Introduction
Cervical cancer is associated with some of the
highest rates of morbidities and mortalities in females worldwide
(1). Although the clinical outcome
of therapy in patients improves through early diagnosis, surgical
resection, and chemotherapy/radiation therapy, the prognosis
remains pessimistic (2). The main
reasons for the death of patients with cervical cancer are cancer
progression, metastasis, and drug resistance (3). Therefore, gaining full understanding of
the molecular mechanisms underlying the tumorigenesis and tumor
progression of cervical cancer and developing novel methods of
treatment for patients with this disease is essential.
MicroRNAs (miRNAs/miRs) are a class of small (~22
nucleotides) non-coding RNAs that function as negative regulators
of gene expression by binding to complimentary sequences mainly in
the 3′-untranslated regions (3′-UTRs) of specific mRNAs (4). Accumulating evidence indicates that
miRNAs play a critical role in regulating various biological
processes, such as cell proliferation, metastasis, and drug
resistance (4,5). miR-584 serves as tumor suppressor in
several tumor types (6–13). However, the same miRNA may either
serve as an oncogene or a tumor suppressor, depending on the
characteristic of its target genes (14). Therefore, the biological function and
molecular mechanism of miR-584 in cervical cancer must be
determined.
Glioma-associated oncogene 1 (GLI1) is an important
transcription factor in the Hedgehog signalling pathway that can
regulate transcription and expression of various target genes, such
as c-MYC and PTCH1, thereby affecting cell proliferation,
apoptosis, migration and invasion (15,16).
GLI1 acts as an oncogene and is regulated by miR-361-3p in cervical
cancer (17). However, to the best
of our knowledge, the association between miR-584 and GLI1 has not
been illustrated.
In the current study, the expression level of
miR-584 was analyzed in cervical cancer to illustrate the
biological functions and the molecular mechanism of miR-584.
Materials and methods
Tissue collection
Cervical cancer tissues and adjacent normal tissues
were obtained from 30 patients who underwent surgical resection
between December 2015 and December 2017 at the Department of
Gynaecology and Obstetrics, Weifang Maternity and Child Care
Hospital. Patients were excluded from this study if they were
receiving any anticancer treatment. Tissues were immediately frozen
in liquid nitrogen and stored at −80°C for further usage. The
experimental protocol was approved by the Ethics Committee of
Weifang Maternity and Child Care Hospital, and a signed informed
consent was obtained from all patients before the study.
Cell culture and cell
transfection
Human cervical cancer cell lines (C33A, CaSki, HeLa
and SiHa) and human immortalized normal cervical cell line
Ect1/E6E7 were obtained from American Type Culture Collection. 293T
cells were obtained from The Cell Bank of Type Culture Collection
of the Chinese Academy of Sciences (Shanghai, China). All cell
lines were grown in DMEM (HyClone; GE Healthcare Life Sciences)
supplemented with 10% FBS (HyClone; GE Healthcare Life Sciences),
100 U/ml penicillin and 100 µg/ml streptomycin (Beyotime Institute
of Biotechnology) in a humidified atmosphere at 37°C containing 5%
CO2. The miR-584 mimics (mimics)
(5′-AGUCAAGGUCCAAUUGGUCCGA-3′), miR-584 mimics negative control
(miR-NC) (5′-ACUACUGAGUGACAGUAGA-3′), miR-584 inhibitors
(inhibitors) (5′-GCUCUGCUACACUCGGUACUA-3′) and miR-584 inhibitors
negative control (anti-NC) (5′-UUCUCCGAACGUGUCACGUTT-3′) were
synthesized by Guangzhou RiboBio, Co., Ltd. Full-length GLI1 from
the human cDNA library was cloned into a pcDNA3.1 vector
(Invitrogen; Thermo Fisher Scientific, Inc.). The pcDNA3.1 vector
alone (empty plasmid) served as a negative control. The HeLa cells
were transfected with miR-NC (50 nM) or miR-584 mimics (50 nM)
and/or pcDNA3.1/GLI1 vector (100 nM) or pcDNA3.1 (100 nM) using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
CaSki cells were transfected with anti-NC (50 nM) or miR-584
inhibitors (50 nM) using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Following transfection for 48 h, the cells
were collected for subsequent experiments.
Cell Counting Kit-8 (CCK-8)
assays
HeLa and CaSki cell viability was detected using
CCK-8 (Beyotime Institute of Biotechnology). HeLa and CaSki cells
(1×103 cells/well) were cultured in 96-well plates for
0, 24, 48, and 72 h. At the indicated timepoints, 10 µl of CCK-8
was added to each well and incubated for 3 h. The absorbance of
each well was then determined using Multiskan MK3 (Thermo Fisher
Scientific, Inc.).
Colony formation assay
HeLa and CaSki cells (400 cells/well) were seeded in
six-well plates and cultured in DMEM (HyClone; GE Healthcare Life
Sciences) supplemented with 10% FBS (HyClone; GE Healthcare Life
Sciences) for 10 days. The cells were then fixed with 1 ml 4%
paraformaldehyde (PFA; Beyotime Institute of Biotechnology) and
stained with crystal violet. The number of colonies containing
>50 cells was counted with a light microscope (magnification,
×100; Olympus Corporation).
Transwell assay
The migration and invasion capability of cells was
detected with Corning Transwell chambers (Corning, Inc.). For
invasion capability detection, the Transwell membrane filter was
precoated with 30 µl of Matrigel (BD Biosciences) at 37°C for 4 h.
