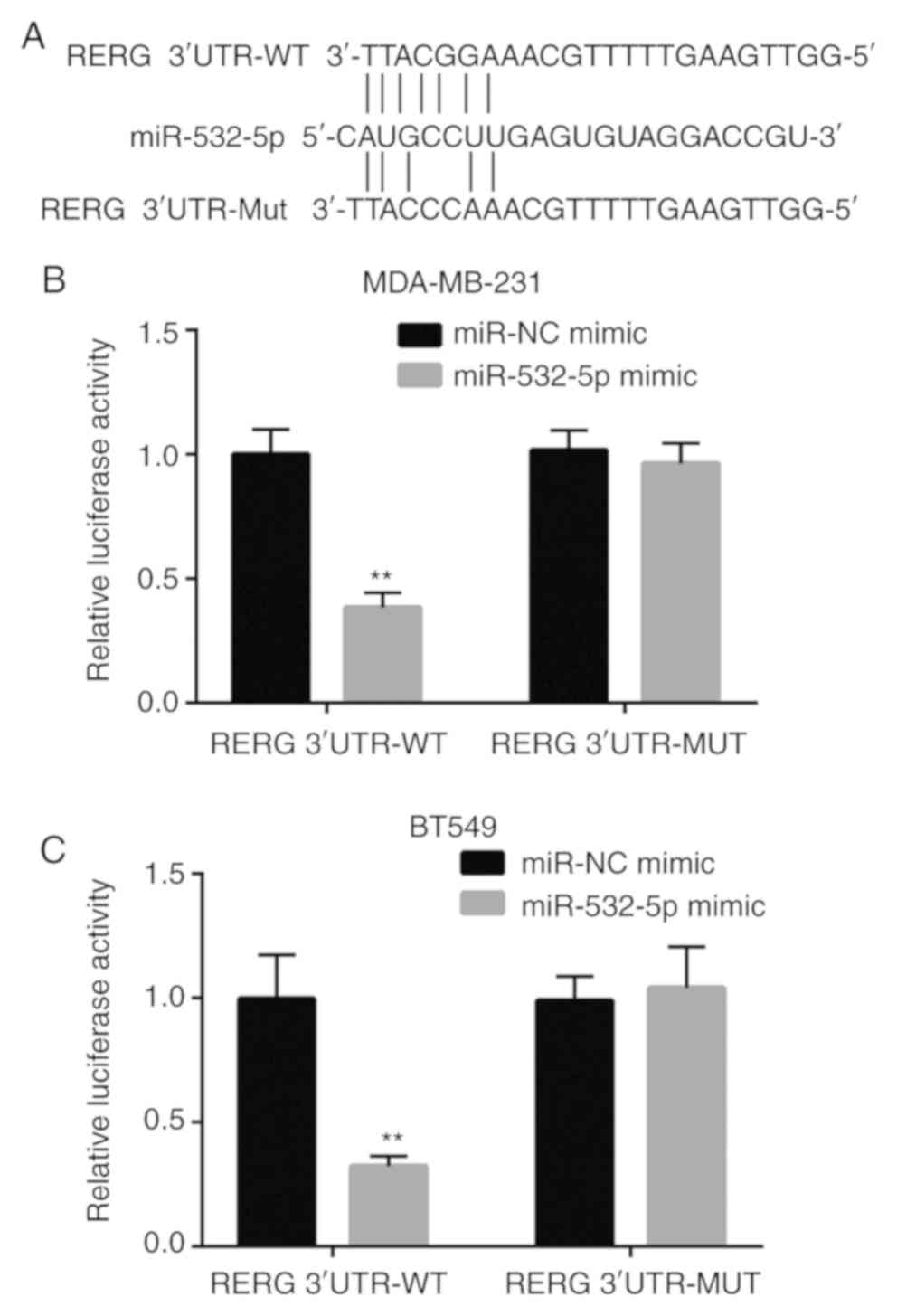
RERG is a target gene of miR-532-5p in
breast cancer cells
To examine whether miR-532-5p directly regulated the
expression of RERG in breast cancer cells, bioinformatic analysis
was applied to predict the potential binding site between
miR-532-5p and the 3′-UTR of RERG mRNA. Sequence alignment showed
that there was a putative binding site between these sequences
(Fig. 3A). Dual luciferase reporter
assays revealed that co-transfection of miR-532-5p mimic and RERG
3′-UTR-WT significantly decreased the luciferase activity while
co-transfection of miR-532-5p mimic and RERG 3′-UTR-Mut did not
change the luciferase activity in MDA-MB-231 cells, compared with
the respective miR-NC mimic groups (Fig.
