Introduction
Based on high-throughput screening techniques, a
novel human gene, chromosome 3 open reading frame 33 (AC3-33),
capable of inhibiting phorbol 12-myristate 13-acetate and AP-1
activation, has been identified from 650 known human genes
(1). Previously, it was demonstrated
that the AC3-33 gene is widely expressed in the adrenal gland and
cervix (2). Further studies on
AC3-33 indicate that it is a secretory protein that inhibits ETS
transcription factor ELK1 transcriptional activity through the
ERK1/2 pathway (2,3). Alternative RNA splicing not only
increases the diversity of mRNA expression, but also plays an
important role in the regulation of gene function (4,5). Wang
et al (6) found that a
single-block intronic expressed sequence tag (EST) containing a
polyadenylation site could form a 3′exon site, thus forming
transcript variants. Although one AC3-33 transcript variant has
previously been reported (1),
previous data suggested that other AC3-33 isoforms may exist.
Breast cancer is the most common cancer in women
worldwide, and its incidence is increasing, making breast cancer a
major public health problem (7).
Numerous signaling pathways can modulate the development of breast
cancer cells, which can affect the cell cycle as well as processes
involving the activation and inhibition of specific genes. The
activation and inhibition of the transcription factor AP-1 can
greatly affect the growth and reproduction of cancer cells,
regulating the development of many deadly cancer types (7,8). AP-1 is
mainly composed of the c-Jun, c-Fos, MAF and activating
transcription factor protein families. In human cells, AP-1
comprises c-Jun and c-Fos, which can activate and affect numerous
signaling pathways, in addition to regulating cell growth and
reproduction (9–15). Previous studies have demonstrated
that infection, growth factors and cancer cells affect the
expression of AP-1-related signaling pathway, leading to the
division, differentiation and apoptosis of cancer cells (16–19).
In the present study, a second AC3-33 transcript
variant was successfully cloned, splice variant (sv)AC3-33. The
data also characterized svAC3-33 and demonstrated the subcellular
localization of the encoded protein. Furthermore, the effect of
raised svAC3-33 expression on cell proliferation was demonstrated.
Our present evidence shows that svAC3-33 may inhibit MCF-7 cell
progression by downregulating c-Jun, which is an important member
of the AP-1 signaling pathway.
Materials and methods
PCR identification
Human breast cancer cell line MCF-7 and human
cervical carcinoma cell line HeLa were purchased from the American
Type Culture Collection and cultured in DMEM (Gibco, Thermo Fisher
Scientific, Inc.) supplied with 10% FBS and penicillin/streptomycin
(Gibco, Thermo Fisher Scientific, Inc.) at 37°C in a humidified 5%
CO2. TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract the total RNA from MCF-7 and
HeLa cells. M-MLV reverse transcriptase (Promega Corporation) was
used for RT-qPCR, and the first strand cDNA was synthesized. For
each sample, cDNA synthesis was performed using 0.25 mg of total
RNA and PrimeScript RT Master Mix Perfect RT (Takara Bio, Inc.).
The cDNA of MCF-7 was used to amplify sv-AC3-33, and the cDNA of
HeLa was used to amplify AC3-33. svAC3-33 and AC3-33 were amplified
by PCR using the following primers: Forward 5′-GAGGAGCTCAGGGCCGC-3′
and reverse 5′-TAAAGCATAAAGAATTCCTTTA-3′. PCR amplification was
conducted using a Sangon Biotech PCR kit (cat. no. B639297; Sangon
Biotech Co., Ltd.). According to the manufacturer's instructions,
DNA template 0.5 µl, primer F 1 µl, primer R 1 µl, Taq PCR Master
Mix 12.5 µl and ddH2O up to 25 µl. Briefly, after an
initial denaturation step at 95°C for 5 min, amplifications were
carried out with 31 cycles, consisting of a melting step at 95°C
for 30 sec, an annealing step at 55°C for 30 sec, and an extension
step at 72°C for 2 min, followed by an extra extension step at 72°C
for 5 min. The PCR product was subjected to electrophoresis on a 1%
agarose gel and sequenced by Sangon Biotech Co., Ltd.
Plasmid construction
A possible novel AC3-33 isoform was identified in
the University of California Santa Cruz Genome Browser sequence
database (http://genome.ucsc.edu/cgi-bin/hgGateway). svAC3-33
and full-length (or wild-type) AC3-33 are two alternatively spliced
transcripts of the AC3-33 gene containing different open reading
frames (ORFs). The full-length fragment of svAC3-33 cDNA was
obtained by RT-PCR from MCF-7 cells using the following primers:
Forward 5′-TATAAGCTTATGGCGGGGCAGCCCGCG-3′ and reverse
5′-TATGGATCCCACCCTTTTCTACGAAAGTTT-3′. BamHI and
HindIII recognition sites were added to the primers. The
purified PCR product was recombined with p-enhanced green
fluorescent protein (EGFP)-C3 vector (BD Biosciences) to form a
recombinant expression vector. To construct pEGFP-C3-svAC3-33, the
corresponding primers were used for fusion with the N-terminal GFP
tag in the PEGFP-C3 vector with BamHI and HindIII. All
clones were confirmed by sequencing using an ABI Prism 3100 Genetic
Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.).
After confirmation, all plasmids were extracted and purified for
transfection using EndoFree Plasmid Maxi Kit (Qiagen, USA). Signal
IP 4.0 Server (http://www.cbs.dtu.dk/services/SignalP) was used to
analyze the signal peptide of the svAC3-33 protein; TMHMM server v
2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) was used to
predict the transmembrane location of proteins.
Preparation of the small interfering
(si)RNA
The siRNA sequence for the svAC3-33 gene was
designed and synthesized. This siRNA sequence was located in the
ORF of the svAC3-33 gene, and the target sequence of the siRNA was
5′-GGACGAUUACGCCGAAUAA-3′. A nonsense sequence siRNA was used as a
control, named si-NC. Sense and antisense strands were designed and
synthesized by Beijing Genomics Institute. According to the
manufacturer's instructions (pSilencer 4.1-CMV neo siRNA Expression
Vector kit), strands were annealed to the corresponding sites of
pSilencer 4.1-CMVneo (Thermo Fisher Scientific, Inc.; www.ambion.com/techlib/msds) HindIII and
BamHI, thus constructing a silent plasmid, named
si-svAC3-33.