In the migration and invasion examination, HeLa and CaSki cells
(5×104 cells) were resuspended in 100 µl DMEM without
FBS and transferred to the upper chambers. A total of 600 µl of
DMEM supplemented with 10% FBS was added to the lower chamber.
Cells were incubated for 12 h before being fixed with 4% PFA,
stained with 0.5% crystal violet at room temperature for 20 min,
and the stained cells from six random fields were counted, and the
images were captured under a light microscope (Olympus Corporation;
magnification, ×100).
Western blotting
Tissue samples and the treated cells were lysed in
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology). Protein concentrations were determined using a BCA
Protein Assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal
amounts of protein extracts (30 µg total protein/lane) were
resolved by 10% SDS-PAGE and transferred onto PVDF membranes,
followed by blocking with 5% nonfat dried milk for 1 h at room
temperature. Subsequently, the PVDF membranes were incubated with
primary GLI1 antibody (cat. no. ab49314; 1:500; Abcam) and β-actin
antibody (cat. no. ab8227; 1:1,000; Abcam) overnight at 4°C with
gentle agitation, and then treated with a Horseradish
peroxidase-labeled goat anti-rabbit secondary antibody (1:1,000;
cat. no. ab150077; Abcam) for 2 h at room temperature. β-actin was
used as a loading control. The protein bands were visualized using
an enhanced chemiluminescence system (Beyotime Institute of
Biotechnology).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA from cells and tissues was extracted with
TRIzol reagent (Thermo Fisher Scientific, Inc.). cDNA was
synthesized using a TaqMan® MicroRNA Reverse
Transcription kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. To quantify the miRNA and mRNA, a qPCR
assay was performed using iQ™ SYBR® Green Supermix
(Bio-Rad Laboratories, Inc.) in an iCycler iQ™ qPCR detection
system (Bio-Rad Laboratories, Inc.). The relative expression levels
of miR-584 and GLI1 were calculated as the inverse log of
ΔΔCq and normalized to the reference gene (18). The thermocycling conditions were as
follows: 95°C for 10 min; followed by 40 cycles of 95°C for 15 sec
and 60°C for 1 min; annealing at 55°C for 30 sec; and elongation at
72°C for 3 min. To analyze GLI1 mRNA expression, β-actin acted as
an internal control. To examine the expression of miR-584, U6 was
used as the internal control. The following primer pairs were used:
miR-584 forward, 5′-TGCAATGTGTGTGTTAGCCA-3′, and reverse,
5′-ATCATTGCTCCTTGGATGGT-3′; GLI1 forward,
5′-TACTCACGCCTCGAAAACCT-3′ and reverse, 5′-AGGACCATGCACTGTCTTGA-3′;
U6 forward, 5′-TGCGGGTGCTCGCTTCGCAGC-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′; β-actin forward,
5′-GATCATTGCTCCTCCTGAGC-3′ and reverse,
5′-ACTCCTGCTTGCTGATCCAC-3′.
Flow cytometry
Transfected HeLa and CaSki cells were treated with
10 µM cisplatin (Jiangsu Hansoh Pharmaceutical Co. Ltd.) for 24 h
at 37°C. The apoptosis rate of cells was then detected using an
Annexin V-PI Assay kit (Beyotime Institute of Biotechnology)
according to the manufacturer's instructions. A flow cytometer
(Cytomics FC 500 MPL; Beckman Coulter, Inc.) was used to analyze
the rate of apoptosis in each sample. Data were analyzed using
ModFit LT 3.0 (Verity Software House, Inc.).
Luciferase reporter assay
The wild-type (WT) or mutant (MUT) GLI1-3′UTR, which
contained the miR-584 binding sites, was inserted into the
psiCHECK2 vector (Promega Corporation) 293T cells (1×105
cells/well) were co-transfected with 0.1 mg psiCHECK2-WT GLI1-3e
miR-5r 0.1 mg psiCHECK2-MUT GLI1-3′-UTR and 10 nM miR-584 mimics or
10 nM miR-584 inhibitors using Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. Cells were cultured at 37°C for 48 h
and luciferase activities were analyzed using dual-luciferase kit
(GeneCopoeia, Inc.) according to the manufacturer's protocol. The
activity of firefly luciferase was normalized to the corresponding
Renilla luciferase activity.
Bioinformatics prediction
To investigate the possible target genes of miR-584,
the online prediction system, TargetScan 7.1 software (http://www.targetscan.org), was used.
Statistical analysis
Results are presented as the mean ± SEM.
Significance was established using the SPSS 13.0 software (SPSS,
Inc). Data were analyzed using a Student's t-test or one-way
analysis of variance followed by Tukey's Honest Significant
Difference test. Pearson's correlation analysis was used to analyze
the correlation between miR-584 and GLI1 mRNA expression. P<0.05
was considered to indicate a statistically significant
difference.