3B). The data indicated that miR-532-5p directly bound to the
3′-UTR of RERG mRNA, leading to the downregulation of RERG
expression.
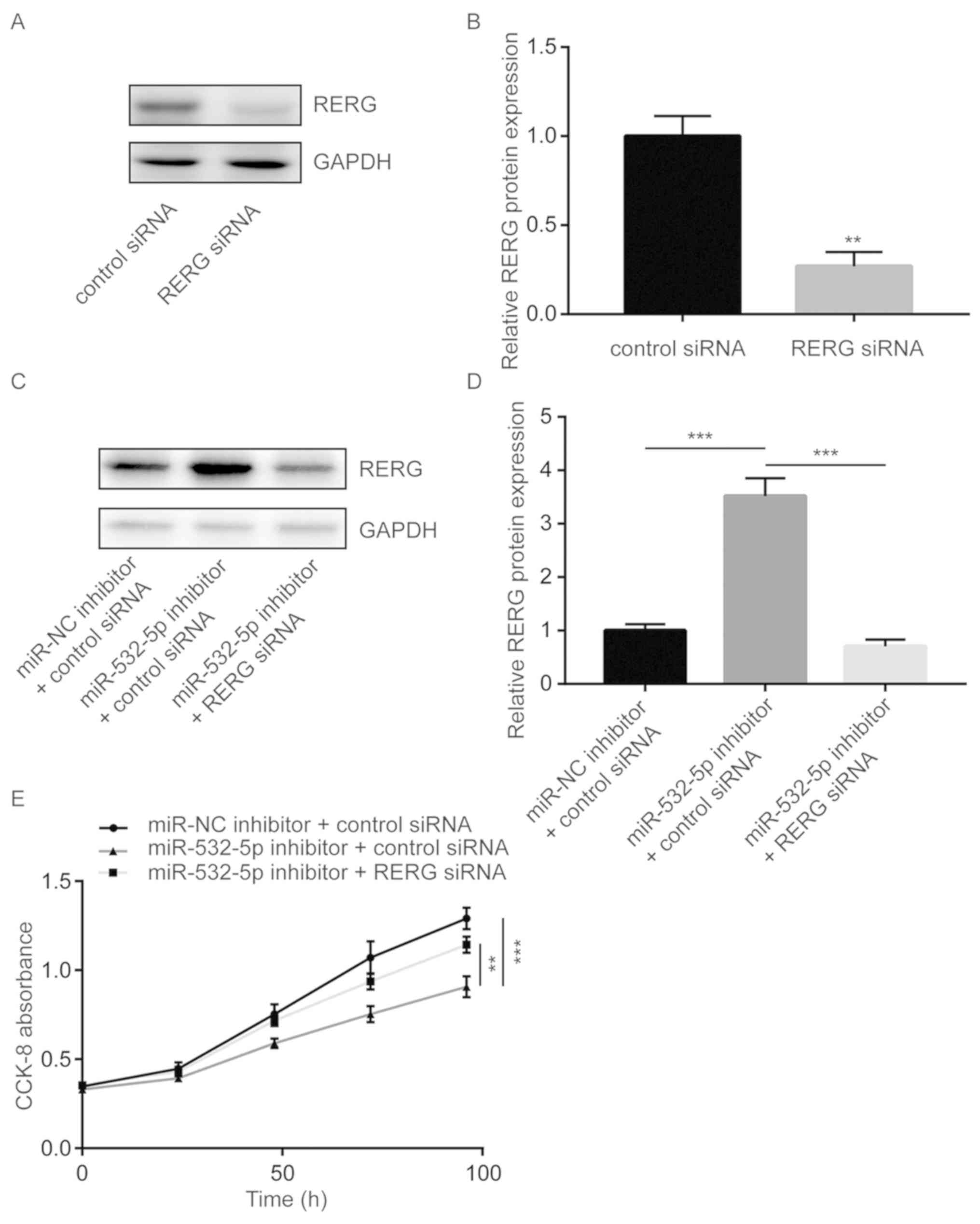
Downregulation of miR-532-5p inhibits
cell proliferation via upregulation of RERG in breast cancer
cells
To further study whether miR-532-5p regulated cell
proliferation via repression of RERG, cell proliferation ability
was detected in MDA-MB-231 cells transfected with miR-532-5p
inhibitor with or without RERG siRNA. Transfection of RERG siRNA
significantly decreased RERG protein expression in MDA-MB-231 cells
(Fig. 4A and B). Transfection of
miR-532-5p inhibitor increased RERG protein expression in