Cell culture and transient
transfection
MCF-7 cells were supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin in
DMEM (Gibco; Thermo Fisher Scientific, Inc.), and maintained at
37°C with 5% CO2. Then 2×104,
5×104 and 1.2×104 MCF-7 cells were seeded
into six-well, 24-well or 96-well plates, respectively, and
transiently transfected with pEGFP-C3, pEGFP-C3-svAC3-33, si-NC or
si-svAC3-33 plasmids using and Lipofectamine® 2000 (all,
Thermo Fisher Scientific, Inc.) was used according to the
manufacturer's protocol.
Subcellular localization
pEGFP-C3 and pEGFP-C3-svAC3-33 plasmids were
transiently transfected into MCF-7 cells in a 6-well plate, and
then each well of the cells were washed twice with PBS. After
transfection MCF-7 cells were incubated at 37°C for 24 h, then 4%
paraformaldehyde was used to fix MCF-7 cells for 30 min at room
temperature, and DAPI was added to stain the nuclei for 2 min at
room temperature. After washing twice with PBS, images of MCF-7
cells were captured (magnification ×400; IX71 inverted fluorescence
microscope; Olympus Corporation) and analyzed using ImageJ (version
1.40; National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
pEGFP-C3, pEGFP-C3-svAC3-33, si-NC or si-svAC3-33
plasmids were transiently transfected into MCF-7 cells as
previously described. After transfection MCF-7 cells were incubated
at 37°C for 24 h, 2,000 cells were seeded into 96-well plate. CCK-8
solution (10 µl; Beijing Zoman Biotechnology Co., Ltd.) was added
to each well, and the cells were incubated at 37°C, 5%
CO2 for 2 h according to the manufacturers protocol. The
absorbance at 450 nm was measured daily with a microplate reader
for 5 days.
EdU cell proliferation assay
MCF-7 cells were transiently transfected with
pEGFP-C3, pEGFP-C3-svAC3-33, si-NC or si-svAC3-33 plasmids in
96-well plates at 50–60% confluence of 5×104. The steps
of cell incubation and staining were based on the manufacturer's
instructions. MCF-7 cells were fixed using 4% paraformaldehyde for
30 min at room temperature, washed with 0.5% Triton X-100 three
times, and images were captured using an IX71 inverted fluorescence
microscope (magnification, ×200, IX71 inverted fluorescence
microscope; Olympus Corporation).
Dual-luciferase assay to measure AP-1
activity
MCF-7 cells were co-transfected with pEGFP-C3 or
pEGFP-C3-svAC3-33; si-NC (nonsense siRNA) or si-svAC3-33 and
pAP-1-Luc plasmid and pGL3-Basic plasmid which contain the
Renilla luciferase gene to normalize the transfection
efficiency (all, Promega Corporation). After transfection the MCF-7
cells were incubated for 24 h at 37°C, and cells were lysed and
assayed. Luciferase activities were tested using the
Dual-Luciferase® Reporter Assay System (Promega
Corporation) following the manufacturer's instructions and measured
using a microplate luminometer reader (Tecan Group, Ltd.). Three
independent repeat tests were performed.
Western blot analysis
MCF-7 cells were transfected with si-NC or
si-svAC3-33, pEGFP-C3 or pEGFP-C3-svAC3-33, and MCF-7 cells were
incubated at 37°C for 48 h, then total cell extracts and the
western blotting was performed as previously described (20). The total protein concentration was
determined using the BCA protein assay kit (Sangon Biotech Co.,
Ltd.). Specific antibodies for the following proteins were used:
Anti-green fluorescent protein (1:5,000; cat. no. D110008; BBI
Solutions), anti-c-Fos (1:500; cat. no. D120415; BBI Solutions),
anti-c-Jun (1:500; cat. no. D155181; BBI Solutions) and
anti-β-actin (1:50,000; cat. no. AC026; ABclonal Biotech Co.,
Ltd.). Bands were visualized using an ECL kit (Sangon Biotech Co.,
Ltd.).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract the total RNA from
MCF-7 cells. M-MLV reverse transcriptase (Promega Corporation) was
used for RT-PCR, and the first strand cDNA was synthesized
(9). Primers were designed and
synthesized by Beijing Genomics Institute, and the primers were as
follows: p15, forward 5′-CTAGTGGAGAAGGTGCGACA-3′ and reverse
5′-ACCAGCGTGTCCAGGAAG-3′; p21, forward 5′-CCCGTGAGCGATGGAACT-3′ and
reverse 5′-AGGCACAAGGGTACAAGACA-3′; p27 forward
5′-TAGAGGGCAAGTACGACGAGTGG-3′ and reverse
5′-CAGGTCGCTTCCTTATTCC-3′; p53 forward 5′-CCTCCTCAGCATCTTATCCG-3′
and reverse 5′-CACAAACACGCACCTCAAA-3′ GAPDH, forward
5′-AAAGGGTCATCATCTCTG-3′ and reverse 5′-GCTGTTGTCATACTTCTC-3′. qPCR
was performed with the designed primers. The reaction was carried
out using a 7900HT Fast Real-Time PCR System (Thermo Fisher
Scientific, Inc.). GAPDH-80 was used as the internal control.
Amplification was performed under the following conditions: 1 min
at 95°C; 5 sec at 95°C; 30 sec at 60°C for 40 cycles; Melt Curve 65
to 95°C. Gene expression levels were calculated as a ratio of the
expression of GAPDH, and data were analyzed using the
2−ΔΔCt method (21).
Statistical analysis
All of the data were collected from three
independent experiments. Statistical analyses were carried out
using one-way ANOVAs followed by post hoc Tukey tests, or Student's
t-tests. P<0.05 was considered to indicate a statistically
significant difference.
