Introduction
Alcohol-use disorder (AUD) is a major health problem
worldwide characterized by a chronic relapsing course of the
disease and it is associated to multiple abnormalities of brain
reward, motivation and memory, along with the inability of AUD
patients to regulate drinking, in spite of multiple negative
consequences of alcohol abuse. AUD is accompanied by significant
comorbidities and mortality, including depression and fetal alcohol
spectrum disorders (FASD) (1). FASD
entails multiple disabilities, including craniofacial and skeletal
abnormalities and it represents the main cause of mental
retardation in USA (1). In addition,
approximately 50% of AUD patients also suffer of post-traumatic
stress disorder (PTSD) (2,3). The patients suffering from AUD and PTSD
manifest greater severity of psychiatric pathologies and are more
resistant to therapeutic interventions (4). AUD therefore represents a major health
problem worldwide entailing social, personal and legal aspects.
The standard of care treatment for AUD consists of
abstinence from alcohol and psychosocial approaches. Adjuvant
pharmacological therapy is also available and recommended (5). The only drugs that are specifically
approved for AUD are acamprosate, naltrexone and nalmefene, while
disulfiram is considered as second line pharmacological approach.
Several drugs are also being considered in clinical development for
AUD patients, including topiramate, gabapentin, diazepam,
ifenprodil, memantine, prazosine and N-acetylcysteine, among others
(6,7). Nonetheless, the efficacy of medications
for alcohol-dependence remains modest, and there are no reliable
laboratoristic and/or clinical predictors of treatment response
(5). It is expected that the limited
pharmacological efficacy could be augmented through the better
understanding of the pathophysiology of alcohol dependence, as well
as through the identification of biomarkers of response to
therapeutic treatments and by modalities that improve medication
adherence (8). Along this line of
research, studies are warranted that aim at identifying pathogenic
and biomolecular mechanisms of alcohol dependence and that may help
design novel tailored approaches for the treatment of this
condition that could be used alone or in combination with existing
treatment and psychotherapeutic approaches.
Inflammation and altered innate immune responses are
hallmarks of alcohol-induced organ damage, affecting the liver,
cardiovascular system, and brain, and immune abnormalities induced
by alcohol exposure have been well documented both in rodent models
and humans with AUD (9). In rodent
models, it has been shown that increased systemic and brain
production of proinflammatory cytokines such as TNFα, IL-1β and
MCP-1 and neuroinflammation occurs when 10-days ethanol
administration is followed by endotoxin challenge. Alcohol exposure
significantly augments the production of proinflammatory cytokines
induced by LPS, with a long-lasting persistence in the brain.
Furthermore, the brains of mice exposed to 10-days alcohol
administration exhibited signs of microglia activation (10). In agreement with these data, it has
been shown that in vivo administration of alcohol to adult male
Wistar rats (1.0 g/kg) once daily for 14 days increased the
concentrations of the proinflammatory cytokines (IL-1β and TNF-α)
in two primary brain regions, such as the hippocampus and frontal
cortex, that are implicated in mood regulation (11). In humans, an increase in serum
proinflammatory cytokines including TNFα, IL-1β occurs upon
prolonged alcohol exposure (12) and
the monocytes isolated from the blood of alcoholics produce greater
amounts of TNFα, both spontaneously and in response to endotoxin
challenge (13). Chronic alcohol
consumption elevates the levels of circulating TNFα, IL-1β and IL-6
both in the woman and in their fetuses (14). It is also known that AUD patients
exhibit abnormal profile of circulating cytokines, including
prototypical Th1, interferon-γ (IFN-γ) (15) and Th-2 cytokines (IL-4, IL-13, IL-10)
(16,17), as well as IL-6 and TNF-α, IL-7 and
GM-CSF (18). In lung, cytokine
production is disrupted by ethanol, exacerbating respiratory
distress syndrome with upregulated expression of the Th3 and
primarily anti-inflammatory cytokine, transforming growth factor-β
(TGF-β) (19,20). In general, AUD patients, and in
particular those with MDD, seem to exhibit higher levels of
proinflammatory cytokines, IL-6, IL-7, TNF-α, GM-CSF, and IFN-γ,
than controls (18,21). One recent study has shown that
high-risk drinkers showed significantly higher mean values of the
proinflammatory cytokine IL-6 than abstainers (22). TNF-α and IL-6 serum levels have been
considered as possible neurobiological markers of alcohol
consumption in AUD patients (23).
It has been clarified that chronic ethanol intake
stimulates the immune system though activation of the NLRP3/ASC
inflammasome. In turn, this leads to activation of caspase-1 and
increase of IL-1β in the cerebellum. Accordingly, selective
blockade of the IL-1/IL-1R signaling suppresses both the activation
of the inflammasome and of neuroinflammation that are provoked by
alcohol (24). In addition, the
altered immune response evoked from alcohol abuse may in turn be
implicated in the generation and maintenance of the mechanisms of
alcohol addiction and regulation of drinking. For example, the
activation of Toll-like receptor (TLR) 3 increases voluntary
alcohol intake in C57BL/6J male (25). The potential clinical relevance of
the interaction between the effects of AUD on the immune system and
the possible role of this latter in regulating drinking has
received further in vivo validation with the observation that
TLR3-TIR-domain-containing adapter-inducing interferon-β (TRIF)
pathway are activated during chronic ethanol consumption. The
IKKε/TBK1 inhibitor Amlexanox down-regulates TRIF-dependent pathway
in the brain and reduces ethanol consumption, suggesting that the
TRIF-dependent pathway is implicated in regulation of drinking
(26). Hence, the NFKB pathway that
lies downstream the activation of TLR signaling may also play a non
canonical non-immune role in mediating complex behaviors, including
processes of learning and memory, stress responses, anhedonia and
drug reward (27). Along this line
of research, in a translational study entailing both mice and
humans, Coleman and coworkers generated strong proof of evidence in
humans that HMGB1 and IL-1β heterocomplexes formed in vivo may
contribute to the pathology of alcoholism. The Authors found that
HMGB1/IL-1β complexes are increased in post-mortem human alcoholic
hippocampus (28). These data have
led to the hypothesis of alcohol-induced neuroimmune gene induction
(29).