MDA-MB-231 cells and co-transfection of RERG siRNA decreased RERG
protein expression (Fig. 4C and D).
Cell proliferation assay showed that downregulation of miR-532-5p
significantly inhibited cell proliferation ability, compared with
the group transfected with miR-NC inhibitor and control siRNA,
which was partially reversed by silencing RERG expression (Fig. 4E), suggesting that miR-532-5p
functioned as a negative regulator of cell growth through
repression of RERG expression.
Downregulation of miR-532-5p inhibits
cell migration via upregulation of RERG in breast cancer cells
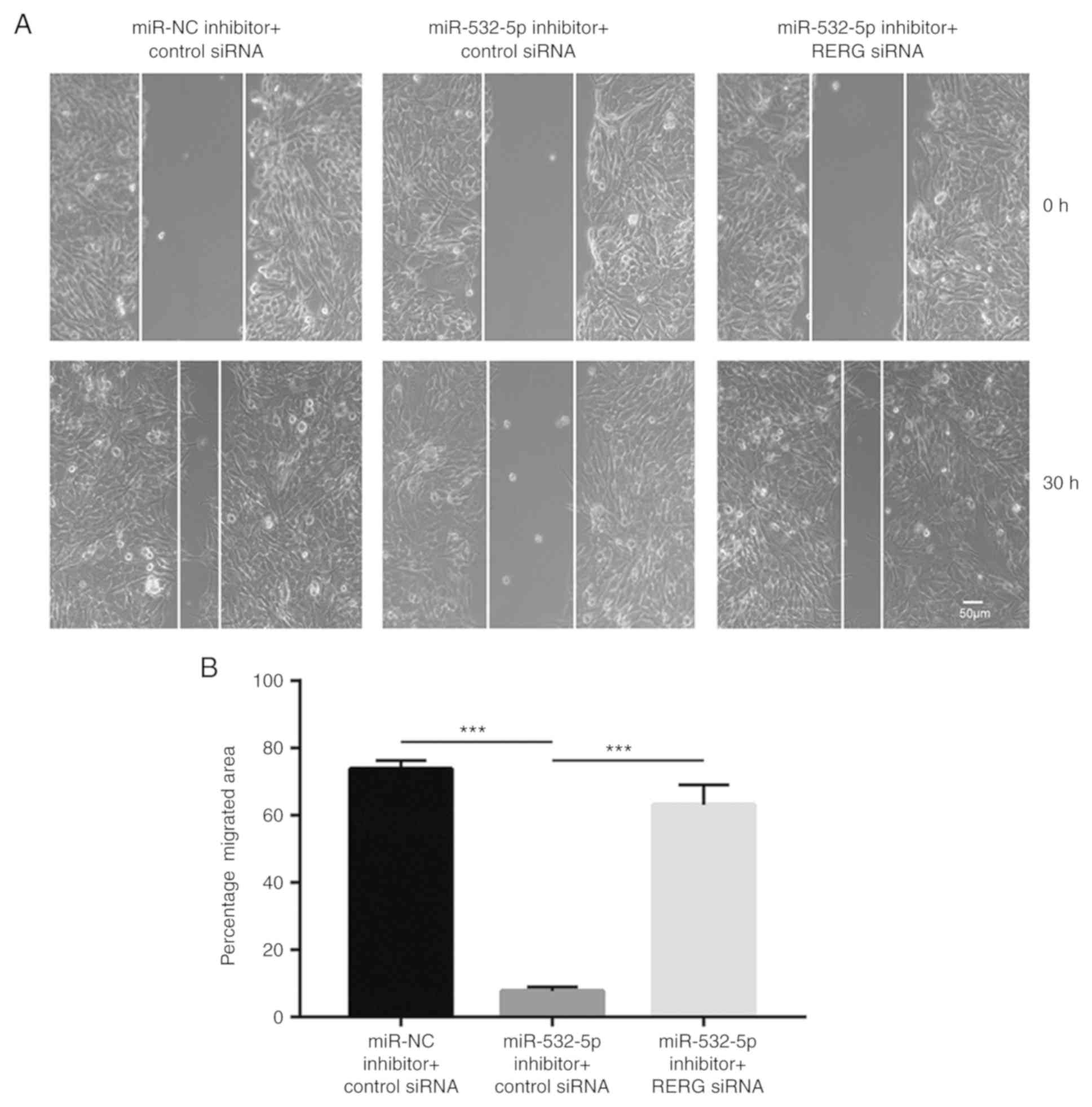
Wound healing assays showed that downregulation of
miR-532-5p reduced the number of cells that migrated towards the