Results
Cloning and characterization of the
svAC3-33 transcript variant
The novel human gene AC3-33 is a secretory protein,
and can suppress the activity of the transcription factor AP-1
(1). In the present study, specific
primers were used based on an EST from AC3-33, to clone a novel
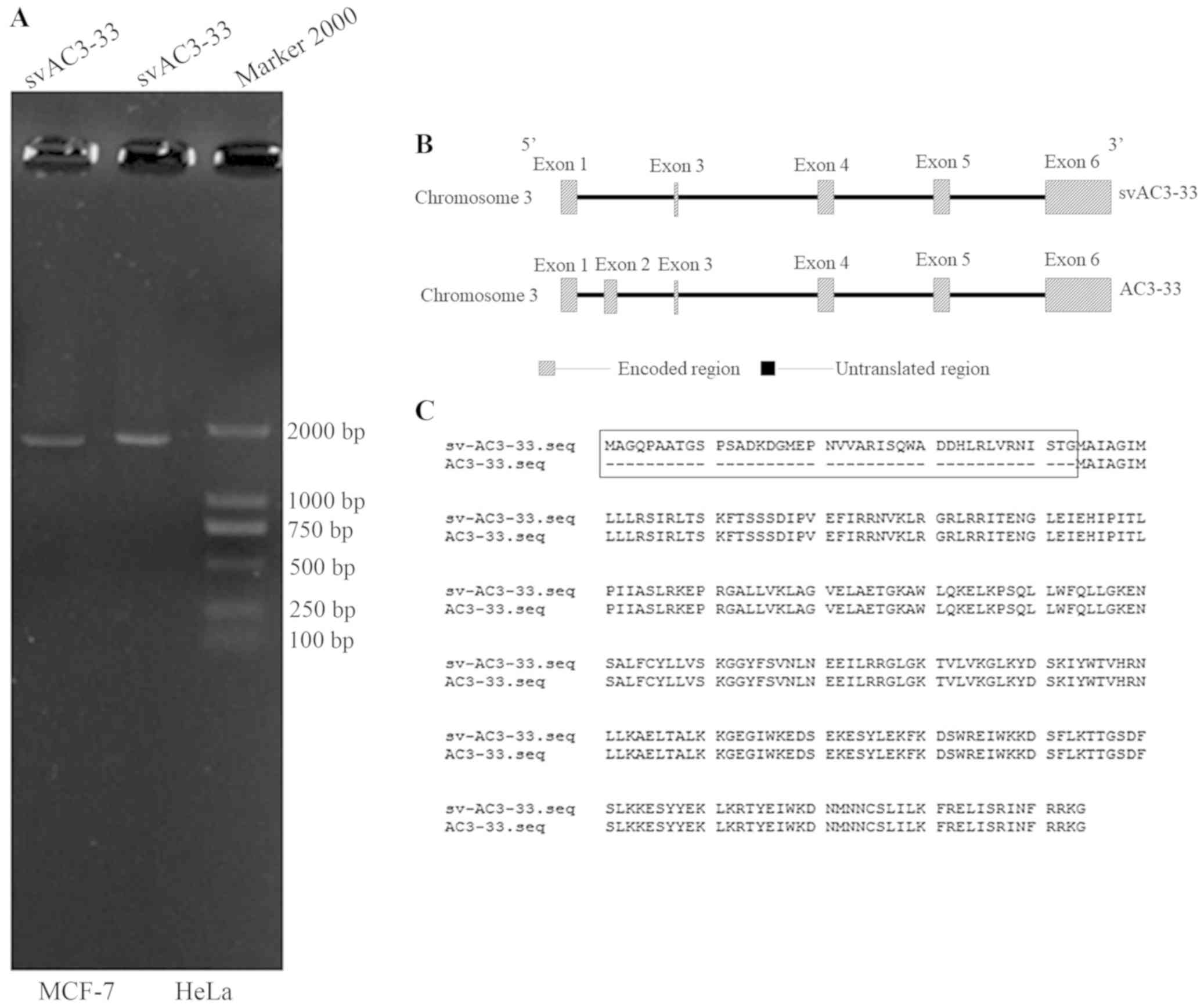
splice variant of AC3-33 cDNA [the PCR product is ~1,825 base pairs
(Fig. 1A)] from MCF-7 cells, which
was designated svAC3-33. Only svAC3-33 was amplified, and no
expression of AC3-33 was noted (Fig.
1A). Therefore, the effects of this siRNA on cell proliferation
were due to the silencing of the svAC3-33 transcript (Fig. 3A). Full-length AC3-33 and svAC3-33
transcripts are the alternatively spliced transcripts of the AC3-33
gene, which contains different ORFs (Fig. 1B). The putative svAC3-33 protein
contains 294 amino acids (Fig. 1C)
and has a predicted molecular mass of 29 kDa. Signal IP 4.0 Server
analysis revealed that the svAC3-33 contained no signal peptides,
whereas TMHMM server v. 2.0 analysis revealed that the svAC3-33
protein does not contain a transmembrane spanning region. The
C3orf33 gene is located on human chromosome 3q25.31. According to
the NCBI database (NM_173657.2 and NM_001308229.1 http://www.ncbi.nlm.nih.gov/nuccore/NM_001308229.2/),
svAC3-33 comprises five exons and four introns. svAC3-33 is very
similar to AC3-33, except that it lacks exon 2 entirely and has a
pre-translation initiation site, resulting in the coding sequence
starting earlier than AC3-33 (Fig.
1C). There are currently no studies indicating the function of
svAC3-33′.
