Introduction
Stem cells are undifferentiated cells defined by
their capacity for self-renewal, in addition to their ability to
differentiate into other cell types (1,2). Dental
pulp stem cells (DPSCs) are a type of mesenchymal stem cell (MSC)
in the cell-rich zone and core of the pulp tissue of teeth
(3). DPSC possess strong
self-renewal capabilities and the potential for multi-directional
differentiation, rendering them with a high therapeutic potential
for repairing damaged and/or defective tissues (4,5). It has
been documented that DPSCs are able to differentiate into multiple
different cell lines, including osteoblasts, odontoblasts,
chondrocytes, adipocytes, hair follicle cells, corneal endothelium
cells, neuronal cells, melanocytes and endothelial cells (6–9).
MicroRNAs (miRs/miRNAs) are a group of non-coding
RNA molecules, with an approximate length of 22–25 nucleotides,
that are able to post-transcriptionally regulate the expression of
their target genes, resulting in the degradation of the target mRNA
(10). An increasing number of
reports have indicated that miRNAs serve an essential function in
numerous molecular and biological processes, including apoptosis,
cell proliferation, migration and necrocytosis (11,12).
Frequently downregulated miR-224-5p (miR-224) expression in human
tumor types has been demonstrated, and may possess fundamental
functions in various cellular processes, particularly in modulating
proliferation and differentiation (13–16). For
instance, Li et al (17)
suggested that miR-224, along with miR-21, may facilitate the
osteogenic differentiation of periodontal ligament cells by
targeting periodontal ligament associated protein-1 (17). miR-224 was also able to promote the
osteogenic differentiation of human bone marrow MSCs, by targeting
the enhancer of zeste 2 polycomb repressive complex 2
subunit/Wnt/β-Catenin cascade (18).
However, further studies are required to examine the underlying
mechanism of miR-224 in regulating the function of human DPSCs.
Rac family small GTPase 1 (Rac1), accompanied by Ras
homolog family member A and cell division cycle 42, belongs to the
GTPase family, which controls the actin cytoskeleton accumulation
and organization in mammalian cells and serves a critical signaling
function in modulating various cellular processes (19,20),
including apoptosis, reactive oxygen species production, membrane
ruffling, lamellipodia formation, the activity of transcriptional
factors, cell cycle control and the integrity of cell-cell
adhesions (21–26). Embade et al (27) reported the first evidence indicating
that Rac1 induced apoptosis by a complex mechanism, involved the
generation of ceramides and induced the de novo synthesis of
Fas ligand (FasL). Another study also suggested that Rac1 mediated
apoptosis via mitogen-activated protein kinase 8 (JNK) and served a
key function in pro-apoptotic pathways in intestinal epithelial
cells (28). The aim of the present
study was to probe the function of miRs in the character of DPSCs,
which may provide a new mechanism for regulating DPSC
viability.
Materials and methods
Cell lines
DPSCs and dental periodontal ligament cells (DPLCs)
were isolated from third molars or premolars extracted from at
least 4 adults under the approved guidelines and protocol
(ethically approved by the Ethics Committee of Shandong University,
Shandong, China) with written informed consent obtained from all
patients. The isolation and cultivation of human DPSCs and DPLCs
were performed according to a previously reported method (9,8).
Briefly, tooth surfaces were cleaned and cut around the
cementum-enamel junction by using sterilized dental fissure burs to
reveal the pulp chamber. The pulp tissue was gently separated from
the crown and root and then digested in a solution of 3 mg/ml
collagenase type I (Worthington Biochemical Corporation, Lakewood,
NJ, USA) and 4 mg/ml dispase (Boehringer Mannheim; Roche Applied
Science, Mannheim, Germany) for 1 h at 37°C. Single-cell
suspensions were obtained by passing the cells through a 70-µm
strainer (Falcon®; Corning, Inc.). Single-cell
suspensions (0.01–1×105/well) of dental pulp and dental
ligament cells were seeded into six-well plates (Costar; Thermo
Fisher Scientific, Inc.) cultured in α-Modified Essential Medium
(α-MEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 15% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc.), 2 mM L-glutamine (Thermo Fisher Scientific,
Inc.), 100 units/ml penicillin (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and 100 mg/ml streptomycin (Sigma-Aldrich;
Merck KGaA) at 37°C under 5% CO2 in air. All methods
were approved by the Research Medical Ethics Committee of Shandong
University (Shandong, China) and were performed in accordance with
the approved guidelines.
For the induction of odentogenic differentiation of
DPSCs, cells were grown in an osteoblast differentiating medium,
consisting of α-MEM supplemented with 10% FBS, 50 µg/ml ascorbic
acid and 10−8 M dexamethasone (Sigma-Aldrich; Merck
KGaA).
miRNA array
Total RNA was extracted using the phenol-chloroform
method using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.).
The quality of the RNA was assessed using capillary electrophoresis
(Agilent Technologies, Inc., Santa Clara, CA, USA). Libraries for
small RNA sequencing were prepared using the NEBNext Multiplex
Small RNA Library Prep Set for Illumina (New England BioLabs, Inc.,
Ipswich, MA, USA) according to the manufacturer's protocol. The
libraries were quantified using the Agilent Bioanalyzer 2100 system
with DNA high-sensitivity chips. The raw sequence files were
subjected to quality control analysis with the Fast QC quality
control tool. To avoid low-quality data, adaptors were removed by
Cutadapt (version 1.2.1; PyPI) and lower-quality sequences were
trimmed. The clean reads were screened at a length of 21–22
nucleotides as miRNA and were located to the reference sequence
using Bowtie software (version 2; CGE Risk Management Solutions
B.V., Leidschendam, The Netherlands). The functions of novel miRNAs
were analyzed using miRDeep2 software (version 2.0.0.8; Max
Delbrück Center). Differential expression sequencing was used to
calculate differential expression levels and to evaluate the
statistical significance of detected alterations between the
control and case samples.
Western blot analysis (WB)
DPSCs and DPLCs were lysed in lysis buffer (30 mM
Tris-HCl, 150 mM NaCl, 1% NP-40 and 0.1% SDS; pH 7.4) supplemented
with a protease inhibitor cocktail (Roche Applied Science). The BCA
Protein Quantitation kit (GenScript) was used to determine protein
concentration. Proteins (20 µg) were separated using 10% SDS-PAGE
and blotted electrophoretically onto polyvinylidene difluoride
(Immobilon) membranes of 0.45 µm pore size. Membranes were blocked
with 5% bovine serum albumin (BSA) in phosphate-buffered saline
containing 0.1% Tween-20 (PBST) for 1 h at room temperature,
followed by incubation with the following primary antibodies
overnight at 4°C: Anti-Rac1 antibody (1:4,000; cat no. ab33186;
Abcam, Cambridge, UK), anti-JNK1 antibody (1:200; cat no. ab201624;
Abcam), anti-caspase-3 antibody (1:1,000; cat no. ab4051; Abcam),
anti-caspase-9 antibody (1:1,000; cat no. ab25758; Abcam),
anti-FasL antibody (1:2,000; cat no. ab15285; Abcam), anti-RUNX
family transcription factor 2 (Runx2) antibody (1:1,500; cat no.
ab23980; Abcam), anti-alkaline phosphatase (ALP) antibody (1:1,000;
cat no. ab67228; Abcam), anti-actin antibody (1:5,000; cat no.
ab1801; Abcam) and anti-GAPDH antibody (1:5,000; cat no. ab9485;
Abcam). Horseradish peroxidase-conjugated goat anti-rabbit
immunoglobulin G secondary antibodies (1:5,000; cat. no. AP132;
Abcam) were incubated with the membranes for 1 h at room
temperature in PBST containing 5% BSA, followed by chemiluminescent
detection. A C-DiGit Blot Scanner and Super Signal West Femto
Maximum Sensitivity Substrate kit (Thermo Fisher Scientific, Inc.)
were used to detect bound antibodies (PhotoShop CS6; Adobe Systems,
Inc.).