wound area, compared with the group transfected with miR-NC
inhibitor and control siRNA, which was antagonized by RERG
silencing (Fig. 5A and B),
suggesting that miR-532-5p functioned as a negative regulator of
cell migration through repression of RERG expression.
Expression of miR-532-5p is negatively
correlated with RERG mRNA expression in breast cancer tissues
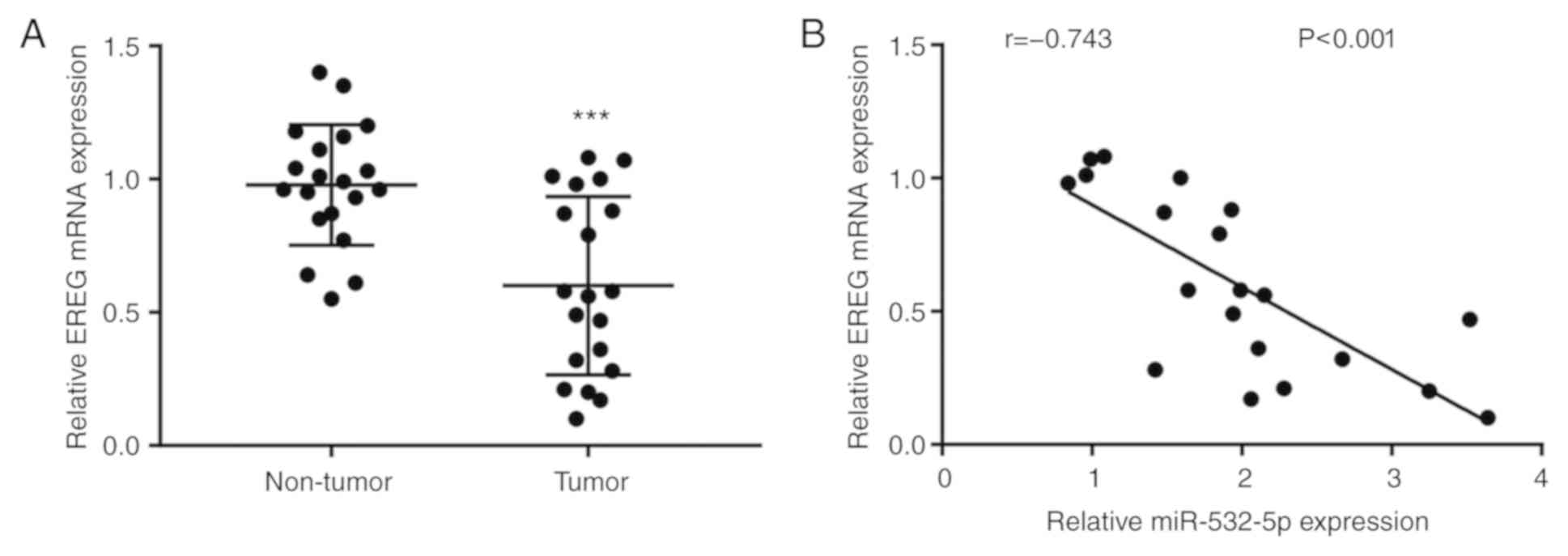
The differences of RERG mRNA level were analyzed
between breast cancer tissues and normal breast tissues from 20
patients with breast cancer. The results showed that RERG mRNA
expression was significantly decreased in breast cancer tissues
compared with normal tissues (Fig.