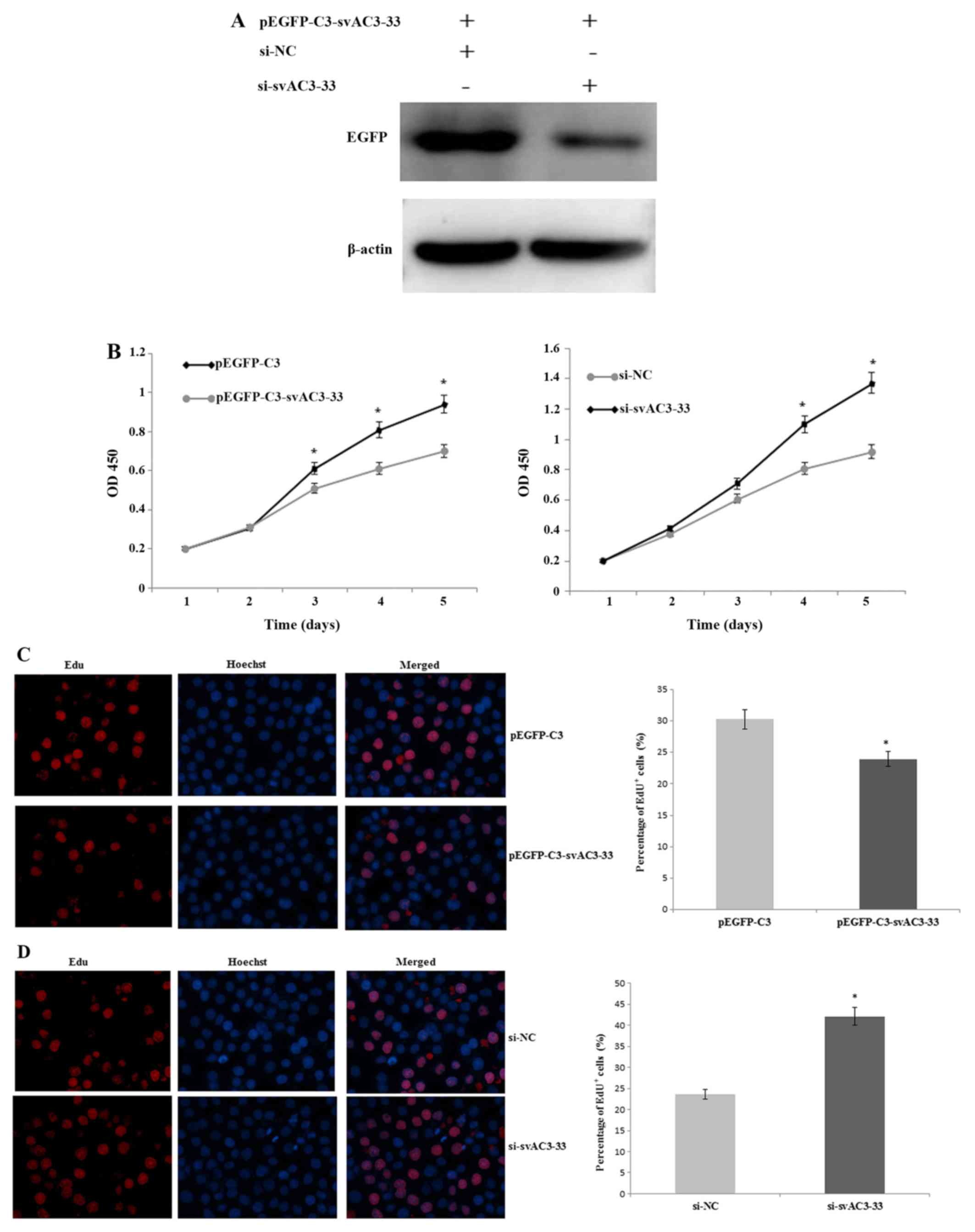 | Figure 3.(A) MCF-7 cells were co-transfected
with pEGFP-C3-svAC3-33 and si-NC, pEGFP-C3-svAC3-33, and
si-svAC3-33 for 48 h. The expression levels of EGFP were measured
by western blotting. β-Actin was used as a loading control. (B)
MCF-7 cells were transfected with pEGFP-C3, pEGFP-C3-svAC3-33,
si-NC, or si-svAC3-33, then assayed with a CCK-8 kit. Cells were
collected and analyzed every 24 h and the absorbance was measured
at 450 nm. MCF-7 cells were transiently transfected with (C)
pEGFP-C3 or pEGFP-C3-svAC3-33 as well as (D) si-NC or si-svAC3-33.
Cell proliferation was measured by calculating the % of EdU+ cells
48 h after transfection (magnification, ×200). Data were collected
from three independent experiments. *P<0.05 vs. respective
control. EGFP, enhanced green fluorescent protein; siRNA, small
interfering RNA; si-NC, nonsense siRNA; si-svAC3-33, siRNA
targeting svAC3-33; sv, splice variant. (E) MCF-7 cells were
transfected with pEGFP-C3 or pEGFP-C3-svAC3-33 for 24 h and the
expression of p15, p21, p27 and p53 was evaluated by RT-qPCR.
However, no significant differences were found of p15, p27 and p53
compared with pEGFP-C3 group (P>0.05). (F) MCF-7 cells were
transfected with pEGFP-C3 or pEGFP-C3-svAC3-33 for 24 h. The
expression of p21 at the mRNA level was measured at different time
points (0, 24, and 48 h) after transfection, using RT-qPCR. GAPDH
was used to normalize the p21 level. (G) si-svAC3-33 was
transfected into MCF-7 cells. Control MCF-7 cells were treated with
a scrambled siRNA. The efficiency of svAC3-33 mRNA knockdown was
examined by RT-semiquantitative PCR. Compared with the si-NC
control group, svAC3-33 was downregulated after transfection of
si-svAC3-33 group. The inhibition of p21 mRNA level was determined
at different time points (0, 24 and 48 h) after transfection, using
RT-qPCR. GAPDH was used to normalize the p21 level, and then values
were further normalized against the corresponding 0 h levels. Data
were collected from three independent experiments. *P<0.05 vs.