RNA extraction and quantitative
polymerase chain reaction (qPCR)
Total RNA was extracted from DPSCs and DPLCs using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol (Invitrogen; Thermo Fisher
Scientific, Inc.). mRNA levels were determined using the
Light-Cycler 480 Real Time PCR system (Roche Applied Science), with
GAPDH used as the internal control. Quantitative PCR was performed
in 20 µl reaction volumes containing SYBR-Green PCR Master Mix
(Invitrogen; Thermo Fisher Scientific, Inc.) for 10 min at 95°C;
95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec, for a total
of 40 cycles. Transcript levels were determined relative to the
endogenous reference gene (GAPDH), and normalized to the untreated
sample (using the 2−ΔΔCq method). The expression of mRNA
was first normalized to GAPDH, and the expression levels of miR-224
were first normalized to the endogenous control U6 small nuclear
RNA. Then, the gene expression levels of genes in the treated group
were compared with the level of gene expression in an untreated
sample (29).
Sequences of the primers used for qPCR detection
were as follows: miR-224 forward, 5′-AGCCCCATCATCTACAGUG-3′ and
reverse, TCACAGTCACTAGGGCACC-3′; Rac1 forward,
5′-AAAATGTCCGTGCAAAGTGGT-3′ and reverse,
5′-CTCGATCGTGTCTTTATCATCCC-3′; insulin like growth factor binding
protein 5 (IGFBP-5) forward, 5′-TTGCCTCAACGAAAAGAGC-3′ and reverse,
5′-AGAATCCTTTGCGGTCACA-3′; JunB proto-oncogene, AP-1 transcription
factor subunit (JunB) forward, 5′-CCAGTCCTTCCACCTCGACGTTTACAA-3′
and reverse, 5′-GACTAAGTGCGTGTTTCTTTTCCACAG-3; nuclear receptor
related-1 protein (NURR1) forward, 5′-GGCATGGTGAAGGAAGTTGT-3′ and
reverse, 5′-CAGGGAAGTGAGGAGATTGG-3′; GAPDH forward,
5′-GGAAGGTGAAGGTCGGAGTCA-3′ and reverse,
5′-GTCATTGATGGCAACAATATCCACT-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
miR-224 mimic/inhibitor
preparation
Mimic/inhibitor of miR-224 and a negative control
(NC) were acquired from Guangzhou RiboBio Co., Ltd. (Guangzhou,
China). miR-224 mimic/inhibitor and NC mimic/inhibitor were
supplemented with 0.9% NaCl to a terminal level of 10 mg/ml for
further use. Transfection of mimic/inhibitor was performed for 36 h
at 37°C.
RNA interference
Short hairpin RNAs (shRNAs) inhibiting Rac1 were
generated via vector pLKO.1 puro and oligonucleotides targeting the
following Rac1 sequence:
5′-GATCCGGAAGGAGATTGGTGCTGTAAAACTCGAGAAAATGTCGTGGTTAGAGGAATTTTTG-3′.
DPSCs and DPLCs cells were transiently transfected with
pLKO.1-shRNA-Rac1 or pLKO.1 empty vector with Lipofectamine
2000® (Thermo Fisher Scientific, Inc.). After 48 h, the
cells were washed with phosphate-buffered saline and harvested in
lysis buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.5
mM dithiothreitol, 0.5% NP-40].
MTT assay
An MTT assay was conducted to evaluate cell
proliferation. Briefly, DPSCs and DPLCs were treated with 20 µl MTT
(0.5 mg/ml), and the supernatant was discarded. Dimethyl sulfoxide
(150 µl) was then added to each well, with rotation for 10 min, to
dissolve the formazan dye. An Infinite M200 microplate reader
(Tecan Group, Ltd., Männedorf, Switzerland) was then used to
measure the absorbance at 490 nm.
Colony formation assay
A colony formation assay was performed by seeding
1,000 of each DPSCs or DPLCs in 10-cm2 flasks and
culturing for 4 days. Cells were then washed with PBS and stained
with 0.1% toluidine blue contained in 1% PFA overnight at 4°C. On
the following day, cells were washed to remove the excess dye.
Stained clusters containing >50 cells were counted as positive
colonies using an inverted light microscope (Leica MICROSYSTEMS;
magnification, ×40).
Terminal
deoxynucleotidyl-transferase-mediated dUTP nick end labeling
(TUNEL) staining
TUNEL staining was used to assess the degree of
apoptosis in DPSCs and DPLCs using a TUNEL fluorescence kit (Roche
Applied Science) according to the manufacturer's protocol. Briefly,
cells were treated with 100 µl TdT reaction buffer (Component A)
and 50 µl TdT reaction cocktail. Cells were then stained with 100
µl Andy Fluor™ 647 (1:100 in staining buffer) for 30 min.
Subsequent to TUNEL staining, the cells were stained with DAPI
solution for 15 min at 4°C (1:5,000; Beyotime Institute of
Biotechnology, Haimen, China) to stain the nuclear DNA in antifade
mouting medium. Fluorescence staining was viewed using a Laser
Scanning Confocal Microscope (SP8; Leica Microsystems, Inc.,
Buffalo Grove, IL, USA). The apoptotic rate was calculated as
TUNEL-positive cells in 10 fields.
Annexin V-fluorescein isothiocyanate
(FITC)/propidium iodide (PI) flow cytometry (FC)
Cell death was evaluated using an Annexin V-FITC/PI
apoptosis detection kit (cat. no. ab14085; Abcam), and the
experiment was performed according to the manufacturer's protocol.
Briefly, following transfection, cells were resuspended in 20 µl
binding buffer and then incubated for 20 min with 5 µl PI and 10 µl
Annexin V-FITC in a dark room at 25°C. Cell death was evaluated
using BD FACSuite™ software (BD Biosciences).
Dual-luciferase reporter assay
(DLRA)
The 3′-untranslated region (UTR) of the Rac1 gene
underwent amplification prior to fusion with the GV126 luciferase
gene of pGL3 (Promega Corporation). The binding site of the Rac1
gene and miR-224 were mutated via site-directed mutagenesis, which
served as a control. Thymidine kinase promoter (Takara Bio, Inc.,
Otsu, Japan; pRL-TK vectors) and plasmids containing Renilla
luciferase were applied to adjust for transfection efficiency using
Lipofectamine 2000® (Thermo Fisher Scientific, Inc.).
293T cells (ATCC®; CRL-3216™) were co-transfected with
miR-224-mimic and NC-mimic with luciferase reporter vectors, and
the luciferase assay was conducted. Approximately 48 h after
transfection, a dual-luciferase reporter gene assay kit (Promega
Corporation) was used for detection before calculating the ratio of
the firefly luciferase activity to Renilla luciferase
activity.
TargetScan prediction
The prediction algorithm TargetScan was used to
nominate targets of miR-224. Using the TargetScan Database
predictions (http://www.targetscan.org) are ranked based on the
predicted efficacy of targeting as calculated using cumulative
weighted context++ scores of the sites (15). As an option, predictions are also
ranked by their probability of conserved targeting (16).
Statistical analysis
All data are presented as the mean ± standard
deviation of three separate experiments. Data were analyzed using a
Student's t-test for comparison between two groups or one way ANOVA
with Tukey's post hoc test for comparison among multiple groups as
appropriate. P<0.05 was considered to indicate a statistically
significant difference. Statistical analyses were performed using
SPSS Statistics 17.0 (Version X; SPSS, Inc., Chicago, IL, USA).