6A). Moreover, Pearson's correlation analysis demonstrated that
there was a strong negative correlation between miR-532-5p levels
and RERG mRNA levels in breast cancer tissues (Fig. 6B).
Discussion
Dysregulation of miRNAs is a critical step for the
initiation, metastasis, angiogenesis, chemoresistance, radiotherapy
resistance and maintenance of stemness in breast cancer (22–26). A
number of studies identified differentially expressed miRNAs
between normal breast tissues and breast cancer tissues (13,17). The
following studies validated that these miRNAs were involved in the
proliferation and metastasis of breast cancer cells through
regulation of their target genes (27–29).
Recently, miR-532-5p was revealed as a markedly upregulated miRNA
in TNBC (17). The present study
investigated the role and mechanism of miR-532-5p in breast cancer
cells. Consistently with the results of a previous study (17), the current RT-qPCR results showed
that miR-532-5p expression was increased in TNBC cells compared
with the immortal breast cells. Downregulation of miR-532-5p
decreased cell proliferation and migration ability of MDA-MB-231
cells. These results demonstrated that miR-532-5p may be a new
oncogenic miRNA in breast cancer cells.
Sustained activation of the MAPK/ERK signaling
promotes proliferation, migration ability and drug resistance of
breast cancer cells (30). RERG was
first identified as a ras-like, growth inhibitory protein induced
by estrogen in breast cancer cells (18). Subsequent studies revealed that RERG
was a negative regulator of multiple oncogenic pathways including
the MAPK/ERK signaling in cancer cells (19,31).
Furthermore, RERG was also regulated by histone deacetylases and
miRNAs (19,32,33). In
the current study, miR-532-5p downregulation led to inactivation of
the MAPK/ERK signaling in MDA-MB-231 cells. Bioinformatic analysis
results indicated that RERG, an upstream negative regulator of the
MAPK/ERK signaling pathway, was a potential target gene of
miR-532-5p. In addition, downregulation of miR-532-5p also
increased RERG mRNA and protein levels in MDA-MB-231 cells.
Additionally, the dual luciferase reporter assay validated that
RERG was a target gene of miR-532-5p. In several cancer types,
including breast cancer, RERG inhibits cell proliferation,
migration, invasion and clonogenicity via the MAPK/ERK pathway
(19,31). In the present study, cell
proliferation and migration assays showed that RERG silencing could
reverse miR-532-5p downregulation-induced cell proliferation and
migration inhibition. Furthermore, in clinical samples, a
significant negative correlation was observed between miR-532-5p
expression and RERG mRNA levels. Thus, the current results suggest
that miR-532-5p acted as an oncogenic miRNA in breast cancer via
repression of RERG. A previous study indicated that in breast
cancer cell lines, including MCF7 and Hs578t, RERG was a target
gene of miR-382-5p. Via suppression of RERG, miR-382-5p promoted
cell growth of breast cancer cells in vitro and in
vivo (33). The current and
previous results suggest that RERG may be regulated by a complex
miRNA network involving miR-382-5p and miR-532-5p in breast cancer
cells.