respective control. EGFP, enhanced green fluorescent protein; RT,
reverse transcription; q, quantitative; siRNA, small interfering
RNA; si-NC, nonsense siRNA; si-svAC3-33, siRNA targeting svAC3-33;
sv, splice variant. |
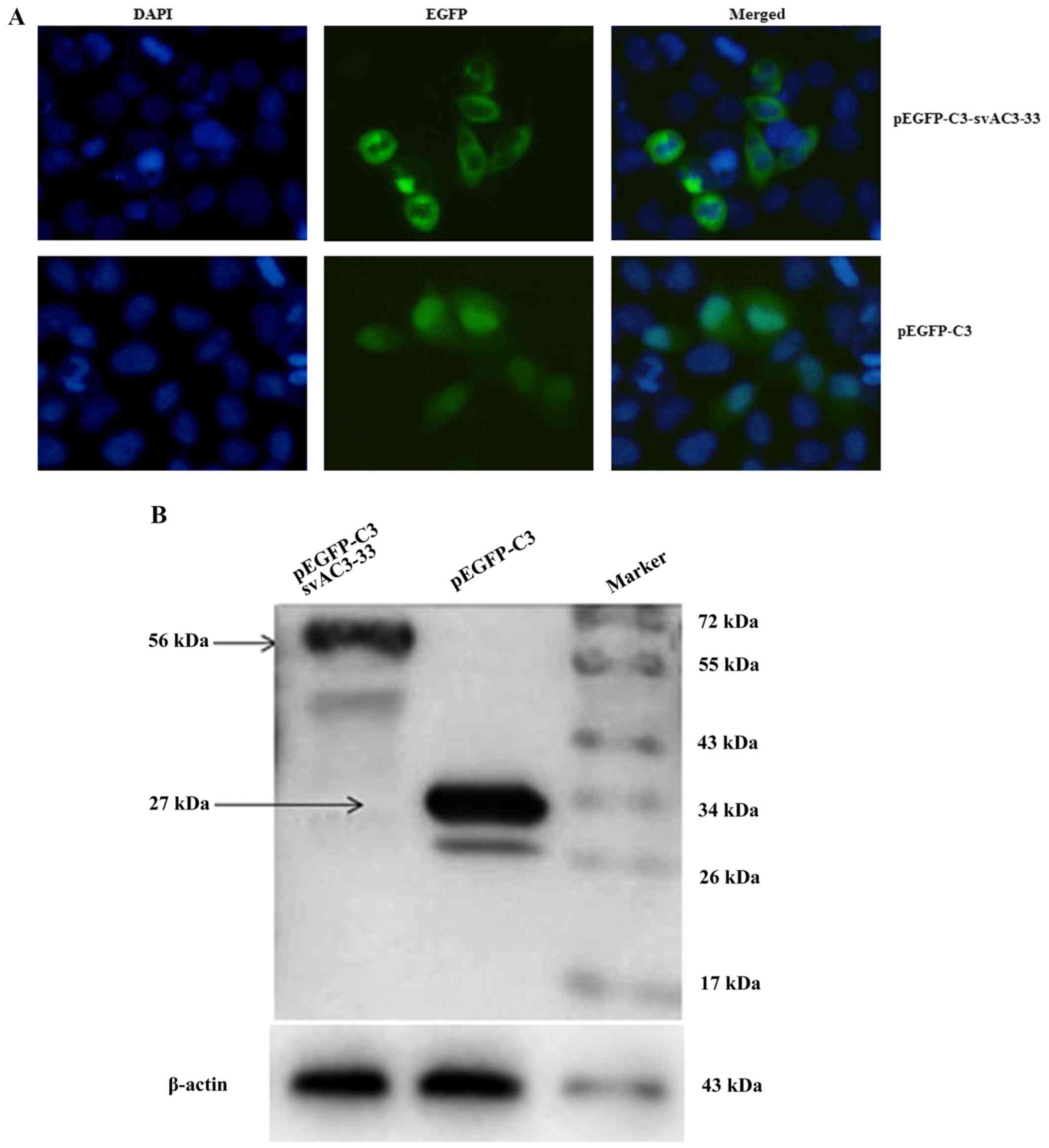
Subcellular localization of
svAC3-33
A pEGFP-C3-svAC3-33 plasmid was constructed and
transiently transfected it into MCF-7 cells. svAC3-33 was mainly
located in the cytoplasm (Fig. 2A)
and the size of the pEGFP-C3-svAC3-33 encoded protein was around 56
k Da (Fig. 2B).
svAC3-33 expression inhibits cell
proliferation
Western blotting indicated that svAC3-33 was
expressed in MCF-7 cells following transfection (Fig. 2B). MCF-7 cells were co-transfected
with pEGFP-C3-svAC3-33 and si-NC, pEGFP-C3-svAC3-33 and si-svAC3-33
for 48 h. pEGFP-C3-svAC3-33 expression was qualitatively observed
to be inhibited by si-svAC3-33 at the protein level (Fig. 3A). Counting of cell numbers with
ССK-8 indicated that the expression of svAC3-33 inhibited the
proliferation of MCF-7 cells compared with those of the negative
control group. Similarly, inhibiting endogenous svAC3-33 expression
with siRNA may reduce the proliferation of MCF-7 cells (Fig. 3B). EdU+ cell numbers in
svAC3-33-expressing cells decreased significantly after 2 days,
compared with those of cells transfected with the empty vector
(Fig. 3C). The EdU cell
proliferation assay revealed a marked decrease in the number of
MCF-7 cells in the S phase following transfection with
pEGFP-C3-svAC3-33 (Fig. 3C). The EdU
assay indicated that the percentage of cells in the S phase
decreased from 30.27% in the transfection with control cells to
23.96% in pEGFP-C3-svAC3-33 cells at 48 h following transfection.
treating pEGFP-C3-svAC3-33+ cells with si-svAC3-33, compared with
si-NC treated cells, increased the percentage of cells in the S
phase from 25.37 to 40.21% at 48 h (Fig.
3D). Taken together, these data suggested that svAC3-33 may
inhibit the cell cycle progression of MCF-7 cells from the S
phase.
To investigate the relationship between svAC3-33 and
cell proliferation, the impact of svAC3-33 on the mRNA expression
of p15, p21, p27, p53 and proliferation-related genes was examined.
It was found that svAC3-33 only regulates p21 expression (Fig. 3E-F). Compared with the pEGFP-C3
transfected negative control group, the expression of p21 mRNA in
the svAC3-33+ group was significantly higher than that in the
control group. After silencing svAC3-33 with si-svAC3-33, compared
with the si-NC negative control group, the mRNA expression levels
of p21 were downregulated by 60% at 48 h (Fig. 3G).
Effects of svAC3-33 on AP-1
signaling
The role of the AP-1 signaling pathways in the
antiproliferative function of svAC3-33 was subsequently examined.
The dual-luciferase reporter assay was used to investigate the
function of svAC3-33. Increased expression of svAC3-33 inhibited
the activity of AP-1 compared with the transfection with empty
vector controls in MCF-7 cells. Compared with the transfection with
the si-NC-treated control, svAC3-33 knockdown by RNA interference
increased the activity of AP-1 (Fig.