Results
miR-224 is upregulated in DPSCs
compared with DPLCs
As the high expression of Runx2 protein is a marker
of DPLC (30), and DPSC typically
displays a high expression of ALP protein subsequent to culturing
(8), the present study examined the
expression of these two proteins for further confirmation. As the
results revealed, in the DPLC group, Runx2 protein expression was
clearly higher compared with those in the DPSC group, while the
DPSC group exhibited a higher ALP expression compared with the DPLC
group (Fig. 1A). To examine the
odentogenic differentiation property of DPSCs, DPSCs were grown in
an osteoblast differentiating medium, consisting of α-MEM
supplemented with 10% FBS, 50 µg/ml ascorbic acid and
10−8 M dexamethasone. The present study then examined
the mRNA levels of IGFBP-5, JunB and NURR1, which are three
reported markers for differentiation. The mRNA levels of these
three genes were significantly increased at day 10 with induction,
compared with the non-induced group (P<0.05; Fig. 1B).
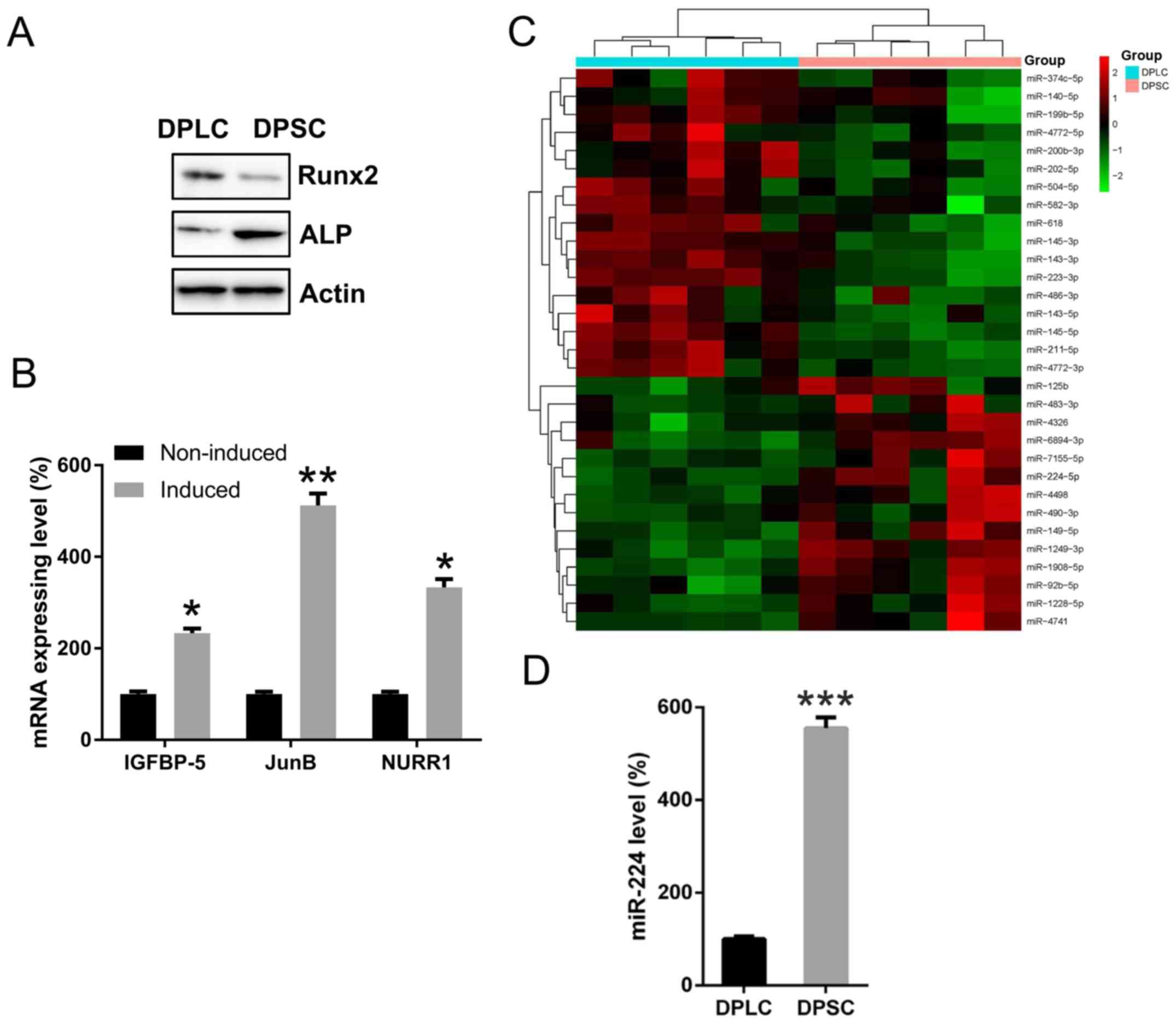 | Figure 1.Expression of Runx2, ALP, IGFBP-5,
JunB, NURR1 and miR-224 in DPLC and DPSC. (A) Western blot analysis
was performed to assess the protein expression of Runx2 and ALP in
DPLC and DPSC to confirm the successful isolation of the two cell
lines. (B) DPSCs were grown in an osteoblast differentiating
medium. A q-PCR was performed to detect the mRNA levels of IGFBP-5,
JunB and NURR1. (C) A miRNA microarray of deregulated miRNAs in the
DPLC and DPSC groups. (D) qPCR detection confirmed the expression
of miR-224 in DPLC and DPSC. Mean ± standard deviation of the
results of three independent experiments was used to describe the
data. n=3. *P<0.05, **P<0.01 and ***P<0.001, vs. the
non-induced or DPLC group. miR/miRNA, microRNA; miR-224,
miR-224-5p; DPSC, Dental pulp stem cells; DPLC, dental periodontal
ligament cells; qPCR, quantitative polymerase chain reaction;
Runx2, RUNX family transcription factor 2; ALP, alkaline
phosphatase; IGFBP-5, insulin like growth factor binding protein 5;
JunB, JunB proto-oncogene, AP-1 transcription factor subunit;
NURR1, nuclear receptor related-1 protein. |
The miRNA microarray analysis revealed that miR-224
was substantially upregulated in DPSC, compared with DPLC (Fig. 1C). To confirm this data, qPCR
analysis was performed to determine miR-224 expression in DPLC and
DPSC. It indicated that the expression levels of miR-224 were
significantly upregulated in DPSC compared with DPLC (P<0.001;
Fig. 1D), suggesting that miR-224
may serve a functional role in DPSC properties.