Results
Expression of miR-584 is downregulated
in human cervical cancer tissues and cells
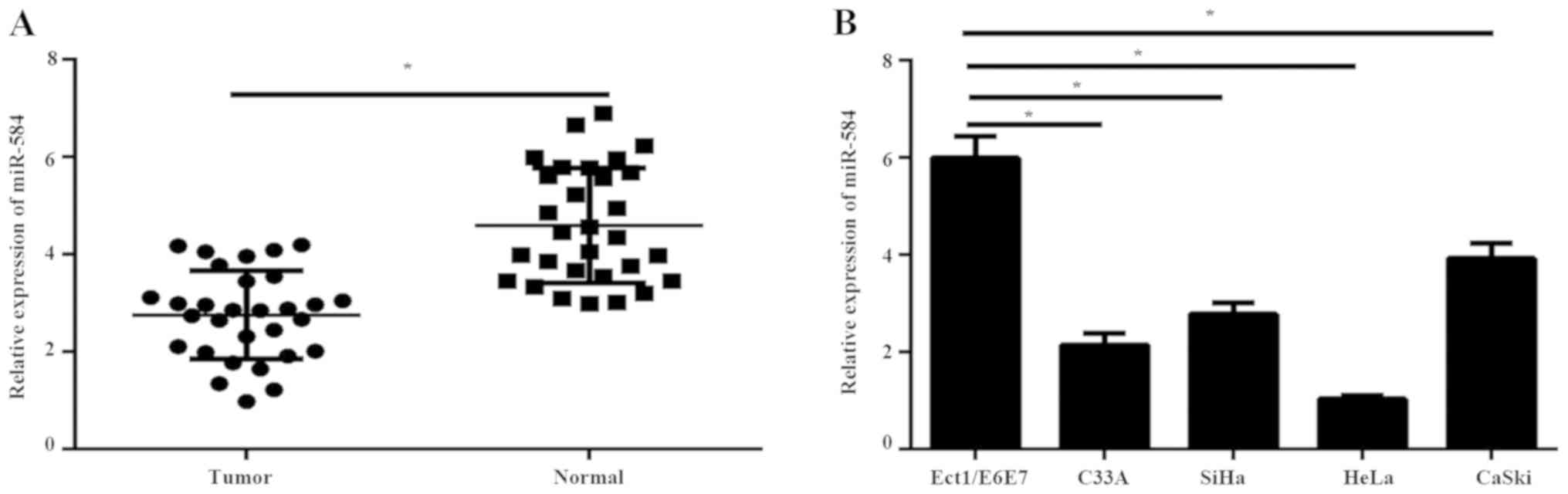
To explore the role of miR-584 in cervical cancer,
miR-584 expression was first detected in 30 pairs of cervical
cancer tissues and adjacent normal tissues by RT-qPCR. RT-qPCR
results illustrated that the expression of miR-584 was
significantly downregulated in tumor tissues compared with normal
tissues (Fig. 1A). In addition, the
expression levels of miR-584 were analyzed in immortalized normal
cervical cell line Ect1/E6E7 and four types of cervical cancer
cells (C33A, SiHa, HeL and CaSki) using RT-qPCR. The results showed
that the expression of miR-584 in cervical cancer cell lines was
significantly reduced compared with Ect1/E6E7 cells (Fig. 1B).
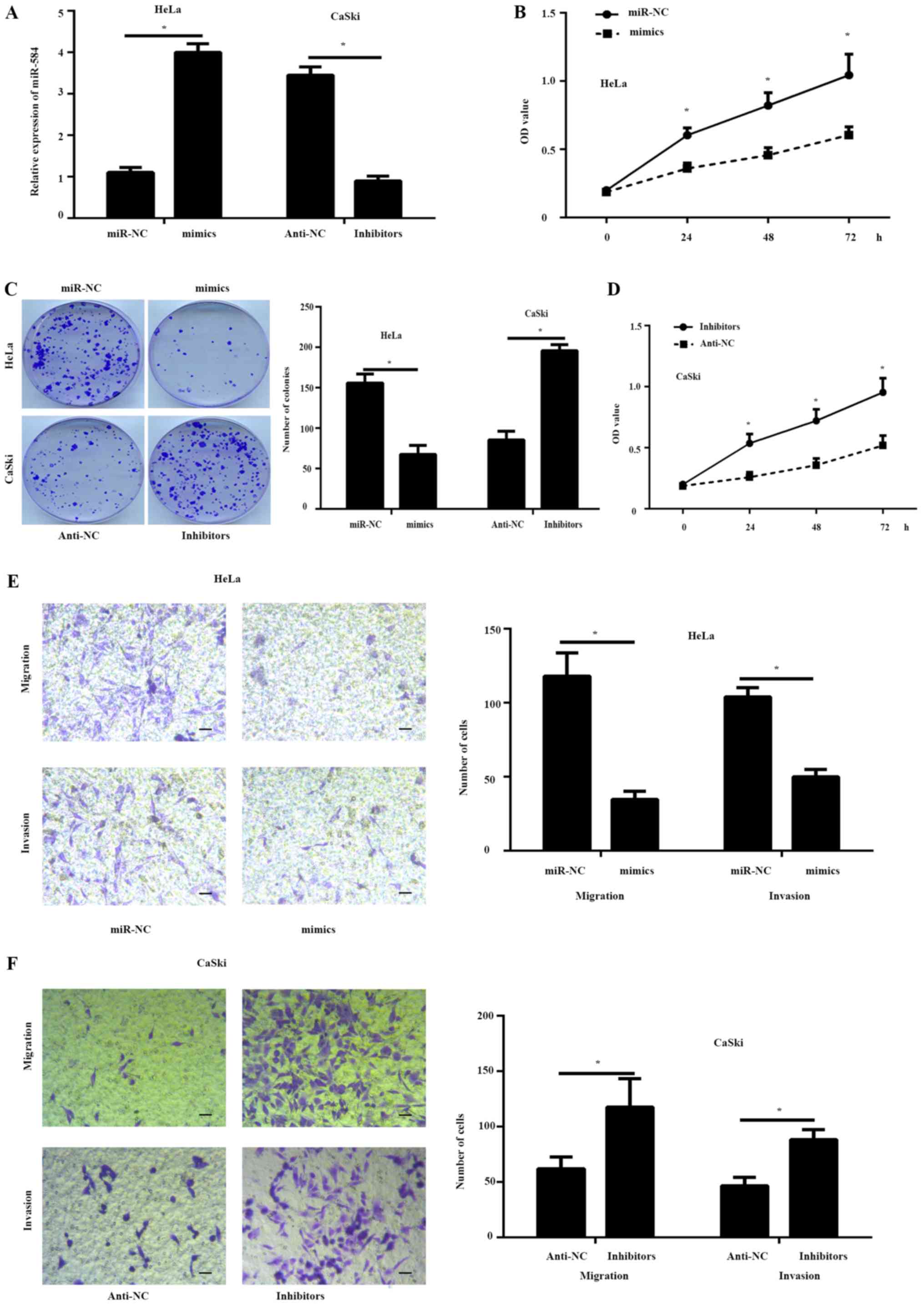
miR-584 inhibits cervical cancer cell
proliferation and metastasis
To study the effects of miR-584 in cervical cancer
progression, miR-584 overexpression or inhibition assays were
performed in HeLa and CaSki cells, which contained the lowest or
highest endogenous miR-584 expression levels, respectively. The
results of the RT-qPCR assay illustrated that miR-584 expression
was significantly increased in HeLa cells and significantly
downregulated in CaSki cells when compared with controls (Fig. 2A). The results of the CCK-8 (Fig. 2B) and colony formation assay