4A). c-Jun and c-Fos are the two important components of the
transcription factor AP-1. Western blot analysis was used to
determine which components of the specific transcription factor
mediated the process wherein svAC3-33 suppressed the AP-1 activity
(Fig. 4B). The expression levels of
c-Jun protein were reduced by the increased expression of svAC3-33
(Fig. 4C), whereas there was no
significantly change in the protein expression levels of c-Fos
(Fig. 4D). These data demonstrate
that svAC3-33 knockdown by RNA interference caused the upregulation
of c-Jun, but not c-Fos.
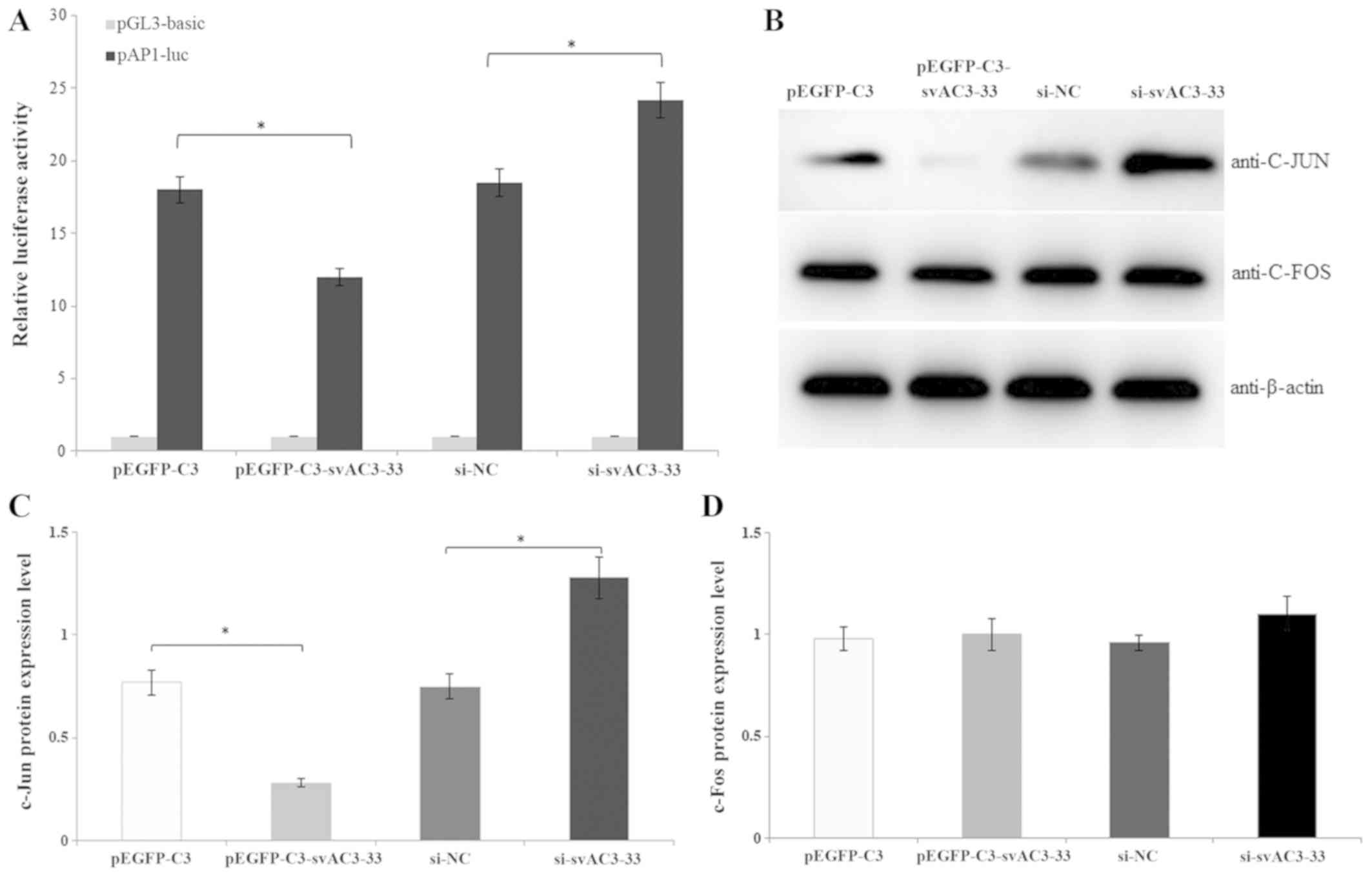 | Figure 4.(A) pEGFP-C3, pEGFP-C3-svAC3-33,
si-NC or si-svAC3-33 was transiently transfected into MCF-7 cells,
and co-transfection with the firefly reporter plasmid and a
Renilla luciferase vector for normalization. Cell lysates
were tested for both firefly and Renilla luciferase
activities with the dual luciferase assay. (B) MCF-7 cells were
transfected with pEGFP-C3, pEGFP-C3-svAC3-33, si-NC and si-svAC3-33
for 48 h. The c-Jun, and c-Fos protein expression levels were
analyzed by western blotting. The protein expression levels of (C)
c-Jun and (D) c-Fos were semi-quantified by western blotting.
*P<0.05. EGFP, enhanced green fluorescent protein; luc,
luciferase; siRNA, small interfering RNA; si-NC, nonsense siRNA;
si-svAC3-33, siRNA targeting svAC3-33; sv, splice variant. |
Discussion
Breast cancer is one of the deadliest cancer types
worldwide. Breast cancer results from cell cycle disorganization
that leads to uncontrolled cellular pro liferation (7). Several studies have reported that in
breast cancer, the expression levels of various growth factors are
altered, which contributes to tumor progression and proliferation.