miR-224 is essential for maintaining
DPSC viability compared with DPLC viability
To elucidate the effect of miR-224 on the properties
of DPLC and DPSC, the cells were transfected with miR-224 inhibitor
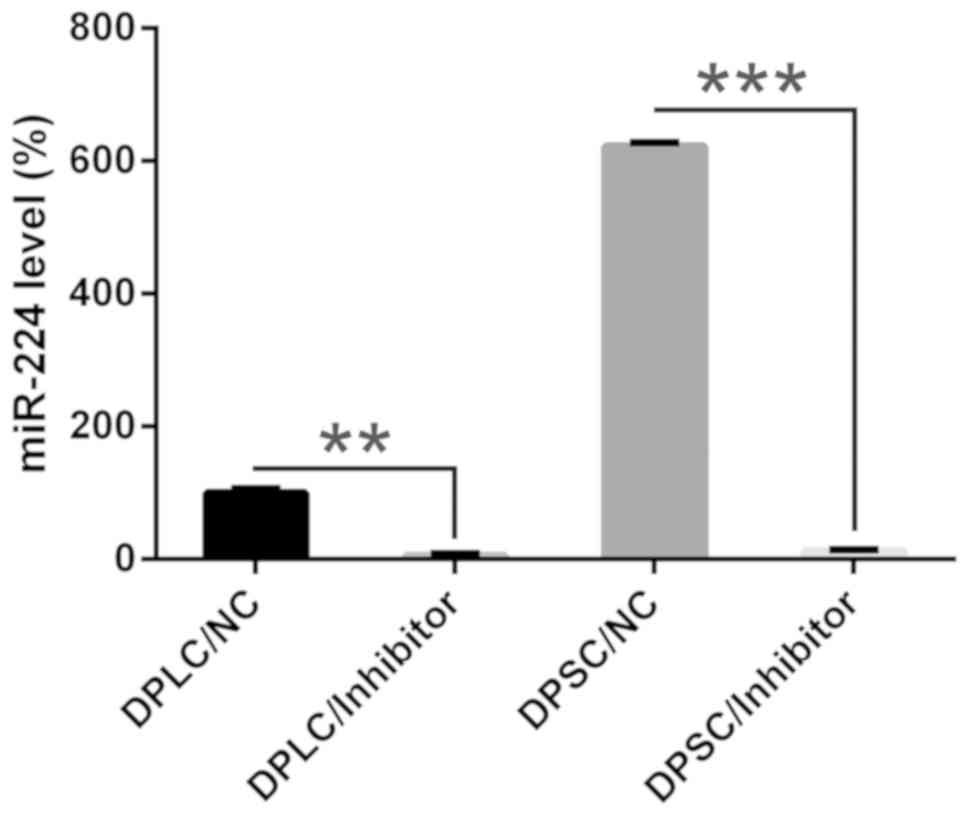
to repress miR expression. The qPCR data indicated that using a
miR-224 inhibitor resulted in significantly decreased miR-224
expression levels in DPLC and DPSC compared with the negative
control (P<0.01; Fig. 2). An MTT
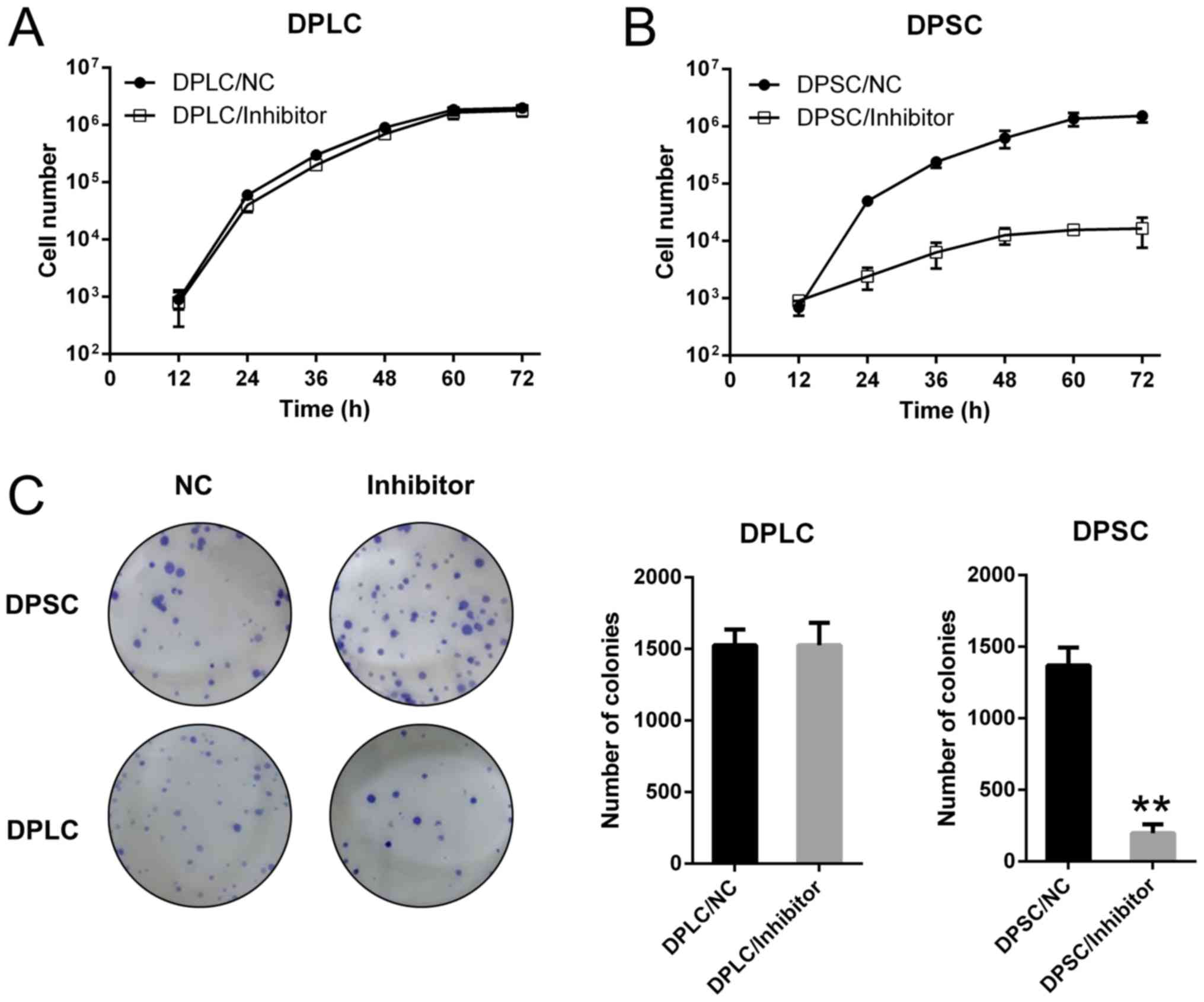
assay was subsequently performed to detect the proliferative rate
of DPLC and DPSC during 72 h post-transfection. For DPLC, the data
indicated that miR-224 depletion did not exert any effect (Fig. 3A), whereas miR-224 inhibition
significantly suppressed the cell proliferation of DPSC during 72 h
post transfection (P=0.0236; Fig.
3B). A colony formation assay was also performed to confirm the
effect of miR-224 on cell viability, and the number of colonies of
DPSC transfected with miR-224 inhibitor were reduced compared with
DPLC transfected with miR-224 inhibitor, and significantly reduced
compared with DPSC transfected with NC inhibitor (P<0.01;
Fig. 3C). These data suggested that
miR-224 served a particularly critical function in sustaining DPSC
viability compared with DPLC viability.
miR-224 inhibition induces the
apoptosis of DPSC
It was hypothesized that miR-224 silencing may
induce the apoptosis of DPSC, therefore causing attenuated DPSC
viability. Thus, TUNEL staining and Annexin V-FITC/PI FC was
conducted in two cell lines following the transfection of miR-224
inhibitor and NC inhibitor. DPSCs transfected with miR-224
inhibitor displayed significantly elevated numbers of cells with
positive TUNEL staining at 36 h post transfection, compared with
the DPSC and DPLC transfected with NC inhibitor (P<0.01;
Fig. 4A). Furthermore, Annexin
V-FITC/PI FC demonstrated that silencing miR-224 expression
significantly increased the percentage of apoptotic DPSC compared
with DPLC (P<0.01; Fig. 4B).
These results suggested that miR-224 depletion resulted in an
increased apoptotic percentage of DPSC.