(Fig. 2C) illustrated that the
proliferation of HeLa cells transfected with miR-584 mimics was
markedly inhibited compared with the miR-NC group. Conversely, a
significant increase in cell proliferation was observed in CaSki
cells transfected with miR-584 inhibitors when compared with
controls (Fig. 2C and D).
Furthermore, the Transwell assay illustrated that the migration and
invasion ability of the HeLa cells transfected with miR-584 mimics
markedly decreased compared to the miR-NC group, while the
silencing of miR-584 increased the migration and the invasion
capability of the CaSki cells (Fig. 2E
and F).
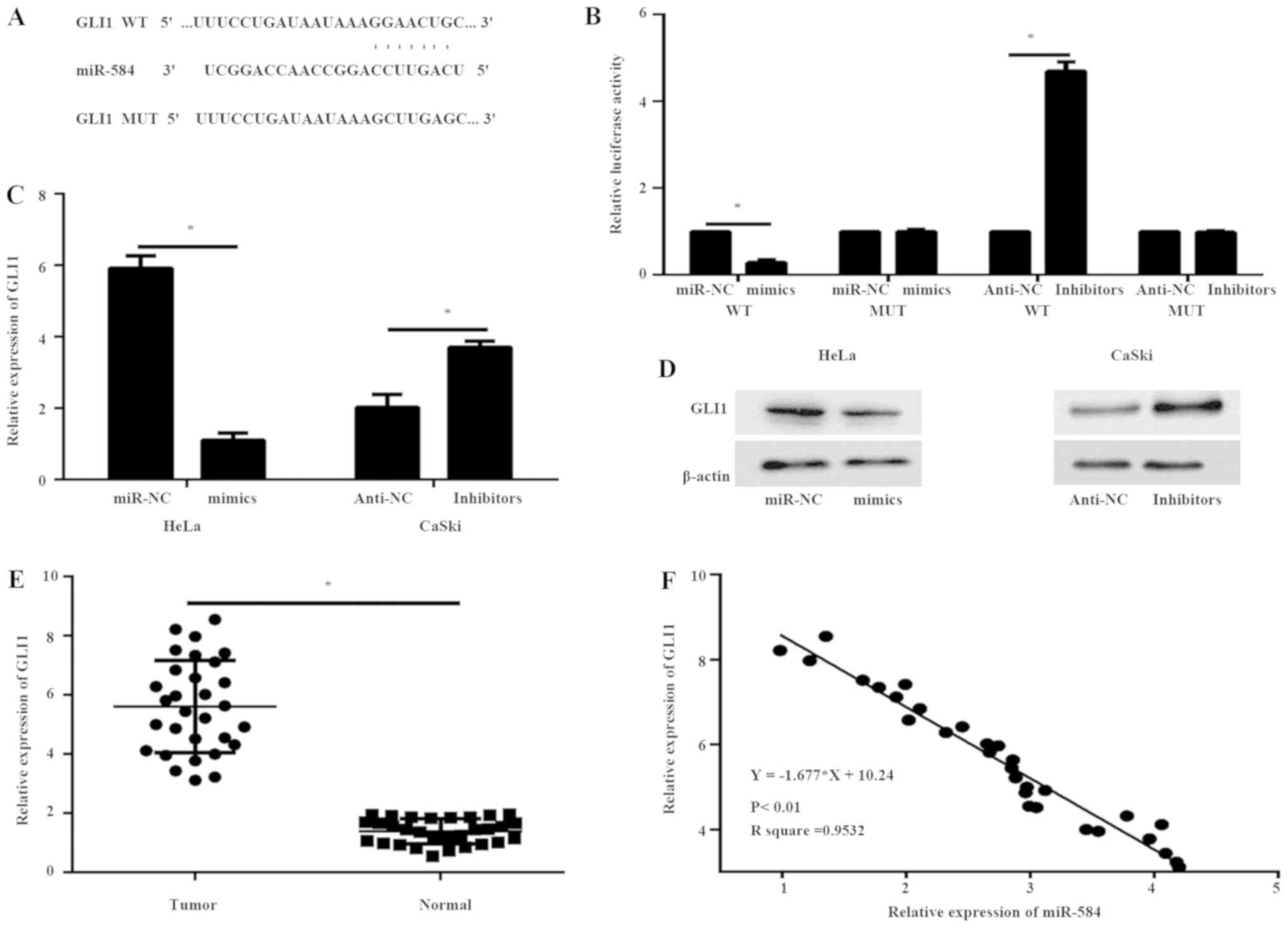
GLI1 is a molecular target gene of
miR-584
To assess the underlying mechanisms of miR-584 in
the progression of cervical cancer cells, Targetscan was used to
predict potential target genes of miR-584. GLI1 mRNA 3′-UTR was
found to contain highly conserved binding sites for miR-584
(Fig. 3A). A luciferase reporter
assay was performed to analyze the association between GLI1 and
miR-584. A miR-584 mimic or inhibitor and a luciferase reporter
plasmid containing a wt or mut 3′-UTR binding site of human GLI1
were co-transfected into 293T cells. miR-584 mimics significantly
decreased the luciferase activity in 293T cells containing the GLI1
wt 3′-UTR but failed to suppress this activity in cells with the