In conclusion, the current study provided new sights
into the role of miR-532-5p and the regulatory mechanism of RERG in
breast cancer cells. The current study demonstrated that miR-532-5p
may be an oncogenic miRNA and may serve as a therapeutic target in
breast cancer cells.
Acknowledgements
Not applicable.
Funding
The current study was funded by Nanjing Medical
Science and Technology Development Project (grant no.
YKK17063).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LH, XT and XS were responsible for clinical sample
collection. LS was responsible for study design and supervision,
and manuscript preparation, review and editing. Data acquisition
and analysis were performed by LH and XT.
Ethics approval and consent to
participate
All patients provided written informed consent
before the study and the Ethic Committee of Nanjing University
approved the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McDermott AM, Miller N, Wall D, Martyn LM,
Ball G, Sweeney KJ and Kerin MJ: Identification and validation of
oncologic miRNA biomarkers for luminal A-like breast cancer. PLoS
One. 9:e870322014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Notas G, Pelekanou V, Kampa M, Alexakis K,
Sfakianakis S, Laliotis A, Askoxilakis J, Tsentelierou E, Tzardi M,
Tsapis A and Castanas E: Tamoxifen induces a pluripotency signature
in breast cancer cells and human tumors. Mol Oncol. 9:1744–1759.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ahmed S, Sami A and Xiang J: HER2-directed
therapy: Current treatment options for HER2-positive breast cancer.
Breast Cancer. 22:101–116. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Karlsson E, Veenstra C, Emin S, Dutta C,
Pérez-Tenorio G, Nordenskjöld B, Fornander T and Stål O: Loss of
protein tyrosine phosphatase, non-receptor type 2 is associated
with activation of AKT and tamoxifen resistance in breast cancer.
Breast Cancer Res Treat. 153:31–40. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boulbes DR, Chauhan GB, Jin Q,
Bartholomeusz C and Esteva FJ: CD44 expression contributes to
trastuzumab resistance in HER2-positive breast cancer cells. Breast
Cancer Res Treat. 151:501–513. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
O'Reilly EA, Gubbins L, Sharma S, Tully R,
Guang MH, Weiner-Gorzel K, McCaffrey J, Harrison M, Furlong F, Kell
M and McCann A: The fate of chemoresistance in triple negative
breast cancer (TNBC). BBA Clin. 3:257–275. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Venkatesh T, Suresh PS and Tsutsumi R:
Non-coding RNAs: Functions and applications in endocrine-related
cancer. Mol Cell Endocrinol. 416:88–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calore F, Lovat F and Garofalo M:
Non-coding RNAs and cancer. Int J Mol Sci. 14:17085–17110. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanwar JR, Mahidhara G and Kanwar RK:
MicroRNA in human cancer and chronic inflammatory diseases. Front
Biosci (Schol Ed). 2:1113–1126. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lowery AJ, Miller N, Devaney A, McNeill
RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G and
Kerin MJ: MicroRNA signatures predict oestrogen receptor,
progesterone receptor and HER2/neu receptor status in breast
cancer. Breast Cancer Res. 11:R272009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cui S, Liao X, Ye C, Yin X, Liu M, Hong Y,
Yu M, Liu Y, Liang H, Zhang CY and Chen X: ING5 suppresses breast
cancer progression and is regulated by miR-24. Mol Cancer.
16:892017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi Y, Luo X, Li P, Tan J, Wang X, Xiang T
and Ren G: miR-7-5p suppresses cell proliferation and induces
apoptosis of breast cancer cells mainly by targeting REGγ. Cancer
Lett. 358:27–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Raychaudhuri M, Bronger H, Buchner T,
Kiechle M, Weichert W and Avril S: MicroRNAs miR-7 and miR-340
predict response to neoadjuvant chemotherapy in breast cancer.