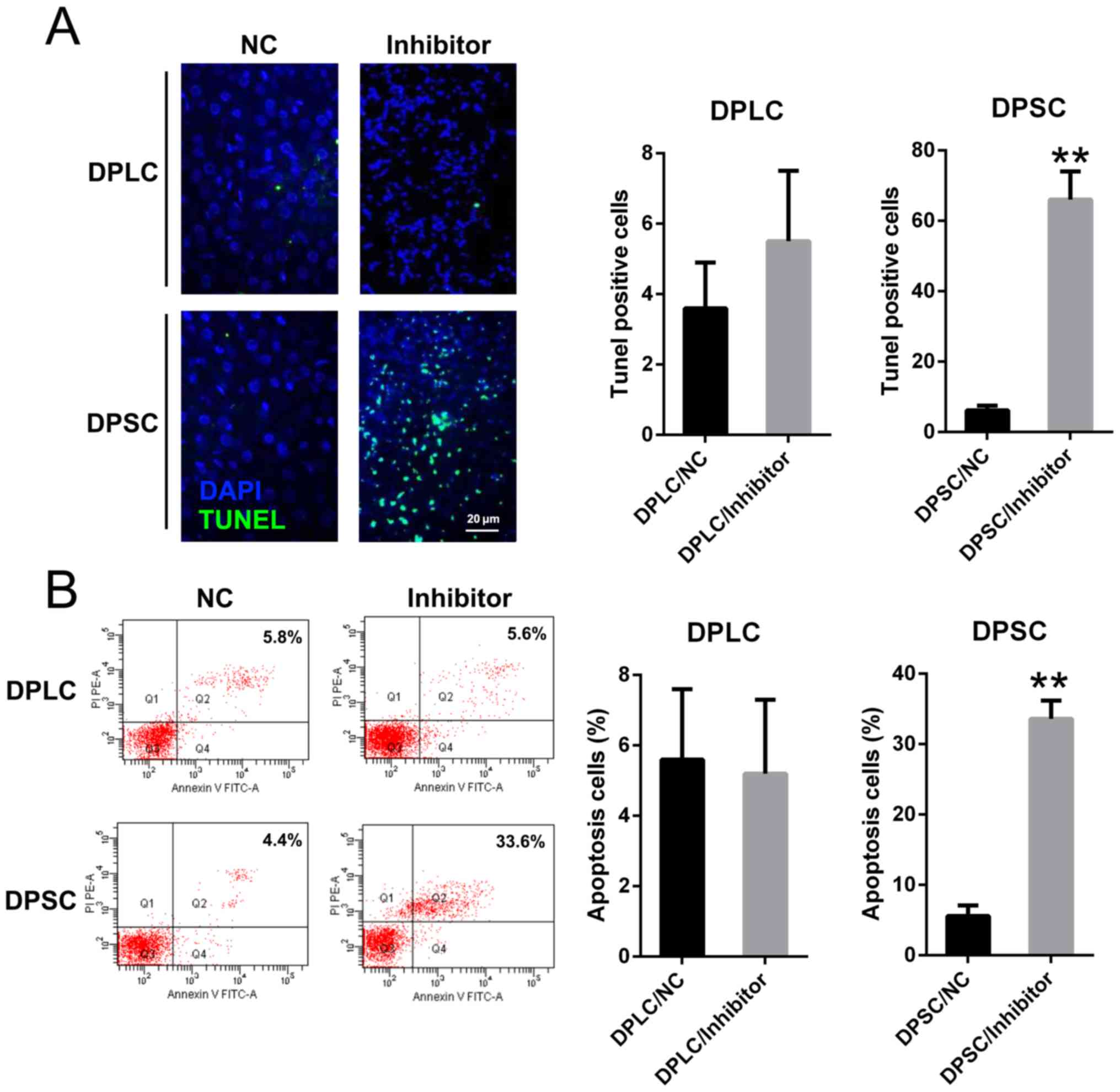 | Figure 4.miR-224 inhibition enhanced the
apoptosis of DPSC cells. (A) TUNEL staining was utilized in each
group of DPLC and DPSC with miR-224 silencing or NC (magnification,
×400). The apoptotic number of positive stained cells is displayed
in the right panel. (B) Number of apoptotic cells, with early
apoptotic cells presented in the right quadrant of each plot.
Analysis of the apoptotic rate of the cells in all groups is
displayed in the right panel. Mean ± standard deviation of the
results of three independent experiments were used to describe the
data. n=3. **P<0.01 vs. the group. miRs, microRNAs; miR-224,
miR-224-5p; DPSC, Dental pulp stem cells; DPLC, dental periodontal
ligament cells; TUNEL, terminal
deoxynucleotidyl-transferase-mediated dUTP nick end labeling; NC,
negative control; FITC, fluorescein isothiocyanate; PI, propidium
idodide. |
miR-224 targets Rac1
It has been reported that the Rac1 sensor serves an
important function in modulating apoptotic cell signaling. Taking
into consideration the key function of Rac1 protein in apoptosis,
its expression was measured using WB and qPCR. The present study
indicated the downregulation of Rac1 in DPSC compared with DPLC
(Fig. 5A). In addition, Rac1 mRNA
expression levels were significantly reduced in DPSC compared with
DPLC (P<0.001; Fig. 5B).
Furthermore, bioinformatics analysis indicated that miR-224 may
target the 3′-UTR of the Rac1 gene (Fig.
5C). Transfection with a miR-224 mimic significantly inhibited
luciferase function compared with the negative control (P<0.01),
which was fused with the Rac1 3′-UTR by 70% compared with the other
control groups (Fig. 5D).
Furthermore, the protein and mRNA expression levels of Rac1 were
upregulated with miR-224 inhibitor transfection in DPSC cells,
whereas Rac1 expression levels were not affected in transfected
DPLC (Fig. 5E and F). These results
suggest that miR-224 may target the 3′-UTR of Rac1.
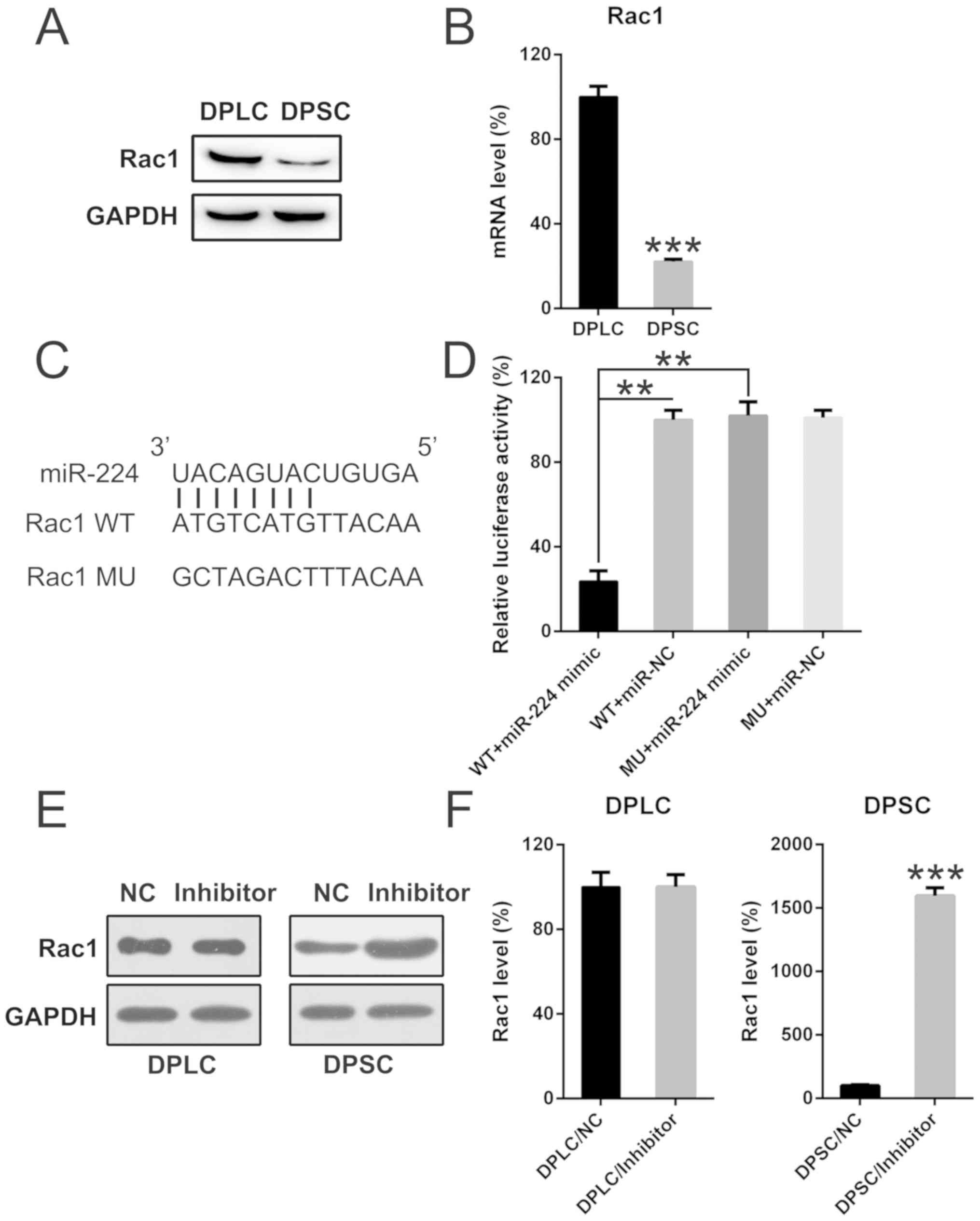 | Figure 5.miR-224 targets the 3′-UTR of the
Rac1 gene in DPSCs. (A) Western blot analysis and (B) quantitative
polymerase chain reaction analysis determined the protein and mRNA
expression levels of Rac1 in DPLCs and DPSCs. ***P<0.001 vs. the
DPLC group (C) Bioinformatics analysis of miR-224 and the 3′-UTR of
the Rac1 gene. (D) A dual-luciferase reporter assay was performed
following co-transfection with a luciferase reporter containing
either a WT or MU 3′-UTR from Rac1, and a miR-224-mimic into 293T
cells. The effect of miR-224-mimic transfection on the luciferase
activities of the WT and MU Rac1 reporter constructs was
determined. **P<0.01 with comparisons shown by lines. Rac1
levels were determined at the (E) protein and (F) mRNA levels in
DPLC and DPSC following transfection with an miR-224 inhibitor or
an NC inhibitor. ***P<0.001 vs. the DPSC/NC group. Mean ±
standard deviation of the results of three independent experiments
were used to describe the data. n=3. Rac1, Rac family small GTPase
1; WT, wild-type; MU, mutant; miRs, microRNAs; miR-224, miR-224-5p;
DPSC, Dental pulp stem cells; DPLC, dental periodontal ligament
cells; UTR, untranslated region; NC, negative control. |
Silencing of Rac1 suppresses the
pro-apoptotic effect of miR-224 transfection in DPSC
To determine the effect of the miR-224 inhibitor and
shRNA-Rac1 on the expression of miR-224 and Rac1 in DPSC, the
present study detected the levels of miR-224 and Rac1 mRNA using
qPCR analysis. The results revealed that miR-224 and Rac1 mRNA