mut GLI1 3′-UTR, while the miR-584 inhibitors enhanced the
luciferase activity in 293T cells containing the GLI1 wt 3′-UTR
(Fig. 3B). These data demonstrated
that GLI1 is a specific target of miR-584 (Fig. 3B). Furthermore, the results of the
RT-qPCR and western blotting illustrated that overexpression of
miR-584 downregulated the expression of GLI1 in HeLa cells both at
mRNA (Fig. 3C) and protein (Fig. 3D) levels compared with miR-NC cells,
while the silencing of miR-584 in CaSki cells resulted in opposite
results, further confirming that GLI1 is a target gene of miR-584.
The mRNA expression levels of GLI1 were subsequently analyzed in
breast cancer samples and corresponding normal tissues using
RT-qPCR. The expression of GLI1 at the mRNA level was significantly
higher in breast cancer tissues than in adjacent normal tissues
(Fig. 3E). In addition, Pearson's
correlation analysis (linear regression analysis) showed that the
mRNA expression level of GLI1 was inversely correlated with the
expression of miR-584 in breast cancer tissues (Fig. 3F).
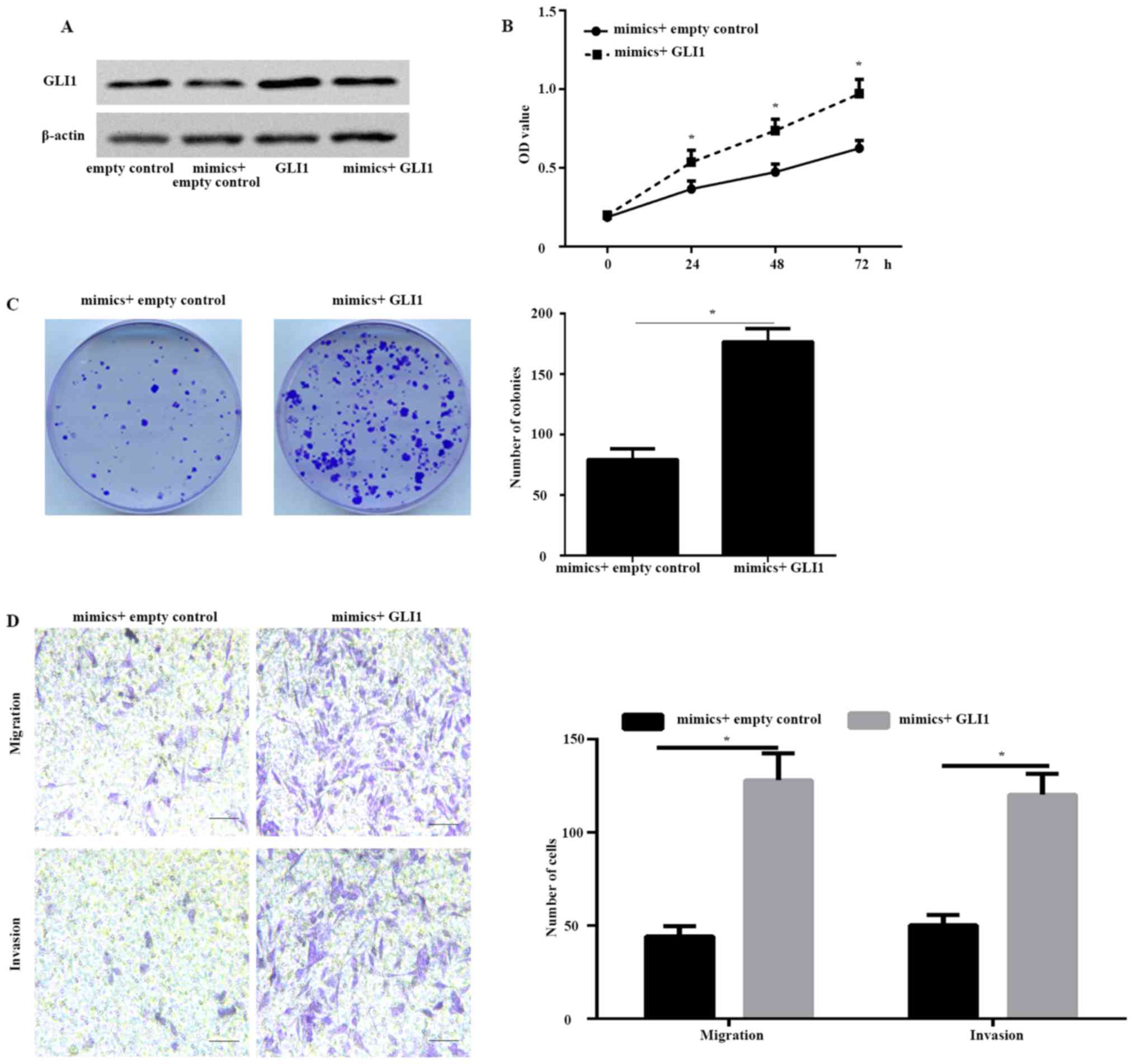
GLI1 is a functional target gene of
miR-584
To further illustrate whether GLI1 is a function
target gene of miR-584, HeLa cells were co-transfected with miR-584
mimic and GLI1 plasmid or empty plasmid. The western blot results
illustrated that the protein expression of GLI1 was markedly