Breast Cancer Res Treat. 162:511–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsai HP, Huang SF, Li CF, Chien HT and
Chen SC: Differential microRNA expression in breast cancer with
different onset age. PLoS One. 13:e01911952018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Finlin BS, Gau CL, Murphy GA, Shao H,
Kimel T, Seitz RS, Chiu YF, Botstein D, Brown PO, Der CJ, et al:
RERG is a novel ras-related, estrogen-regulated and
growth-inhibitory gene in breast cancer. J Biol Chem.
276:42259–42267. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ho JY, Hsu RJ, Liu JM, Chen SC, Liao GS,
Gao HW and Yu CP: MicroRNA-382-5p aggravates breast cancer
progression by regulating the RERG/Ras/ERK signaling axis.
Oncotarget. 8:22443–22459. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kasinski AL and Slack FJ: miRNA-34
prevents cancer initiation and progression in a therapeutically
resistant K-ras and p53-induced mouse model of lung adenocarcinoma.
Cancer Res. 72:5576–5587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tung SL, Huang WC, Hsu FC, Yang ZP, Jang
TH, Chang JW, Chuang CM, Lai CR and Wang LH: miRNA-34c-5p inhibits
amphiregulin-induced ovarian cancer stemness and drug resistance
via downregulation of the AREG-EGFR-ERK pathway. Oncogenesis.
6:e3262017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kong W, He L, Richards EJ, Challa S, Xu
CX, Permuth-Wey J, Lancaster JM, Coppola D, Sellers TA, Djeu JY and
Cheng JQ: Upregulation of miRNA-155 promotes tumour angiogenesis by
targeting VHL and is associated with poor prognosis and
triple-negative breast cancer. Oncogene. 33:679–689. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Allen KE and Weiss GJ: Resistance may not
be futile: microRNA biomarkers for chemoresistance and potential
therapeutics. Mol Cancer Ther. 9:3126–3136. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu L, Xue F, Xu X, Xu J, Hu S, Liu S, Cui
Y and Gao C: MicroRNA-198 inhibition of HGF/c-MET signaling pathway
overcomes resistance to radiotherapy and induces apoptosis in human
non-small-cell lung cancer. J Cell Biochem. 119:7873–7886. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Blenkiron C, Goldstein LD, Thorne NP,
Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE,
Green AR, Ellis IO, et al: MicroRNA expression profiling of human
breast cancer identifies new markers of tumor subtype. Genome Biol.
8:R2142007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Volinia S, Galasso M, Sana ME, Wise TF,
Palatini J, Huebner K and Croce CM: Breast cancer signatures for
invasiveness and prognosis defined by deep sequencing of microRNA.
Proc Natl Acad Sci USA. 109:3024–3029. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mirzoeva OK, Das D, Heiser LM,
Bhattacharya S, Siwak D, Gendelman R, Bayani N, Wang NJ, Neve RM,
Guan Y, et al: Basal subtype and MAPK/ERK kinase
(MEK)-phosphoinositide 3-kinase feedback signaling determine
susceptibility of breast cancer cells to MEK inhibition. Cancer
Res. 69:565–572. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao W, Ma N, Wang S, Mo Y, Zhang Z, Huang
G, Midorikawa K, Hiraku Y, Oikawa S, Murata M and Takeuchi K: RERG
suppresses cell proliferation, migration and angiogenesis through
ERK/NF-κB signaling pathway in nasopharyngeal carcinoma. J Exp Clin
Cancer Res. 36:942017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang AG, Fang W, Han YH, Cho SM, Choi JY,
Lee KH, Kim WH, Kim JM, Park MG, Yu DY, et al: Expression of the
RERG gene is gender-dependent in hepatocellular carcinoma and
regulated by histone deacetyltransferases. J Korean Med Sci.
21:891–896. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Ren S, Yuan F, Zhang K, Fan Y,
Zheng S, Gao Z, Zhao J, Mu T, Zhao S, et al: miR-135 promotes
proliferation and stemness of oesophageal squamous cell carcinoma
by targeting RERG. Artif Cells Nanomed Biotechnol. 46
(Suppl):1210–1219. 2018. View Article : Google Scholar : PubMed/NCBI
|