expression levels were significantly inhibited by transfections
with miR-224 inhibitor and shRNA-Rac1, as compared with their
respective negative control groups (P<0.01; Fig. 6A and B). To determine whether Rac1
silencing may suppress the effect of miR-224 on apoptosis in DPSC,
Rac1 was silenced in DPSCs transfected with miR-224 inhibitor or NC
inhibitor. WB analysis was utilized in order to verify the changes
in Rac1 expression levels (Fig. 6C).
It was further indicated that the miR-224 inhibitor upregulated the
expression of apoptotic factors, including caspase-3, caspase-9 and
particularly FasL, whereas the reduced expression of Rac1 decreased
the pro-apoptotic effect of miR-224 inhibition in DPSC, as
indicated by the reduced protein expression of caspase-3, caspase-9
and FasL. It was reported that Rac1 promoted cell apoptosis through
the JNK pathway; therefore, JNK1 expression was also detected in
each group (31). WB data indicated
that miR-224 inhibition or Rac1 upregulation resulted in increased
JNK1 expression, while Rac1 downregulation decreased JNK1
expression. Annexin V-FITC/PI FC was performed to evaluate the
apoptotic rate in DPSC with different treatments. Inhibition of
miR-224 in DPSC resulted in a significant elevation in apoptotic
cell proportion compared with the negative control (P<0.01),
whereas Rac1 silencing significantly reduced the apoptosis of DPSC
to a normal level compared with the miR-224 inhibitor treated group
(P<0.01; Fig. 6D).
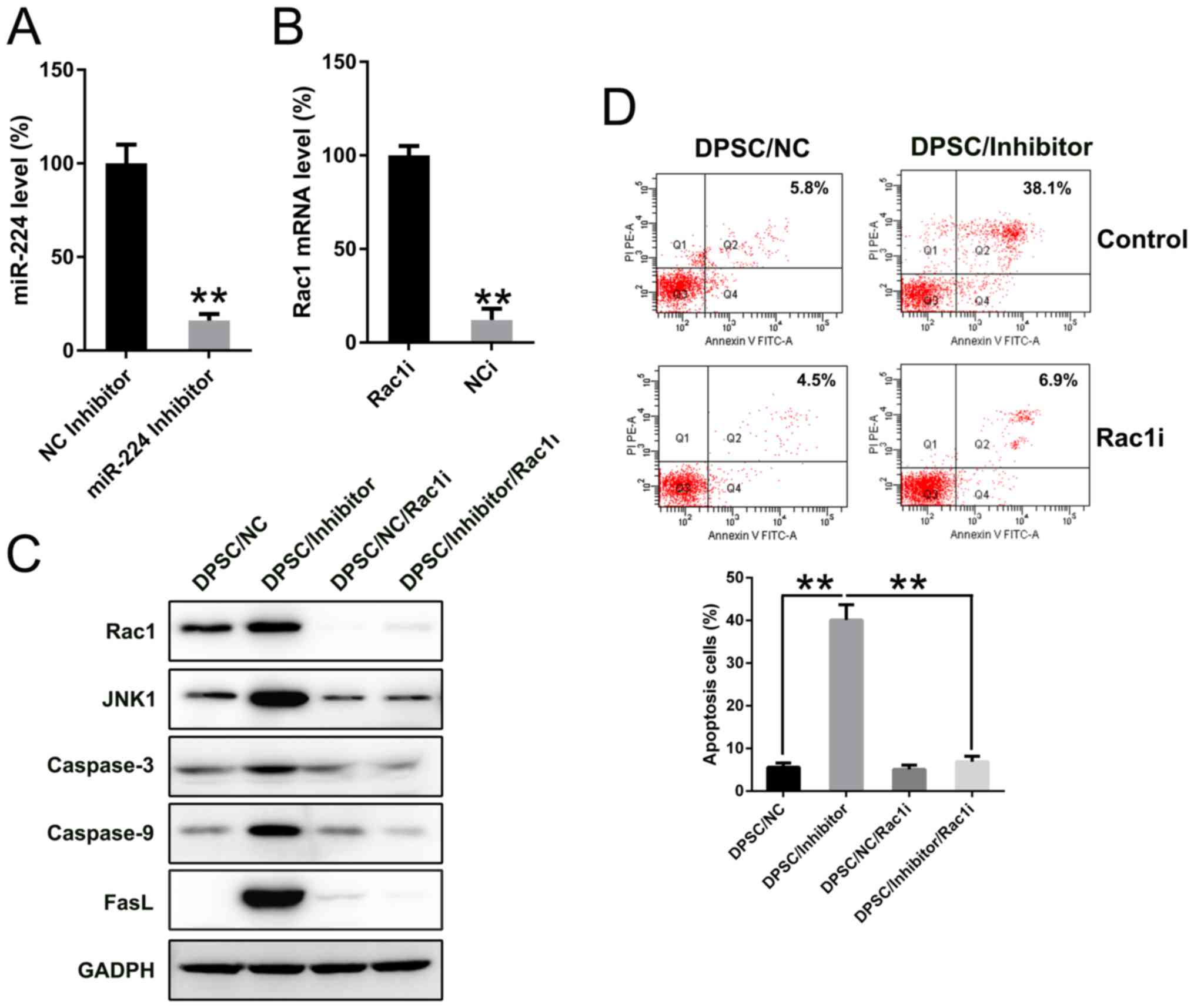 | Figure 6.Rac1 silencing restored miR-224
inhibition impaired cell viability. DPSCs were transfected with
either miR-224 inhibitor or shRNA-Rac1, or their controls. mRNA
expression levels of (A) miR-224 and (B) Rac1 in each group were
detected. **P<0.01 vs. the NC inhibitor group or the Rac1
inhibitor group, respectively. DPSCs were co-transfected with
miR-224 inhibitor and shRNA-Rac1, or their controls. (C) Protein
expression of Rac1, JNK1, caspase-3, caspase-9 and FasL in each
group was detected. (D) Number of apoptotic cells. Early apoptotic
cells are presented in the right quadrant of each plot. Analysis of
the apoptotic rate of DPSCs in all groups is displayed in the lower
panel. **P<0.01 with comparisons shown by lines. Mean ± standard
deviation of the results of three independent experiments were used
to describe the data. n=3. Rac1, Rac family small GTPase 1; WT,
wild-type; MU, mutant; miRs, microRNAs; miR-224, miR-224-5p; DPSC,
Dental pulp stem cells; JNK1, mitogen-activated protein kinase 8;
FasL, Fas ligand; shRNA, short hairpin RNA; FITC, fluorescein
isothiocyanate; PI, propidium iodide. |
Discussion
The aim of the present study was to examine the
effect of miR-224 in human DPSCs, particularly its effect on cell
proliferation and apoptosis. The expression of miR-224 in DPSCs was
higher compared with that in DPLCs. In addition, miR-224 silencing
in DPSCs, in contrast with DPLCs, caused impaired cell viability
and promoted cell apoptosis. In silico prediction and DLRA
data suggested that miR-224 targeted the 3′-UTR of the Rac1 gene,
and the transfection of miR-224 mimic in DPSCs attenuated Rac1
expression at the protein and mRNA expression levels. The data also
indicated that Rac1 silencing restored the expression of
pro-apoptotic factors, including caspase-3 and FasL, and increased
the apoptotic cell number, which were induced by miR-224
inhibition, thus serving as an essential target for miR-224 in
maintaining cell viability.