Taken as a whole, all these data strongly support
the hypothesis that AUD can be characterized by generalized immune
system dysfunction, caused by chronic alcohol exposure and supports
the concept that alcoholism may represent a systemic
proinflammatory condition (30).
These data have attracted attention on the possible interaction
between immunoinflammatory events that are induced by AUD and the
possible role of immunoinflammatory events in the initiation and
maintenance of AUD.
Along this line of research, a recent study has
shown that the dual inhibitor of phosphodiesterase-4 and −10 and of
the pleiotropic cytokine macrophage migration inhibitory factor
(MIF), Ibudilast (IBUD), exerts beneficial effects both in animal
models of alcohol dependence (31)
and in humans with AUD (32). On the
basis of these data, we have presently studied eventual differences
in transcriptomic levels of MIF and its homologue DDT along with
their receptors CD74, CXCR2, CXCR4 and CXCR7 in the brain and
peripheral blood of patients with AUD. The transcriptomic analysis
of the present study demonstrates a trend to increased expression
of MIF, DDT and CD74 in AUD patients. However, when expression
levels of MIF and DDT were merged the expression of this
superfamily was significantly higher than in healthy controls.
These data indicate that dysregulation of MIF family members may
occur during AUD and that specific tailored treatments with
antagonists of MIF superfamily for AUD is worthy of further
investigation.
Materials and methods
Microarray datasets selection and
analysis
Analysis of prefrontal cortex
transcriptomic data
In order to investigate the expression levels of MIF
and related genes in alcohol-related disorder, we interrogated the
publicly-available whole-genome transcriptomic microarray datasets,
GSE123114, GSE53808 and GSE49376, obtained from the Gene Expression
Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database. GSE123114
included whole genome transcriptomic data from an animal model of
chronic alcohol self-administration (33). Briefly, for the generation of this
dataset, mice were given free access to three bottles containing 5
and 10% of ethanol and water for 16 weeks. On the last two weeks,
both ethanol solutions were adulterated with the aversive
substance, quinine. Following this procedure, mice were classified
into two groups, based on the ethanol intake and preference: ‘Light
Drinkers’ (preference for water at all experimental stages); and
‘Inflexible Drinkers’ (preference for ethanol at all experimental
stages, even after addition of quinine to the ethanol solution). At
the end of the experimental period, total RNA was extracted from
the Prefrontal cortex and the Affymetrix Mouse Genome 430 2.0 Array
was used for the generation of the dataset. Details on the
GSE123114 dataset can be obtained from the relative publication
(33).
GSE53808 was used for the analysis of gene
expression patterns in the prefrontal cortex of three alcoholics
without hepatic encephalopathy and three neurologically normal
controls (34). Brain tissues were
obtained from the New South Wales Tissue Research Centre (NSW TRC),
part-funded by the National Institute on Alcohol Abuse and
Alcoholism (R24AA012725) (35). The
Affymetrix Human Genome U219 Array was used for the generation of
the dataset. For the analysis, technical replicates were averaged.
Detailed description of the GSE53808 can be obtained from (35).
The GSE49376 dataset included whole-genome
transcriptomic levels of the prefrontal cortex from 46 European
Australians, obtained from the NSW TRC. Among the 46 subjects, 23
(16 males and 7 females) had alcohol use disorders, divided in
alcohol-dependence (n=9) and alcohol abuse (n=14). The male and
female samples did not significantly differ in age, postmortem
intervals, brain pH and alcohol daily use. All subjects were not
affected by illicit drug abuse and dependence or major psychotic
disorders. The clinical information of all the patients are
available in (36).
Analysis of blood transcriptomic
data
GSE59206 included whole-genome blood transcriptomic
data from two groups of subjects (n=11/group): Heavy drinkers (HD,
defined as regular alcohol use over the past year of at least 8
standard drinks/week for women and at least 15 standard drinks/week
for men), and moderate drinkers (MD, defined as up to 7 standard
drinks/week for women and 14 standard drinks/week for men), as
defined by the NIAAA. All subjects were between 21–50 years-old,
had no history of any drug dependence (including alcohol
dependence), and did not meet criteria for Axis I DSM-IV
psychiatric diagnoses. Exclusion criteria included dependence on
psychoactive substances; current or past use of opiates; current
use of psychoactive drugs; psychotic disorders; neurological,
cardiovascular, endocrine, renal, liver, thyroid diseases; subjects
on medications for any medical condition; oral contraceptives; and
pregnant and lactating women. Blood was collected at baseline and 1
h following exposure to an alcohol-related cue. Alcohol-related cue
scripts were based on individual situations that included
alcohol-related stimuli and resulted in subsequent alcohol use
(e.g. buying alcohol and being at a bar, watching others drink
alcohol). Microarray analysis was performed on blood cells using
the Illumina HumanHT-12 V4.0 expression beadchip (37).
A summary of all the microarray datasets used in
this study is presented in Table
I.
 | Table I.Characteristics of the microarray
datasets analyzed in the study. |
Table I.