increased in HeLa cells co-transfected with both miR-584 mimic and
GLI1 plasmid compared with cells co-transfected with miR-584 mimic
and empty plasmid (Fig. 4A). The
CCK-8 (Fig. 4B) and colony formation
assay (Fig. 4C) illustrated that the
overexpression of GLI1 significantly rescued the proliferation rate
decreased by miR-584 mimics. Furthermore, the Transwell assay
illustrated that the metastatic capability of HeLa cells
co-transfected with both miR-584 mimics and GLI1 plasmid was
enhanced compared with HeLa cells co-transfected with miR-584 mimic
and empty plasmid (Fig. 4D). These
data illustrated that GLI1 was a functional target gene of
miR-584.
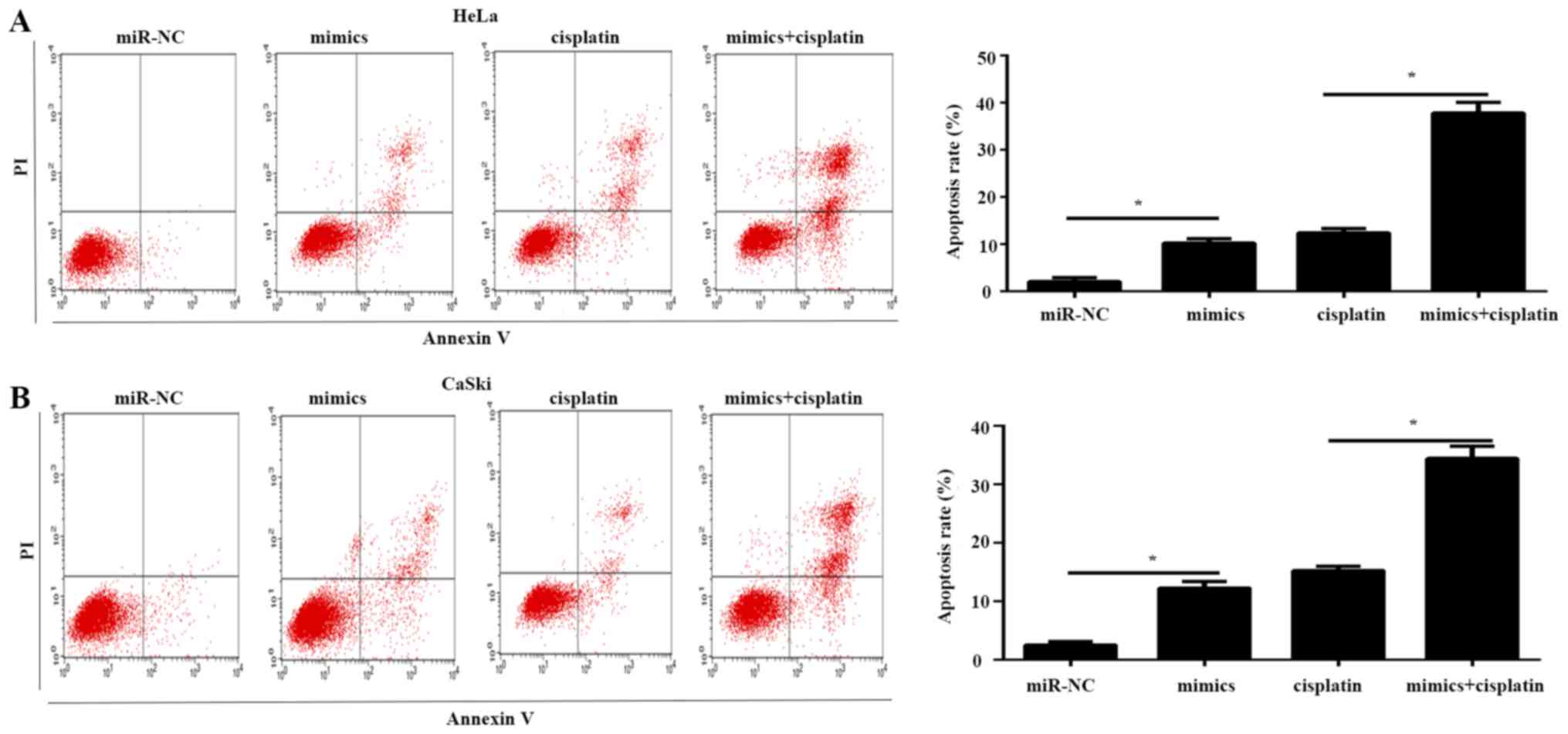
miR-584 promotes drug sensitivity to
cisplatin in cervical cancer cells
The association between miR-584 and cisplatin was
determined. Flow cytometry was used to analyze the effects of
miR-584 on the drug sensitivity of HeLa and CaSki cells. The
results illustrated that overexpression of miR-584 increased the
proportion of apoptotic cells compared with the miR-NC group in
HeLa and CaSki cells (Fig. 5A and
B). In addition, the combination of miR-584 and cisplatin
significantly enhanced the apoptosis rate of HeLa and CaSki cells
compared with cisplatin or miR-584 mimics, respectively (Fig. 5A and B). These data suggested that
miR-584 enhanced the sensitivity of cervical cancer cells to
cisplatin.