Studies have shown that AP-1 plays a critical role in mediating the
proliferation of breast cancer cells. It is estimated that 40–60%
of human genes have alternatives vs. Alternative RNA splicing is
able to modulate the functions of genes by increasing the number of
transcripts, and therefore the number of proteins, that come from a
single gene (22–25). The process of alternative splicing is
regulated by a number of splicing factors, whose expression and
activity are tightly regulated during development, cell cycle
progression and cell differentiation. Previous studies have
demonstrated that intron removal from a pre-mRNA by RNA splicing
was initially thought to be controlled by intron splicing signals
(22,26), the regulation of alternative splicing
is closely related to the function of cells. Alternative RNA
splicing is mainly mediated and executed by exonic splicing
enhancers as well as suppressors or silencers (27,28).
p21 is an important member of the family of
cyclin-dependent kinase inhibitors. It is closely related to tumor
inhibition and can coordinate the relationship between the cell
cycle, and DNA replication and repair, thus closely linking tumor
suppression with the cell cycle. Under cellular stress conditions,
p53 positively modulates p21. In turn, p21 exerts an inhibitory
effect on cell-dependent kinases, thereby preventing cell cycle
progression through the detection of DNA damage. It has been
reported that the overexpression of estrogen receptor α in MCF-7
cells not only directly suppresses the activity of the p21
promoter, but also reduces the expression of p21 at both the mRNA
and protein levels (29). It has
been reported that the sustained arrest on the cell cycle caused by
raised p21 expression could only be achieved when p53 was present
in the cell and capable of transcriptionally activating the cyclin
dependent kinase inhibitor 1A or p21 (30). Downregulating certain cyclins and
upregulating of p21 and p27 at the transcript level may promote
cell cycle arrest, and in turn promote growth inhibition (31).
Many studies have shown that p21 is a potent
regulator of cell proliferation in different cell cultures. For
example, in NIH3T3 cells, the hepatitis C virus (HCV) core protein
inhibits the transcription of p21 by the transforming growth factor
(TGF)-β response element on the p21 promoter, indicating that the
HCV core protein inhibits transcription by the inhibition of the
TGF-β pathway, thereby promoting cell cycle progression (32). c-Jun, a member of the transcription
factor AP-1 family, is a key inducer of hepatocyte proliferation,
which regulates liver regeneration by inhibiting p21 activity
(33). These results are consistent
with the present findings that svAC3-33 may contribute to the
upregulation of p21 at the mRNA level in MCF-7 breast cancer
cells.
In this present study, a novel splice variant of
AC3-33 was successfully cloned, designated svAC3-33. Notably,
svAC3-33 and AC3-33 have different structures but have the same
subcellular localization; they are both predominantly expressed in
the cytoplasm (2). In the present
study, the contribution of svAC3-33 to the regulation of MCF-7 cell
proliferation was identified. The present study also examined the
effect of svAC3-33 on the transcriptional activity of AP-1. It was
demonstrated in this present study that increasing the expression
of svAC3-33 caused the inhibition of AP-1. This also promoted the
translocation of AP-1 into the nucleus and the binding of AP-1 with
DNA in MCF-7 cells. A Previous study indicated that the
transcriptional activity of AP-1 is mainly mediated by c-Jun, and
c-Fos (34). Additionally, this
present study identified that svAC3-33 exerted its functional role
through the c-Jun signaling pathway, but not through c-Fos. It
would be valuable to investigate and elucidate the further
mechanisms of action that svAC3-33 functions through.
In conclusion, a novel splice variant of AC3-33 was
successfully cloned, designated svAC3-33, which has a different
structure and expression profile than AC3-33. Notably, svAC3-33 was
predominantly expressed in the cytoplasm. Moreover, svAC3-33
inhibits MCF-7 cell cycle progression and the transcription of the
AP-1 reporter gene by downregulating the expression of c-Jun, but
not c-Fos, inhibiting MCF-7 cell proliferation.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Hebei Province (grant nos. C2017209062 and
H2018209140), and the National Natural Science Foundation of China
(grant no. 81302323), and General Higher Education Young Talents
Program of Hebei Province (grant no. BJ2014027).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LY, FH, YZ and XZ performed the RT-qPCR and western
blot analysis. LM, TA and YC performed the in vitro
experiment. LY, FH and XZ were major contributors in writing the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang X, Ma X, Xue Y, Meng L, He B, He S,
Zhao J, Wang Y and Yang W: Prokaryotic expression and
characterization of human AC3-33 protein. Front Biosci (Elite Ed).
2:1134–1142. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu P, Deng WW, Gao P, Lu Y, Sun B, Li M,
Zhao J, Shi TP and Zhang XJ: Molecular cloning and preliminary
function study of a novel human gene AC3-33 related to suppress
AP-1 activity. Yi Chuan. 30:575–582. 2008.(In Chinese). View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hao D, Gao P, Liu P, Zhao J, Wang Y, Yang
W, Lu Y, Shi T and Zhang X: AC3-33, a novel secretory protein,
inhibits Elk1 transcriptional activity via ERK pathway. Mol Biol
Rep. 38:1375–1382. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang ET, Sandberg R, Luo S, Khrebtukova I,
Zhang L, Mayr C, Kingsmore SF, Schroth GP and Burge CB: Alternative
isoform regulation in human tissue transcriptomes. Nature.
456:470–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Matlin AJ, Clark F and Smith CW:
Understanding alternative splicing: Towards a cellular code. Nat
Rev Mol Cell Bio. 6:386–398. 2005. View
Article : Google Scholar
|
|
6
|
Wang P, Yu P, Gao P, Shi T and Ma D:
Discovery of novel human transcript variants by analysis of
intronic single-block EST with polyadenylation site. BMC Genomics.
10:5182009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Valachovicova T, Slivova V, Bergman H,
Shuherk J and Sliva D: Soy isoflavones suppress invasiveness of
breast cancer cells by the inhibition of NF-kappaB/AP-1-dependent
and -independent pathways. Int J Oncol. 25:1389–1395.
2004.PubMed/NCBI
|
|
8
|
Zhou Y, Yau C, Gray JW, Chew K, Dairkee
SH, Moore DH, Eppenberger U, Eppenberger-Castori S and Benz CC:
Enhanced NF kappaB and AP-1 transcriptional activity associated
with antiestrogen resistant breast cancer. BMC Cancer. 7:592007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Q, Song YJ, Meng LJ, Hu F, Gou LX, Jia
CH, Tang HM, Wang WJ, Li M, Zhang XJ and Jia MC: Role of LM23 in
cell proliferation and apoptosis and its expression during the
testis development. Asian J Androl. 15:539–544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo L, Chen C, Shi M, Wang F, Chen X, Diao
D, Hu M, Yu M, Qian L and Guo N: Stat3-coordinated
Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain
oncostatin M-driven epithelial-mesenchymal transition. Oncogene.