The properties of human stem cells are regulated by
intracellular or extracellular mechanical and molecular signals,
and are a promising target for stem cell-based therapies for
various types of human disease (32). However, the regulatory mechanisms of
miRs on DPSC viability remain unclear. The results of the present
study suggested that the high expression of miR-224 in DPSC
promoted cell viability, as indicated by the increased number of
apoptotic cells in the group transfected with an miR-224 inhibitor.
However, a number of previous studies have reported contradictory
results, suggesting that miR-224 overexpression induces apoptosis
in various cell lines. For instance, a retrospective report
indicated that in neuroendocrine neoplasms, miR-224 agomir may
promote apoptosis and suppress the proliferation and invasion of
BON-1 cells (33). In
SW480/adrenomedullin (ADM) cells, the transfection of an miR-224
inhibitor substantially increased glycogen synthase kinase-3β
expression and decreased β-catenin and Survivin expression,
resulting in enhanced cell apoptosis and suppressed ADM resistance
(34). Nevertheless, additional
studies have reported contradictory evidence and suggest that
miR-224 inhibits the apoptosis of cells. Systemic lupus
erythematosus (SLE) is a systemic autoimmune disease with abnormal
T cell immune responses (35,36).
Chang et al (37) reported
that miR-145 and miR-224 were expressed aberrantly in SLE T cells
that modulated the protein expression of their target genes, signal
transducer and activator of transcription 1 (STAT1) and apoptosis
inhibitor 5 (API5), respectively. These miRNA aberrations
accelerated T cell activation-induced cell death by suppressing
API5 expression and were associated with lupus nephritis by
enhancing STAT1 expression (37).
Another study demonstrated that amiodarone enhanced reactive oxygen
species production and increased cell apoptosis, while it reduced
DNA damage. Meanwhile, amiodarone suppressed miR-224 and increased
its target cyclooxygenase-2 expression (37). For cervical cancer, Cong et al
(38) indicated that the Notch1 gene
is able to increase cervical cancer cell proliferation by
regulating the miR-224/leucine rich repeats and immunoglobulin like
domains 2 signal pathway. These controversial experimental evidence
and conclusions may be attributed to the different cell lines
applied in the studies, therefore indicating that miR-224 possesses
a tissue- or cell-specific function in mediating apoptosis.
Previous evidence has indicated that Rac1 is able to
regulate injury, damage, apoptosis and necrosis in various cell
lines (28,39,40). Jin
et al (28) revealed that
Rac1 is activated during tumor necrosis factor-α (TNF-α)-induced
apoptosis in intestinal epithelial cells, and that the inhibition
of Rac1, prior to the administration of apoptotic stimuli,
substantially prevents apoptosis via the TNF-α-induced activation
of JNK1/2. Another study suggested that Rac 1 and Rac 2 stimulation
mediated apoptosis through the apoptotic extrinsic pathway
(Fas/FasL) (41). In the present
study, inhibition of miR-224 increased the apoptotic level of DPSC,
in addition to the expression levels of JNK1 and FasL, while Rac1
silencing restored the apoptosis and expression of these two
proteins to normal levels. This suggests that the JNK pathway and
Fas/FasL pathway participated in the apoptosis triggered by miR-224
inhibition.
In conclusion, these data indicate that miR-224
participates in the regulation of the cell viability of DPSC by
directly repressing Rac1 levels and also indirectly, by inhibiting
the expression of JNK and FasL, which may function as pro-apoptotic
factors. The present study, therefore, suggests that miR-224 is a
key modulator in maintaining the DPSC phenotype.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Shandong Province (grant no.
ZR2014HZ001).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JG conceived and designed the study. WQ analyzed,
interpreted data and drafted the manuscript. WQ, DL, QS, HuW and
HaW performed the experiments. All authors participated in the
writing and revision of the manuscript. DL and JG supervised the
project. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Shandong University (Shandong, China) with written informed consent
obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sonoyama W, Liu Y, Fang D, Yamaza T, Seo
BM, Zhang C, Liu H, Gronthos S, Wang CY, Wang S and Shi S:
Mesenchymal stem cell-mediated functional tooth regeneration in
swine. PLoS One. 1:e792006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gronthos S, Brahim J, Li W, Fisher LW,
Cherman N, Boyde A, DenBesten P, Robey PG and Shi S: Stem cell
properties of human dental pulp stem cells. J Dent Res. 81:531–535.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Estrela C, Alencar AH, Kitten GT, Vencio
EF and Gava E: Mesenchymal stem cells in the dental tissues:
Perspectives for tissue regeneration. Braz Dent J. 22:91–98. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iohara K, Nakashima M, Ito M, Ishikawa M,
Nakasima A and Akamine A: Dentin regeneration by dental pulp stem
cell therapy with recombinant human bone morphogenetic protein 2. J
Dent Res. 83:590–595. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kerkis I, Kerkis A, Dozortsev D,
Stukart-Parsons GC, Gomes Massironi SM, Pereira LV, Caplan AI and
Cerruti HF: Isolation and characterization of a population of
immature dental pulp stem cells expressing OCT-4 and other
embryonic stem cell markers. Cells Tissues Organs. 184:105–116.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gronthos S, Mankani M, Brahim J, Robey PG
and Shi S: Postnatal human dental pulp stem cells (DPSCs) in vitro
and in vivo. Proc Natl Acad Sci USA. 97:13625–13630. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kawashima N: Characterisation of dental
pulp stem cells: A new horizon for tissue regeneration? Arch Oral
Biol. 57:1439–1458. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mori G, Brunetti G, Oranger A, Carbone C,
Ballini A, Lo Muzio L, Colucci S, Mori C, Grassi FR and Grano M:
Dental pulp stem cells: Osteogenic differentiation and gene
expression. Ann N Y Acad Sci. 1237:47–52. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mori G, Centonze M, Brunetti G, Ballini A,
Oranger A, Mori C, Lo Muzio L, Tetè S, Ciccolella F, Colucci S, et
al: Osteogenic properties of human dental pulp stem cells. J Biol
Regul Homeost Agents. 24:167–175. 2010.PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang XL, Zhang T, Wang J, Zhang DB, Zhao
F, Lin XW, Wang Z, Shi P and Pang XN: MiR-378b promotes
differentiation of keratinocytes through NKX3.1. PLoS One.