Discussion
A number of studies have illustrated the
relationship between cancer progression and deregulated miRNA
expression (19). Several miRNAs act
as either tumor suppressors or oncogenes involved in cervical
cancer progression (19). Decreased
expression of tumor suppressor miRNAs leads to enhanced oncogene
translation, which in turn enhances tumor development. Similar
effects are caused by oncogenic miRNA overexpression, which
contributes to the inhibition of tumor suppressor genes (20).
miR-584 acts as a tumor suppressor and is
downregulated in some types of cancer (6–13).
However, to the best of our knowledge, the expression and functions
of miR-584 in cervical cancer have not been elucidated. In the
present study, the expression of miR-584 was markedly downregulated
in cervical cancer tissues and cell lines compared with adjacent
normal cervical tissues and Ect1/E6E7 cells. The current study
illustrated the role of miR-584 in cervical cancer cell
proliferation, metastasis and apoptosis. The results suggested that
miR-584 overexpression inhibited the viability, proliferation,
migration and invasion of these cells and enhanced the apoptosis
rate in vitro.
Recent studies confirmed that miRNAs inhibit the
expression of specific target genes, which leads to tumor
occurrence. In medulloblastoma, miR-584 was downregulated in tumor
tissues compared with normal tissues, and histone deacytelase 1 and
eIF4E3 were the direct target genes of miR-584 (21). In lung cancer, miR-584 was
downregulated and inhibited cell growth and metastasis by directly
targeting metadherin (6). In gastric
cancer, miR-584 directly targeted the matrix metalloproteinase-14
(MMP-14) promoter to repress YY1-facilitated MMP-14 expression and
inhibited gastric cancer progression (7). In the current study, miR-584 directly
targeted GLI1 and negatively regulated the expression of GLI1 to
inhibit cervical cancer progression. GLI proteins, including GLI1,
GLI2 and GLI3, are zinc finger transcription factors and are the
main effectors of the Hedgehog signalling (22). In primary cilium, GLI1 dissociates
from the negative regulator suppressor of fused, is converted into
its activated form and translocates to the nucleus (22). The translocation of GLI1 enhances the
downstream target oncogene expression, such as cyclin D1 and
homeobox protein NANOG (22). GLI1
is overexpressed in numerous types of cancer, such as non-small
cell lung cancer, gastric cancer, pancreatic cancer and colon
cancers, and acts as an oncogene in these tumors (15,23). The
current study confirmed that GLI1 is upregulated in cervical cancer
tissues, as reported previously (24). miR-584 decreased the expression of
GLI1 through the direct binding of the 3′-UTR of GLI1. In addition,
the inverse correlation between miR-584 expression and mRNA
expression of GLI1 in cervical cancer tissues further supported
this conclusion. Moreover, GLI1 overexpression reduced the effects
of miR-584 on cell survival and metastasis in cervical cancer.
Together, these findings suggested that GLI1 was indeed a direct
target gene of miR-584.
Chemoresistance in cancer is the main cause of
treatment failure (3). Recent data
indicated that aberrant miRNA expression was closely linked to
chemoresistance by targeting genes related to chemosensitivity or
chemoresistance (25). However,
specific chemoresistance-related miRNAs are largely unknown.
Cisplatin resistance is a major obstacle to the successful
treatment of cervical cancer (3).
The current study further illustrated the association between the
sensitivity of cells to cisplatin and miR-584 in cervical cancer.
miR-584 had a negative impact on cisplatin resistance in HeLa
cells. In the current study, it was indicated that GLI1 was a
direct target gene of miR-584. Therefore, the underlying mechanism
of miR-584 to drug resistance may be through GLI1.
In conclusion, the current study demonstrated that
miR-584 is markedly downregulated in human cervical cancer tissues
and cell lines. Overexpression of miR-584 inhibited proliferation,
invasion, and migration, while enhancing cell apoptosis rates and
chemosensitivity to cisplatin in cervical cells. GLI1 was
identified as the molecular and biological target gene of miR-584.