Characteristics of the microarray
datasets analyzed in the study.
| Author, year | GEO identifier | Sample type | Samples | Platform | (Refs.) |
|---|
| da Silva e Silva
et al, 2016 | GSE123114 | Prefrontal
cortex | 3 inflexible
drinker mice and 3 light drinker mice | Affymetrix Mouse
Genome 430 2.0 Array | (33) |
| Sutherland et
al, 2014 | GSE53808 | Prefrontal cortex
(frozen) | 3 alcoholics
without hepatic encephalopathy and 3 neurologically normal
controls | Affymetrix Human
Genome U219 Array | (34) |
| Xu et al,
2014 | GSE49376 | Prefrontal cortex
(frozen) | Prefrontal cortex
from 46 subjects, 23 (16 males and 7 females) with alcohol use
disorders, divided in alcohol-dependence (n=9) and alcohol abuse
(n=14) and 23 controls | Illumina HumanHT-12
V4.0 expression beadchip | (36) |
| Beech et al,
2014 | GSE59206 | Peripheral
blood | 11 heavy drinkers
and 11 light drinkers | Illumina HumanHT-12
V4.0 expression beadchip | (37) |
Statistical analysis
Expression data are presented as mean ± standard
deviation (SD). The limma (Linear Models for Microarray Analysis) R
package was used for identifying differentially expressed genes. An
adjusted P-value <0.05 and a |log(fold change)|>1 was
considered as threshold of significance.
Fisher's Inverse χ2 test was used as integrative
analysis for MIF and DDT modulation in blood. This methods computes
a combined statistic from the P-values obtained from the individual
genes, as follows:
s=-2log∑i=0n(pi)
which follows a χ2 distribution with 2n degrees of
freedom under the null hypothesis. Graphs were constructed using
the GraphPad Prism 8 software.
Results
Analysis of brain MIF, DDT and
receptors in murine and human alcohol abuse
Analysis of the transcriptomic levels of MIF, DDT
and CD74 and the coreceptors, CXCR2, CXCR4 and CXCR7, in a murine
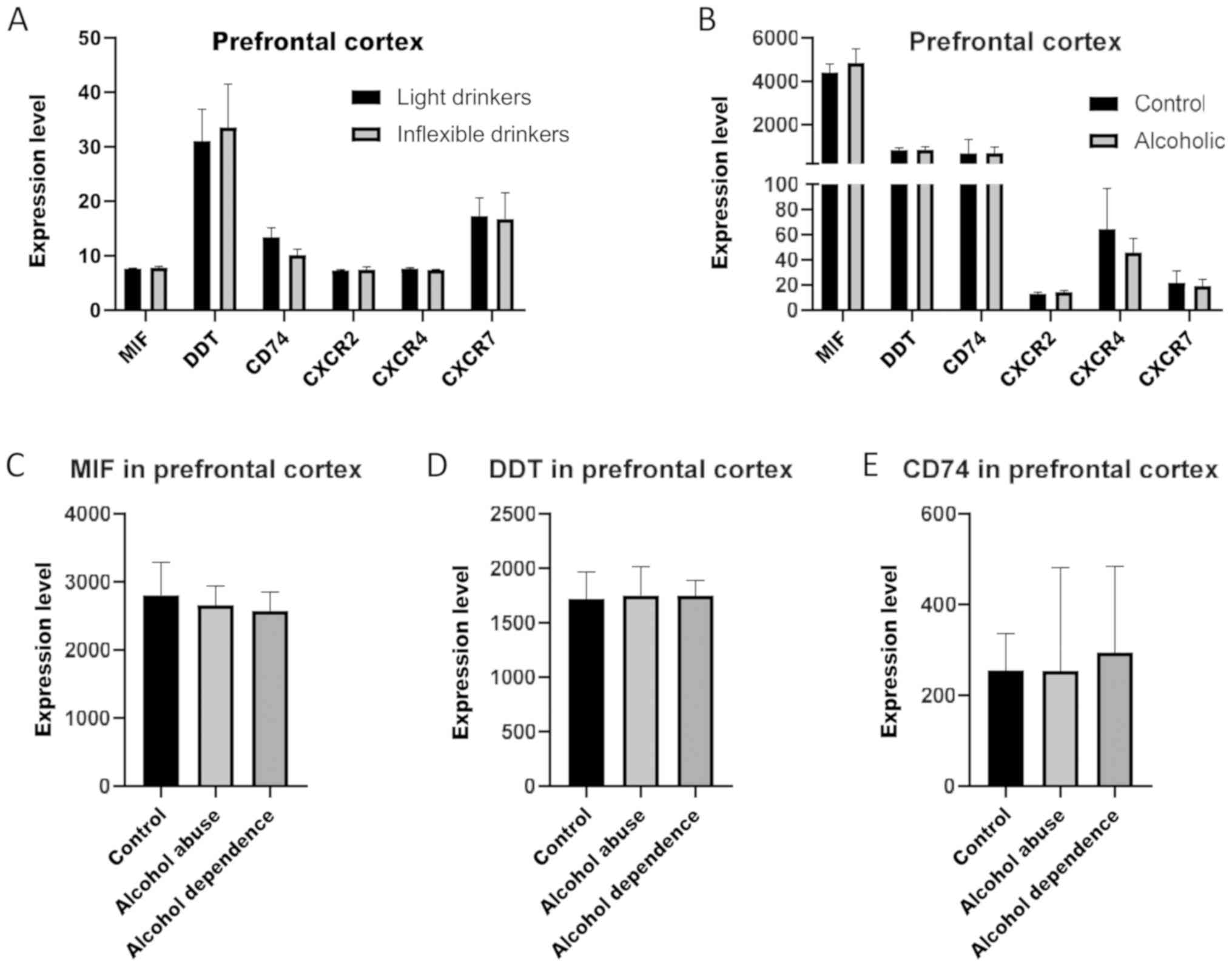
model of alcohol dependence (GSE123114), revealed no significant
modulation in Inflexible Drinkers as compared to Light Drinkers
(Fig. 1A). In a similar manner, as
determined in the GSE53808 dataset, in the prefrontal cortex, no
significant differences between healthy control subjects and
alcoholics were observed for MIF, DDT and their receptors (Fig. 1B). Similarly, as shown on Fig. 1C, alcohol abuse and dependence was
not associated to significant alterations in the RNA levels of MIF,
DDT and CD74 in the prefrontal cortex, as compared to healthy
subjects (GSE49376). The expression levels of the coreceptors could
not be evaluated in the latter dataset, as they were below the
threshold of detection.