10:e01360492015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu X, Gong J and Xu B: miR-143
down-regulates TLR2 expression in hepatoma cells and inhibits
hepatoma cell proliferation and invasion. Int J Clin Exp Pathol.
8:12738–12747. 2015.PubMed/NCBI
|
|
13
|
Lei Q, Shen F, Wu J, Zhang W, Wang J and
Zhang L: MiR-101, downregulated in retinoblastoma, functions as a
tumor suppressor in human retinoblastoma cells by targeting EZH2.
Oncol Rep. 32:261–269. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Strillacci A, Valerii MC, Sansone P,
Caggiano C, Sgromo A, Vittori L, Fiorentino M, Poggioli G, Rizzello
F, Campieri M and Spisni E: Loss of miR-101 expression promotes
Wnt/β-catenin signalling pathway activation and malignancy in colon
cancer cells. J Pathol. 229:379–389. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thu KL, Chari R, Lockwood WW, Lam S and
Lam WL: miR-101 DNA copy loss is a prominent subtype specific event
in lung cancer. J Thorac Oncol. 6:1594–1598. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang C, Lu S, Jiang J, Jia X, Dong X and
Bu P: Hsa-microRNA-101 suppresses migration and invasion by
targeting Rac1 in thyroid cancer cells. Oncol Lett. 8:1815–1821.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li C, Li C, Yue J, Huang X, Chen M, Gao J
and Wu B: miR-21 and miR-101 regulate PLAP-1 expression in
periodontal ligament cells. Mol Med Rep. 5:1340–1346.
2012.PubMed/NCBI
|
|
18
|
Wang H, Meng Y, Cui Q, Qin F, Yang H, Chen
Y, Cheng Y, Shi J and Guo Y: MiR-101 Targets the EZH2/Wnt/β-catenin
the pathway to promote the osteogenic differentiation of human bone
marrow-derived mesenchymal stem cells. Sci Rep. 6:369882016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hall A: G proteins and small GTPases:
Distant relatives keep in touch. Science. 280:2074–2075. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hall A: Rho GTPases and the actin
cytoskeleton. Science. 279:509–514. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Coso OA, Chiariello M, Yu JC, Teramoto H,
Crespo P, Xu N, Miki T and Gutkind JS: The small GTP-binding
proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK
signaling pathway. Cell. 81:1137–1146. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Knaus UG, Heyworth PG, Evans T, Curnutte
JT and Bokoch GM: Regulation of phagocyte oxygen radical production
by the GTP-binding protein Rac 2. Science. 254:1512–1515. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Minden A, Lin A, Claret FX, Abo A and
Karin M: Selective activation of the JNK signaling cascade and
c-Jun transcriptional activity by the small GTPases Rac and
Cdc42Hs. Cell. 81:1147–1157. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moore KA, Sethi R, Doanes AM, Johnson TM,
Pracyk JB, Kirby M, Irani K, Goldschmidt-Clermont PJ and Finkel T:
Rac1 is required for cell proliferation and G2/M progression.
Biochem J. 326:17–20. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ridley AJ, Paterson HF, Johnston CL,
Diekmann D and Hall A: The small GTP-binding protein rac regulates
growth factor-induced membrane ruffling. Cell. 70:401–410. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takaishi K, Sasaki T, Kotani H, Nishioka H
and Takai Y: Regulation of cell-cell adhesion by rac and rho small
G proteins in MDCK cells. J Cell Biol. 139:1047–1059. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Embade N, Valerón PF, Aznar S,
López-Collazo E and Lacal JC: Apoptosis induced by Rac GTPase
correlates with induction of FasL and ceramides production. Mol
Biol Cell. 11:4347–4358. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jin S, Ray RM and Johnson LR: Rac1
mediates intestinal epithelial cell apoptosis via JNK. Am J Physiol
Gastrointest Liver Physiol. 291:G1137–G1147. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
An S, Huang X, Gao Y, Ling J, Huang Y and
Xiao Y: FGF-2 induces the proliferation of human periodontal
ligament cells and modulates their osteoblastic phenotype by
affecting Runx2 expression in the presence and absence of
osteogenic inducers. Int J Mol Med. 36:705–711. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang Y, Dai Q, Zeng F and Liu H: MALAT1
promotes the proliferation and metastasis of osteosarcoma cells by
activating the Rac1/JNK pathway via targeting miR-509. Oncol Res.
Apr 27–2018.(Epub ahead of print). doi:
10.3727/096504017X14957939026111. View Article : Google Scholar
|
|
32
|
Hassan MQ, Tye CE, Stein GS and Lian JB:
Non-coding RNAs: Epigenetic regulators of bone development and
homeostasis. Bone. 81:746–756. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bai J, Na H, Hua X, Wei Y, Ye T, Zhang Y,
Jian G, Zeng W, Yan L and Tang Q: A retrospective study of NENs and
miR-224 promotes apoptosis of BON-1 cells by targeting PCSK9
inhibition. Oncotarget. 8:6929–6939. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liang CQ, Fu YM, Liu ZY, Xing BR, Jin Y
and Huang JL: The effect of miR-224 down-regulation on SW80 cell
proliferation and apoptosis and weakening of ADM drug resistance.
Eur Rev Med Pharmacol Sci. 21:5008–5016. 2017.PubMed/NCBI
|
|
35
|
Suárez-Fueyo A, Bradley SJ and Tsokos GC:
T cells in systemic lupus erythematosus. Curr Opin Immunol.
43:32–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu MC, Lai NS, Chen HC, Yu HC, Huang KY,
Tung CH, Huang HB and Yu CL: Decreased microRNA(miR)-145 and
increased miR-224 expression in T cells from patients with systemic
lupus erythematosus involved in lupus immunopathogenesis. Clin Exp
Immunol. 171:91–99. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chang YL, Liu ST, Wang YW, Lin WS and
Huang SM: Amiodarone promotes cancer cell death through elevated
truncated SRSF3 and downregulation of miR-224. Oncotarget.
9:13390–13406. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cong L, Zhang F and Shang H: Notch1
targeted regulation of mir-224/LRIG2 signaling for the
proliferation and apoptosis of cervical cancer cells. Oncol Lett.
13:2304–2308. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shen E, Li Y, Li Y, Shan L, Zhu H, Feng Q,
Arnold JM and Peng T: Rac1 is required for cardiomyocyte apoptosis
during hyperglycemia. Diabetes. 58:2386–2395. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoshida T, Zhang Y, Rivera Rosado LA, Chen
J, Khan T, Moon SY and Zhang B: Blockade of Rac1 activity induces
G1 cell cycle arrest or apoptosis in breast cancer cells through
downregulation of cyclin D1, survivin, and X-linked inhibitor of
apoptosis protein. Mol Cancer Ther. 9:1657–1668. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gulbins E, Coggeshall KM, Brenner B,
Schlottmann K, Linderkamp O and Lang F: Fas-induced apoptosis is
mediated by activation of a ras and rac protein-regulated signaling
pathway. J Biol Chem. 271:26389–26394. 1996. View Article : Google Scholar : PubMed/NCBI
|