These data illustrated that miR-584 may serve as a novel
therapeutic target in cervical cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TW and AZ conceived and designed the experiments.
TW, JF and AZ conducted all of the experiments. TW and JF wrote and
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Weifang Maternity and Child Care Hospital. Prior written informed
consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Burki TK: Cervical cancer: Screening and
risk with age. Lancet Oncol. 15:e1072014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Waggoner SE: Cervical cancer. Lancet.
361:2217–2225. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen J, Stass SA and Jiang F: MicroRNAs as
potential biomarkers in human solid tumors. Cancer Lett.
329:125–136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Wang Y and Wang J: MicroRNA-584
inhibits cell proliferation and invasion in non-small cell lung
cancer by directly targeting MTDH. Exp Ther Med. 15:2203–2211.
2018.PubMed/NCBI
|
|
7
|
Zheng L, Chen Y, Ye L, Jiao W, Song H, Mei
H, Li D, Yang F, Li H, Huang K and Tong Q: miRNA-584-3p inhibits
gastric cancer progression by repressing Yin Yang 1-facilitated
MMP-14 expression. Sci Rep. 7:89672017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Q, Li Z, Wei S, Wang W, Chen Z, Zhang
L, Chen L, Li B, Sun G, Xu J, et al: Overexpression of miR-584-5p
inhibits proliferation and induces apoptosis by targeting WW
domain-containing E3 ubiquitin protein ligase 1 in gastric cancer.
J Exp Clin Cancer Res. 36:592017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Orlandella FM, Di Maro G, Ugolini C,
Basolo F and Salvatore G: TWIST1/miR-584/TUSC2 pathway induces
resistance to apoptosis in thyroid cancer cells. Oncotarget.
7:70575–70588. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xue H, Guo X, Han X, Yan S, Zhang J, Xu S,
Li T, Guo X, Zhang P, Gao X, et al: MicroRNA-584-3p, a novel tumor
suppressor and prognostic marker, reduces the migration and
invasion of human glioma cells by targeting hypoxia-induced ROCK1.
Oncotarget. 7:4785–4805. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xiang J, Wu Y, Li DS, Wang ZY, Shen Q, Sun
TQ, Guan Q and Wang YJ: miR-584 suppresses invasion and cell
migration of thyroid carcinoma by regulating the target oncogene
ROCK1. Oncol Res Treat. 38:436–440. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang XP, Deng XL and Li LY: MicroRNA-584
functions as a tumor suppressor and targets PTTG1IP in glioma. Int
J Clin Exp Pathol. 7:8573–8582. 2014.PubMed/NCBI
|
|
13
|
Ueno K, Hirata H, Shahryari V, Chen Y,
Zaman MS, Singh K, Tabatabai ZL, Hinoda Y and Dahiya R: Tumor
suppressor microRNA-584 directly targets oncogene Rock-1 and
decreases invasion ability in human clear cell renal cell
carcinoma. Br J Cancer. 104:308–315. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang L, Wang B, Fang M, Guo F and Cui M:
Identification of microRNAs and target genes involved in serous
ovarian carcinoma and their influence on survival. Eur J Gynaecol
Oncol. 35:655–661. 2014.PubMed/NCBI
|
|
15
|
Didiasova M, Schaefer L and Wygrecka M:
Targeting GLI transcription factors in cancer. Molecules. 23(pii):
E10032018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mastrangelo E and Milani M: Role and
inhibition of GLI1 protein in cancer. Lung Cancer. 9:35–43.
2018.PubMed/NCBI
|
|
17
|
Zhao D and Cui Z: MicroRNA-361-3p
regulates retinoblastoma cell proliferation and stemness by
targeting hedgehog signaling. Exp Ther Med. 17:1154–1162.
2019.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hasanzadeh M, Movahedi M, Rejali M, Maleki
F, Moetamani-Ahmadi M, Seifi S, Hosseini Z, Khazaei M, Amerizadeh
F, Ferns GA, et al: The potential prognostic and therapeutic
application of tissue and circulating microRNAs in cervical cancer.
J Cell Physiol. 234:1289–1294. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: Oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Abdelfattah N, Rajamanickam S, Panneerdoss
S, Timilsina S, Yadav P, Onyeagucha BC, Garcia M, Vadlamudi R, Chen
Y, Brenner A, et al: MiR-584-5p potentiates vincristine and
radiation response by inducing spindle defects and DNA damage in
medulloblastoma. Nat Commun. 9:45412018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sabol M, Trnski D, Musani V, Ozretic P and
Levanat S: Role of GLI transcription factors in pathogenesis and
their potential as new therapeutic targets. Int J Mol Sci. 19(pii):
E25622018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Skoda AM, Simovic D, Karin V, Kardum V,
Vranic S and Serman L: The role of the Hedgehog signaling pathway
in cancer: A comprehensive review. Bosn J Basic Med Sci. 18:8–20.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chaudary N, Pintilie M, Hedley D, Fyles
AW, Milosevic M, Clarke B, Hill RP and Mackay H: Hedgehog pathway
signaling in cervical carcinoma and outcome after chemoradiation.
Cancer. 118:3105–3115. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Çalışkan M, Güler H and Bozok Çetintaş V:
Current updates on microRNAs as regulators of chemoresistance.
Biomed Pharmacother. 95:1000–1012. 2017. View Article : Google Scholar : PubMed/NCBI
|