Analysis of blood MIF, DDT and
receptors in heavy drinkers
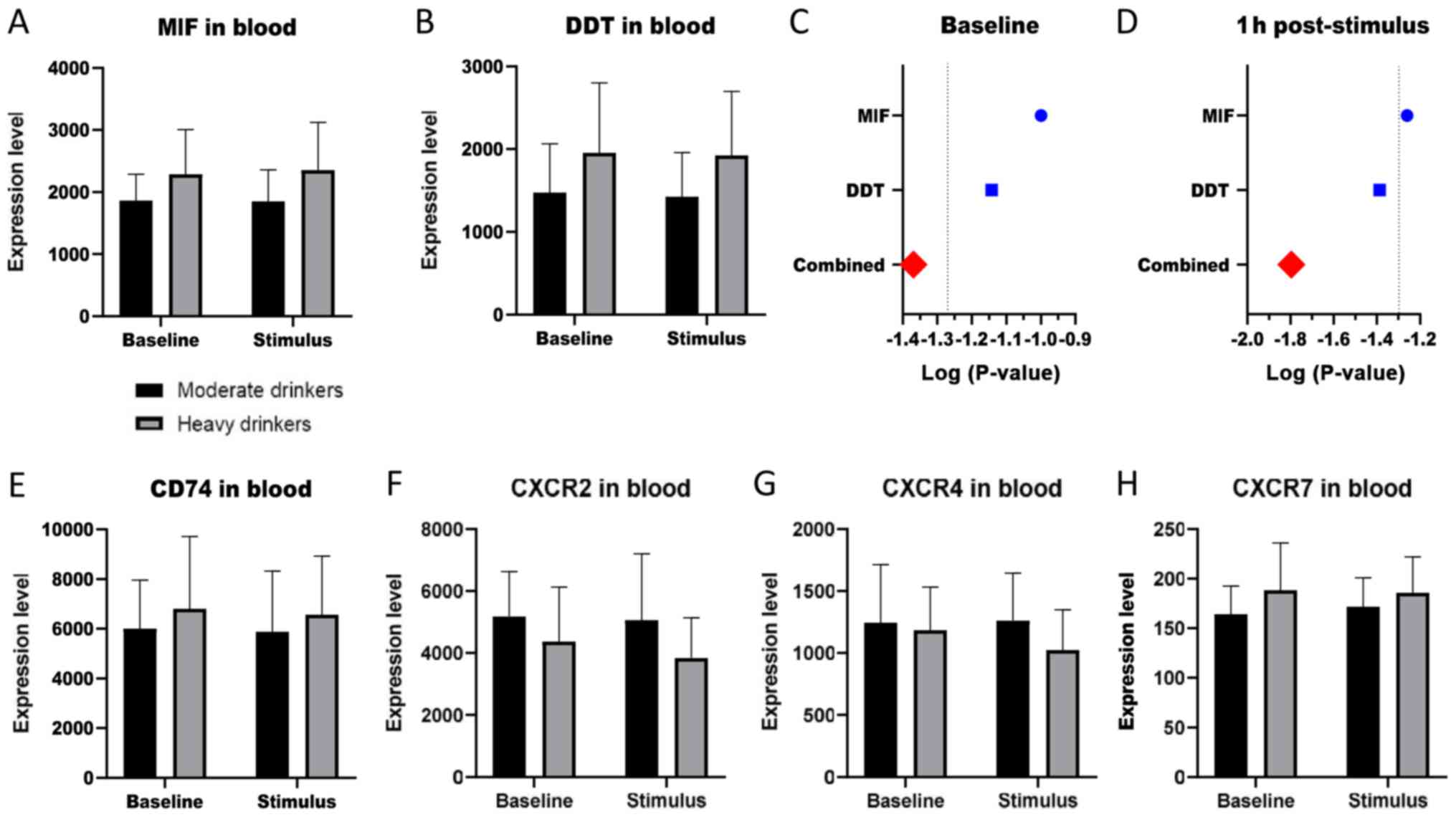
The GSE59206 dataset was interrogated to evaluate
the levels of MIF, DDT and their receptors/coreceptors in subjects
at risk for alcohol dependence. As shown in Fig. 2A and B, heavy drinkers showed a
moderate trend of increase in the levels of MIF and DDT, both at
baseline and upon exposure to alcohol-related cues. Interestingly,
alcohol-related cues did not, in the short term (1 h), determined a
modulation in the levels of these two genes. Although the
statistical significance was not reached, and in consideration of
the overlapping effects of MIF and DDT (38), an inverse Fisher's chi square test
was performed on unadjusted P-values to evaluate whether the
combined effect of MIF and DDT modulation could be statistically
significant. As expected, a combined p value of 0.042 was obtained
at baseline and 0.016 at 1 h post-exposure (Fig. 2C and D). No significant modulation
was observed for the receptor CD74 and the coreceptors, CXCR2,
CXCR4 and CXCR7 (Fig. 2E-H).
Discussion
Increasing experimental and clinical evidence
indicate that immunoinflammatory events are induced by alcohol
exposure. It has been shown that microglia, the innate immune cells
of the brain, and neurons respond to alcohol through receptors and
molecules of the innate immune system, including, but not limited
to, proinflammatory cytokines and their receptors (39,40). The
pathogenic relevance of immunoinflammatory events to the
pathogenesis and maintenance of AUD has gained translational
relevance with the demonstration of the beneficial effect obtained
with minocycline in rodent models of AUD. However, clinical studies
with this drug in human patients failed to confirm these
preclinical findings (41).
Nonetheless, the multiple data supporting a role for
neuroinflammation in the pathogenesis of AUD have warranted further
studies aimed at identifying other cells, molecules and receptors
of the immune system that may play a role in the pathogenesis of
the disorder and may eventually represent suitable therapeutic
target.