32:5272–5282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Y, Ludes-Meyers J, Zhang Y,
Munoz-Medellin D, Kim HT, Lu C, Ge G, Schiff R, Hilsenbeck SG,
Osborne CK and Brown PH: Inhibition of AP-1 transcription factor
causes blockade of multiple signal transduction pathways and
inhibits breast cancer growth. Oncogene. 21:7680–7689. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang J, Shi T, Ma T, Zhang Y, Ma X, Lu Y,
Song Q, Liu W, Ma D and Qiu X: CCDC134, a novel secretory protein,
inhibits activation of ERK and JNK, but not p38 MAPK. Cell Mol Life
Sci. 65:338–349. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Byun HJ, Hong IK, Kim E, Jin YJ, Jeoung
DI, Hahn JH, Kim YM, Park SH and Lee H: A splice variant of CD99
increases motility and MMP-9 expression of human breast cancer
cells through the AKT-, ERK- and JNK-dependent AP-1 activation
signaling pathways. J Biol Chem. 281:34833–473. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sunters A, Madureira PA, Pomeranz KM,
Aubert M, Brosens JJ, Cook SJ, Burgering BM, Coombes RC and Lam EW:
Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer
cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res.
66:212–220. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Philips A, Chalbos D and Rochefort H:
Estradiol increases and anti-estrogens antagonize the growth
factor-induced activator protein-1 activity in MCF7 breast cancer
cells without affecting c-fosand c-jun synthesis. J Biol Chem.
268:14103–14108. 1993.PubMed/NCBI
|
|
16
|
Hu F, Meng X, Tong Q, Liang L, Xiang R,
Zhu T and Yang S: BMP-6 inhibits cell proliferation by targeting
microRNA-192 in breast cancer. Biochim Biophys Acta.
1832:2379–2390. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zang S, Chen F, Dai J, Guo D, Tse W, Qu X,
Ma D and Ji C: RNAi-mediated knockdown of Notch-1 leads to cell
growth inhibition and enhanced chemosensitivity in human breast
cancer. Oncol Rep. 23:893–899. 2010.PubMed/NCBI
|
|
18
|
Sun B, Nishihira J, Yoshiki T, Kondo M,
Sato Y, Sasaki F and Todo S: Macrophage migration inhibitory factor
promotes tumor invasion and metastasis via the Rho-dependent
pathway. Clin Cancer Res. 11:1050–1058. 2005.PubMed/NCBI
|
|
19
|
Das R, Mahabeleshwar GH and Kundu GC:
Osteopontin induces AP-1-mediated secretion of urokinase-type
plasminogen activator through c-Src-dependent epidermal growth
factor receptor transactivation in breast cancer cells. J Biol
Chem. 279:11051–11064. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang P, Sun B, Hao D, Zhang X, Shi T and
Ma D: Human TMEM174 that is highly expressed in kidney tissue
activates AP-1 and promotes cell proliferation. Biochem Biophys Res
Commun. 394:993–999. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lewis BP, Green RE and Brenner SE:
Evidence for the widespread coupling of alternative splicing and
nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA.
100:189–192. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Modrek B and Lee C: A genomic view of
alternative splicing. Nat Genet. 30:13–19. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Graveley BR: Alternative splicing:
Increasing diversity in the proteomic world. Trends Genet.
17:100–117. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brett D, Hanke J, Lehmann G, Haase S,
Delbrück S, Krueger S, Reich J and Bork P: EST comparison indicates
38% of human mRNAs contain possible alternative splice forms. FEBS
Lett. 474:83–86. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ladd AN and Cooper TA: Finding signals
that regulate alternative splicing in the post-genomic era. Genome
Biol. 3:reviews00082002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zheng ZM: Regulation of alternative RNA
splicing by exon definition and exon sequences in viral and
mammalian gene expression. J Biomed Sci. 11:278–294. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Unoki M, Shen JC, Zheng ZM and Harris CC:
Novel splice variants of ING4 and their possible roles in the
regulation of cell growth and motility. J Biol Chem.
281:34677–34686. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li
YQ, Yan TB, Sun XG, Hu P and Zhang TC: Estrogen receptor a mediates
proliferation of breast cancer MCF-7 cells via a
p21/PCNA/E2F1-dependent pathway. FEBS J. 281:927–942. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Al-Saran N, Subash-Babu P, Al-Nouri DM,
Alfawaz HA and Alshatwi AA: Zinc enhances CDKN2A, pRb1 expression
and regulates functional apoptosis via upregulation of p53 and p21
expression in human breast cancer MCF-7 cell. Environ Toxicol
Pharmacol. 47:19–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang L, Wang G, Yang D, Guo X, Xu Y, Feng
B and Kang J: Euphol arrests breast cancer cells at the G1 phase
through the modulation of cyclin D1, p21 and p27 expression. Mol
Med Rep. 8:1279–1285. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee MN, Jung EY, Kwun HJ, Jun HK, Yu DY,
Choi YH and Jang KL: Hepatitis C virus core protein represses the
p21 promoter through inhibition of a TGF-beta pathway. J Gen Virol.
83:2145–2151. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stepniak E, Ricci R, Eferl R, Sumara G,
Sumara I, Rath M, Hui L and Wagner EF: c-Jun/AP-1 controls liver
regeneration by repressing p53/p21 and p38 MAPK activity. Genes
Dev. 20:2306–2314. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gallo A, Cuozzo C, Esposito I, Maggiolini
M, Bonofiglio D, Vivacqua A, Garramone M, Weiss C, Bohmann D and
Musti AM: Menin uncouples Elk-1, JunD and c-Jun phosphorylation
from MAP kinase activation. Oncogene. 21:6434–6445. 2002.
View Article : Google Scholar : PubMed/NCBI
|