MIF is a protein that has been discovered at the end
of the 1960s, and acquired its name from its ability to inhibit the
migration of macrophages. The receptor of MIF is composed of the
cell surface receptor CD74, that is associated with CD44, and of
additional 3 non-cognate ligand for C-X-C chemokine receptor type 2
(CXCR2), type 4 (CXCR4) and type 7 (CXCR7) (38). A second member of the MIF family has
also been identified in 2011, named D-dopachrome tautomerase
(D-DT), that exhibits multiple synergisms with MIF [reviewed in
(38)].
MIF, and more recently DDT, have been implicated in
the pathogenesis of several pathologies, including
immunoinflammatory and autoimmune diseases (38,42–44) and
cancer (45–47). As anticipated above, that MIF may
represent an important pathogenic molecule in the pathogenesis of
AUD emerges from recent studies indicating that the dual inhibitor
of phosphodiesterase-4 and −10 and MIF, named IBUD, has beneficial
effects of AUD both in rodent models and human patients (31,32). In
particular, this trial consisted of a randomized, crossover,
double-blind, placebo-controlled laboratory study of IBUD in non
treatment-seeking individuals that had suffered with mild-to-severe
AUD, within the last month. The trial evaluated the safety,
tolerability, and initial human laboratory efficacy of IBUD (50 mg
b.i.d.) on primary measures of subjective response to alcohol, as
well as secondary measures of cue- and stress-induced changes in
craving and mood. IBUD was well tolerated but it failed to
significantly influence primary measures. However, it significantly
improved mood on the secondary measures of stress exposure and
reduced tonic levels of craving. Furthermore, exploratory analyses
indicated that IBUD preferentially influenced the stimulant and
mood altering effects of alcohol as compared to placebo in those
patients exhibiting the most severe depressive symptoms (39).
However, since IBUD is a dual inhibitor of PDE 4–10
and MIF, it is difficult to understand whether the effects in AUD
is due to the combined synergistic pharmacological properties of
IBUD or whether either of them accounts for by the beneficial
effects of IBUD. It is therefore important to better identify which
of the two pharmacological properties may be eventually relevant
for the efficacy of drug in this condition. In addition, since it
has recently been demonstrated that the MIF superfamily is composed
from at least 2 different homologues, such as MIF and DDT, that
most often, but not always, act synergistically. It is also
important to shed light on the possible combined action that MIF,
DDT and their receptors, play in development and maintenance of
AUD. For this reason, we carried out a transcriptomic study that
evaluated the expression of MIF, DDT and their receptors in the
brains of AUD patients and in peripheral blood cells of people at
risk of developing AUD, under basal conditions and upon exposure to
alcohol-related cues. In addition, we also evaluated the
transcriptomic analysis of MIF, DDT and their receptors in the
brains of a mouse model of alcoholism. This analysis was carried
out by DNA microarray analysis that represents a useful in silico
toll for the better understanding of pathogenic pathways and the
possible prediction of new diagnostic therapeutic approaches in
several clinical settings, including immunoinflammatory and
autoimmune diseases and fibrotic diseases (45,46,48–59). Our
analysis first demonstrated that no differences in the expression
of MIF, DDT and their receptors can be found in brains of both
rodent models of AUD and patients with this condition. However,
when peripheral blood of heavy drinkers was studied, it was
possible to observe a consistent, though not statistically
significant, trend, of increased expression of MIF and DDT in the
blood of heavy drinkers as compared to healthy controls, both at
baseline and upon exposure to alcohol-related cues. Interestingly,
knowing the overlapping effects of MIF and DDT in several
biological responses (38) we
performed the inverse Fisher's χ2 test on unadjusted p
values and found out that the combined effect of MIF and DDT
modulation was statistically significant as compared to controls.
Along with the trend of augmented expression of the CD74 receptors
in the peripheral blood of heavy drinkers as compared to controls
these data seem to indicate that a hyperactivation of the MIF
superfamily may also occur in AUD patients. This finding is of
particular relevance for its possible translation, as it has
recently been demonstrated that dual specific antagonists of MIF
and DDT, such as 4-iodo-6-phenylpyrimidine (4-IPP), have been
developed (60). Hence, this new
class of drugs could be worthy being studied in rodent models of
AUD in a head to head comparison with single MIF inhibitors and
eventually IBUD. This is of further particular relevance as MIF has
been implicated in the pathogenesis of other forms of dependence
and addiction that could benefit from specific dual inhibitors of
MIF and DDT. Although our data are the first to demonstrate that a
moderate upregulated transcriptomic expression of the MIF-DDT axis
occurs in the blood of people at risk of developing AUD, other
studies are necessary to fully understand the exact role of MIF and
DDT in development and maintenance of AUD. In particular, it will
be important to prove whether selected groups of AUD patients can
be identified on the basis of their MIF-DDT secretory capacity and
whether those with highest expression profile are eventually those
that may respond better to tailored anti MIF-DDT therapies. Studies
of genetic polymorphism of MIF and DDT in AUD patients may be
warranted. It will also be important to determine on larger number
of patients whether in a manner similar to progressive multiple
sclerosis, MIF and DDT are more abundantly produced in males than
in females (61), as this may
further facilitate identification of individual potentially
responders to anti-MIF-DDT. In addition, it will be relevant to
measure circulating levels of MIF and DDT in AUD patients at
different time points during their development and maintenance of
addiction, as our transcriptomic analysis do not take into account
the amount of MIF and DDT that can be secreted from resident cells,
such as epithelial and endothelial cells that are not represented
in peripheral blood samples. Nonetheless, in spite of the needs of
these further studies, our present analysis provides in vivo
evidence for the upregulated synthesis of MIF and DDT in AUD and
highlights the possibility that these molecules may offer novel
important diagnostic and therapeutic opportunities in prevention
and treatment of AUD along with psychotherapeutic approaches.
Acknowledgements
Not applicable.
Funding
The present study was supported by the current
research funds 2019 of IRCCS ‘Centro Neurolesi ‘Bonino Pulejo’
(Messina-Italy).
Availability of data and materials
The datasets analyzed in the current study are
available from the Gene Expression Omnibus website (GEO; http://www.ncbi.nlm.nih.gov/geo/) with accession
nos. GSE123114, GSE53808, GSE49376 and GSE59206.
Authors' contributions
MCP, EM, FN and EC conceived and designed the
present study. KM, PF, RDM and LF analyzed the data and prepared
the figures. MCP, KM, MSB and EC interpreted the data and wrote the
manuscript. EM, RDM and FN critically revised the manuscript. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang X, Zhang K, Yang F, Ren Z, Xu M,
Frank JA, Ke ZJ and Luo J: Minocycline protects developing brain
against ethanol-induced damage. Neuropharmacology. 129:84–99. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Debell F, Fear NT, Head M, Batt-Rawden S,
Greenberg N, Wessely S and Goodwin L: A systematic review of the
comorbidity between PTSD and alcohol misuse. Soc Psychiatry
Psychiatr Epidemiol. 49:1401–1425. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pietrzak RH, Goldstein RB, Southwick SM
and Grant BF: Prevalence and Axis I comorbidity of full and partial
posttraumatic stress disorder in the United States: Results from
Wave 2 of the national epidemiologic survey on alcohol and related
conditions. J Anxiety Disord. 25:456–465. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Neupane SP, Bramness JG and Lien L:
Comorbid post-traumatic stress disorder in alcohol use disorder:
Relationships to demography, drinking and neuroimmune profile. BMC
Psychiatry. 17:3122017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Franck J and Jayaram-Lindström N:
Pharmacotherapy for alcohol dependence: Status of current
treatments. Curr Opin Neurobiol. 23:692–699. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Soyka M and Müller CA: Pharmacotherapy of
alcoholism-an update on approved and off-label medications. Expert
Opin Pharmacother. 18:1187–1199. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hillemacher T and Frieling H:
Pharmacotherapeutic options for co-morbid depression and alcohol
dependence. Expert Opin Pharmacother. 20:547–569. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garbutt JC: The state of pharmacotherapy
for the treatment of alcohol dependence. J Subst Abuse Treat.
36:S15–S25. 2009.PubMed/NCBI
|
|
9
|
Szabo G, Mandrekar P, Petrasek J and
Catalano D: The unfolding web of innate immune dysregulation in
alcoholic liver injury. Alcohol Clin Exp Res. 35:782–786. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Qin L, He J, Hanes RN, Pluzarev O, Hong JS
and Crews FT: Increased systemic and brain cytokine production and
neuroinflammation by endotoxin following ethanol treatment. J
Neuroinflammation. 5:102008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kalejaiye O, Getachew B, Ferguson CL,
Taylor RE and Tizabi Y: Alcohol-induced increases in inflammatory
cytokines are attenuated by nicotine in region-selective manner in
male rats. J Drug Alcohol Res. 6(pii): 2360362017.PubMed/NCBI
|
|
12
|
McClain C, Barve S, Deaciuc I, Kugelmas M
and Hill D: Cytokines in alcoholic liver disease. Semin Liver Dis.
19:205–219. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McClain CJ and Cohen DA: Increased tumor
necrosis factor production by monocytes in alcoholic hepatitis.
Hepatology. 9:349–351. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ahluwalia B, Wesley B, Adeyiga O, Smith
DM, Da-Silva A and Rajguru S: Alcohol modulates cytokine secretion
and synthesis in human fetus: an in vivo and in vitro study.
Alcohol. 21:207–213. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nicoletti F, Zaccone P, Di Marco R,
Lunetta M, Magro G, Grasso S, Meroni P and Garotta G: Prevention of
spontaneous autoimmune diabetes in diabetes-prone BB rats by
prophylactic treatment with antirat interferon-gamma antibody.
Endocrinology. 138:281–288. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nicoletti F, Mancuso G, Cusumano V, Di
Marco R, Zaccone P, Bendtzen K and Teti G: Prevention of
endotoxin-induced lethality in neonatal mice by interleukin-13. Eur
J Immunol. 27:1580–1583. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marchant A, Bruyns C, Vandenabeele P,
Ducarme M, Gérard C, Delvaux A, De Groote D, Abramowicz D, Velu T
and Goldman M: Interleukin-10 controls interferon-gamma and tumor
necrosis factor production during experimental endotoxemia. Eur J
Immunol. 24:1167–1171. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
González-Reimers E, Santolaria-Fernández
F, Medina-García JA, González-Pérez JM, de la Vega-Prieto MJ,
Medina-Vega L, Martín-González C and Durán-Castellón MC: TH-1 and
TH-2 cytokines in stable chronic alcoholics. Alcohol Alcohol.
47:390–396. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nicoletti F, Di Marco R, Patti F, Reggio
E, Nicoletti A, Zaccone P, Stivala F, Meroni PL and Reggio A: Blood
levels of transforming growth factor-beta 1 (TGF-beta1) are
elevated in both relapsing remitting and chronic progressive
multiple sclerosis (MS) patients and are further augmented by
treatment with interferon-beta 1b (IFN-beta1b). Clin Exp Immunol.
113:96–99. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Crews FT, Bechara R, Brown LA, Guidot DM,
Mandrekar P, Oak S, Qin L, Szabo G, Wheeler M and Zou J: Cytokines
and alcohol. Alcohol Clin Exp Res. 30:720–730. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nikou T, Ioannidis A, Zoga M, Tzavellas E,
Paparrigopoulos T, Magana M, Pliatsika P, Nikolaou C and
Chatzipanagiotou S: Alteration in the concentrations of
Interleukin-7 (IL-7), Interleukin-10 (IL-10) and Granulocyte Colony
Stimulating Factor (G-CSF) in alcohol-dependent individuals without
liver disease, during detoxification therapy. Drug Alcohol Depend.
163:77–83. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Archer M, Kampman O, Bloigu A, Bloigu R,
Luoto K, Kultti J, Hämäläinen M, Moilanen E, Leinonen E and Niemelä
O: Assessment of alcohol consumption in depression follow-up using
self-reports and blood measures including inflammatory biomarkers.
Alcohol Alcohol. 54:243–250. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heberlein A, Käser M, Lichtinghagen R,
Rhein M, Lenz B, Kornhuber J, Bleich S and Hillemacher T: TNF-α and
IL-6 serum levels: Neurobiological markers of alcohol consumption
in alcohol-dependent patients? Alcohol. 48:671–676. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lippai D, Bala S, Petrasek J, Csak T,
Levin I, Kurt-Jones EA and Szabo G: Alcohol-induced IL-1β in the
brain is mediated by NLRP3/ASC inflammasome activation that
amplifies neuroinflammation. J Leukoc Biol. 94:171–182. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Warden AS, Azzam M, DaCosta A, Mason S,
Blednov YA, Messing RO, Mayfield RD and Harris RA: Toll-like
receptor 3 activation increases voluntary alcohol intake in
C57BL/6J male mice. Brain Behav Immun. 77:55–65. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McCarthy GM, Warden AS, Bridges CR,
Blednov YA and Harris RA: Chronic ethanol consumption: Role of
TLR3/TRIF-dependent signaling. Addict Biol. 23:889–903. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nennig SE and Schank JR: The role of NFkB
in drug addiction: Beyond inflammation. Alcohol Alcohol.
52:172–179. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Coleman LG Jr, Zou J, Qin L and Crews FT:
HMGB1/IL-1β complexes regulate neuroimmune responses in alcoholism.
Brain Behav Immun. 72:61–77. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Crews FT and Vetreno RP: Mechanisms of
neuroimmune gene induction in alcoholism. Psychopharmacology
(Berl). 233:1543–1557. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
González-Reimers E, Santolaria-Fernández
F, Martín-González MC, Fernández-Rodríguez CM and Quintero-Platt G:
Alcoholism: A systemic proinflammatory condition. World J
Gastroenterol. 20:14660–14671. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bell RL, Lopez MF, Cui C, Egli M, Johnson
KW, Franklin KM and Becker HC: Ibudilast reduces alcohol drinking
in multiple animal models of alcohol dependence. Addict Biol.
20:38–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ray LA, Bujarski S, Shoptaw S, Roche DJ,
Heinzerling K and Miotto K: Development of the neuroimmune
modulator ibudilast for the treatment of alcoholism: A randomized,
placebo-controlled, human laboratory trial.
Neuropsychopharmacology. 42:1776–1788. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
da Silva e Silva DA, Frozino Ribeiro A,
Damasceno S, Rocha CS, Berenguer de Matos AH, Boerngen-Lacerda R,
Correia D and Brunialti Godard AL: Inflexible ethanol intake: A
putative link with the Lrrk2 pathway. Behav Brain Res. 313:30–37.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sutherland GT, Sheedy D, Sheahan PJ,
Kaplan W and Kril JJ: Comorbidities, confounders, and the white
matter transcriptome in chronic alcoholism. Alcohol Clin Exp Res.
38:994–1001. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sheedy D, Garrick T, Dedova I, Hunt C,
Miller R, Sundqvist N and Harper C: An Australian brain bank: A
critical investment with a high return! Cell Tissue Bank.
9:205–216. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu H, Wang F, Liu Y, Yu Y, Gelernter J and
Zhang H: Sex-biased methylome and transcriptome in human prefrontal
cortex. Hum Mol Genet. 23:1260–1270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Beech RD, Leffert JJ, Lin A, Hong KA,
Hansen J, Umlauf S, Mane S, Zhao H and Sinha R: Stress-related
alcohol consumption in heavy drinkers correlates with expression of
miR-10a, miR-21, and components of the TAR-RNA-binding
protein-associated complex. Alcohol Clin Exp Res. 38:2743–2753.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Günther S, Fagone P, Jalce G, Atanasov AG,
Guignabert C and Nicoletti F: Role of MIF and D-DT in
immune-inflammatory, autoimmune, and chronic respiratory diseases:
From pathogenic factors to therapeutic targets. Drug Discov Today.
24:428–439. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Crews FT, Lawrimore CJ, Walter TJ and
Coleman LG: The role of neuroimmune signaling in alcoholism.
Neuropharmacology. 122:56–73. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Alfonso-Loeches S, Pascual-Lucas M, Blanco
AM, Sanchez-Vera I and Guerri C: Pivotal role of TLR4 receptors in
alcohol-induced neuroinflammation and brain damage. J Neurosci.
30:8285–8295. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Petrakis IL, Ralevski E, Gueorguieva R,
Sloan ME, Devine L, Yoon G, Arias AJ and Sofuoglu M: Targeting
neuroinflammation with minocycline in heavy drinkers.
Psychopharmacology (Berl). 236:3013–3021. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nicoletti F, Mazzon E, Fagone P, Mangano
K, Mammana S, Cavalli E, Basile MS, Bramanti P, Scalabrino G, Lange
A and Curtin F: Prevention of clinical and histological signs of
MOG-induced experimental allergic encephalomyelitis by prolonged
treatment with recombinant human EGF. J Neuroimmunol. 332:224–232.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fagone P, Mazzon E, Cavalli E, Bramanti A,
Petralia MC, Mangano K, Al-Abed Y, Bramati P and Nicoletti F:
Contribution of the macrophage migration inhibitory factor
superfamily of cytokines in the pathogenesis of preclinical and
human multiple sclerosis: In silico and in vivo evidences. J
Neuroimmunol. 322:46–56. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cavalli E, Mazzon E, Basile MS, Mangano K,
Di Marco R, Bramanti P, Nicoletti F, Fagone P and Petralia MC:
Upregulated expression of macrophage migration inhibitory factor,
its analogue D-dopachrome tautomerase, and the CD44 receptor in
peripheral CD4 T cells from clinically isolated syndrome patients
with rapid conversion to clinical defined multiple sclerosis.
Medicina (Kaunas). 55(pii): E6672019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lombardo SD, Presti M, Mangano K, Petralia
MC, Basile MS, Libra M, Candido S, Fagone P, Mazzon E, Nicoletti F
and Bramanti A: Prediction of PD-L1 expression in neuroblastoma via
computational modeling. Brain Sci. 9(pii): E2212019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Presti M, Mazzon E, Basile M, Petralia MC,
Bramanti A, Colletti G, Bramanti P, Nicoletti F and Fagone P:
Overexpression of macrophage migration inhibitory factor and
functionally-related genes, D-DT, CD74, CD44, CXCR2 and CXCR4, in
glioblastoma. Oncol Lett. 16:2881–2886. 2018.PubMed/NCBI
|
|
47
|
Mangano K, Mazzon E, Basile MS, Di Marco
R, Bramanti P, Mammana S, Petralia MC, Fagone P and Nicoletti F:
Pathogenic role for macrophage migration inhibitory factor in
glioblastoma and its targeting with specific inhibitors as novel
tailored therapeutic approach. Oncotarget. 9:17951–17970. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fagone P, Mangano K, Mammana S, Pesce A,
Pesce A, Caltabiano R, Giorlandino A, Portale TR, Cavalli E,
Lombardo GA, et al: Identification of novel targets for the
diagnosis and treatment of liver fibrosis. Int J Mol Med.
36:747–752. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lombardo SD, Mazzon E, Basile MS, Campo G,
Corsico F, Presti M, Bramanti P, Mangano K, Petralia MC, Nicoletti
F and Fagone P: Modulation of tetraspanin 32 (TSPAN32) expression
in T cell-mediated immune responses and in multiple sclerosis. Int
J Mol Sci. 20(pii): E43232019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Candido S, Lupo G, Pennisi M, Basile MS,
Anfuso CD, Petralia MC, Gattuso G, Vivarelli S, Spandidos DA, Libra
M and Falzone L: The analysis of miRNA expression profiling
datasets reveals inverse microRNA patterns in glioblastoma and
Alzheimer's disease. Oncol Rep. 42:911–922. 2019.PubMed/NCBI
|
|
51
|
Petralia MC, Mazzon E, Fagone P, Falzone
L, Bramanti P, Nicoletti F and Basile MS: Retrospective follow-up
analysis of the transcriptomic patterns of cytokines, cytokine
receptors and chemokines at preconception and during pregnancy, in
women with post-partum depression. Exp Ther Med. 18:2055–2062.
2019.PubMed/NCBI
|
|
52
|
Mangano K, Lanteri R, Basile MS, Bellavia
N, Latino R, Messina D, Fagone P, Colletti G, Nania R, Caltabiano
R, et al: Effects of GIT-27NO, a NO-donating compound, on hepatic
ischemia/reperfusion injury. Int J Immunopathol Pharmacol.
33:20587384198627362019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lombardo SD, Mazzon E, Basile MS, Cavalli
E, Bramanti P, Nania R, Fagone P, Nicoletti F and Petralia MC:
Upregulation of IL-1 receptor antagonist in a mouse model of
migraine. Brain Sci. 9(pii): E1722019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mammana S, Bramanti P, Mazzon E, Cavalli
E, Basile MS, Fagone P, Petralia MC, McCubrey JA, Nicoletti F and
Mangano K: Preclinical evaluation of the PI3K/Akt/mTOR pathway in
animal models of multiple sclerosis. Oncotarget. 9:8263–8277. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mangano K, Cavalli E, Mammana S, Basile
MS, Caltabiano R, Pesce A, Puleo S, Atanasov AG, Magro G, Nicoletti
F and Fagone P: Involvement of the Nrf2/HO-1/CO axis and
therapeutic intervention with the CO-releasing molecule CORM-A1, in
a murine model of autoimmune hepatitis. J Cell Physiol.
233:4156–4165. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Petralia MC, Mazzon E, Fagone P, Russo A,
Longo A, Avitabile T, Nicoletti F, Reibaldi M and Basile MS:
Characterization of the pathophysiological role of CD47 in uveal
melanoma. Molecules. 24(pii): E24502019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Basile MS, Fagone P, Mangano K, Mammana S,
Magro G, Salvatorelli L, Li Destri G, La Greca G, Nicoletti F,
Puleo S and Pesce A: KCNMA1 expression is downregulated in
colorectal cancer via epigenetic mechanisms. Cancers (Basel).
11(pii): E2452019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Basile MS, Mazzon E, Russo A, Mammana S,
Longo A, Bonfiglio V, Fallico M, Caltabiano R, Fagone P, Nicoletti
F, et al: Differential modulation and prognostic values of
immune-escape genes in uveal melanoma. PLoS One. 14:e02102762019.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fagone P, Caltabiano R, Russo A, Lupo G,
Anfuso CD, Basile MS, Longo A, Nicoletti F, De Pasquale R, Libra M
and Reibaldi M: Identification of novel chemotherapeutic strategies
for metastatic uveal melanoma. Sci Rep. 7:445642017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Rajasekaran D, Zierow S, Syed M, Bucala R,
Bhandari V and Lolis EJ: Targeting distinct tautomerase sites of
D-DT and MIF with a single molecule for inhibition of neutrophil
lung recruitment. FASEB J. 28:4961–4971. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Benedek G, Meza-Romero R, Jordan K, Zhang
Y, Nguyen H, Kent G, Li J, Siu E, Frazer J, Piecychna M, et al: MIF
and D-DT are potential disease severity modifiers in male MS
subjects. Proc Natl Acad Sci USA. 114:E8421–E8429. 2017. View Article : Google Scholar : PubMed/NCBI
|