Introduction
Hepatocellular carcinoma (HCC) is a leading cause of
cancer-associated mortality worldwide. Although the clinical
application of sorafenib, regorafenib and nivolumab has achieved
significant progress in the treatment of HCC, the survival rate of
patients with HCC remains low, as most patients with HCC are
diagnosed at an advanced stage (1).
Cisplatin and various platinum agents have become standard drugs
for the treatment of HCC (2).
However, as cisplatin resistance may occur, the survival benefit
for patients with advanced HCC is unsatisfactory (3). To improve the survival of patients with
HCC, novel biomarkers for early diagnosis and the development of
novel therapeutic targets for cisplatin-resistant HCC are required
(4).
Liver cancer is associated with hepatitis B virus
(HBV), HCV and non-alcoholic fatty liver disease (5). However, the molecular mechanisms of the
genesis of HCC remain poorly understood. Recently, new evidence has
demonstrated that different types of non-coding RNAs (ncRNAs),
including microRNAs (miRNAs), long ncRNAs and partially circular
RNAs (circRNAs), have a role in HCC (6). High-throughput next-generation
sequencing analysis has identified a large number of circRNAs that
are involved in liver cancer through interactions with miRNAs or
proteins (7,8).
CircRNAs are generated from backsplicing of exons
and introns, forming a circular exonic circRNA, a circular intronic
RNA and an exon-intron circRNA. CircRNAs have a covalently closed
continuous loop structure. They cannot be degraded by RNA
exonuclease or RNase R. Therefore, circRNAs are suitable as
diagnostic biomarkers for tumors, including HCC (9). CircRNAs are abundantly expressed in
tissues, blood and microvesicles and are highly conserved among
different species. CircRNAs are able to regulate gene expression at
the transcriptional or post-transcriptional level (10). CircRNA is involved in various
biological processes, including HCC cell proliferation, apoptosis
and metastasis (11). For instance,
overexpression of Homo sapiens circRNA_0001649 inhibits the
proliferation and invasion of HCC cells (12). CircRNA circMTO1 (mitochondrial tRNA
translation optimization 1) inhibits HCC progression by acting as a
sponge for miRNA-9. Reduced levels of circMTO1 in HCC tissue may be
used as a predictor of poor survival (13).
In the present study, a circRNA array was performed
to screen for differentially expressed circRNAs in HCC tissues. The
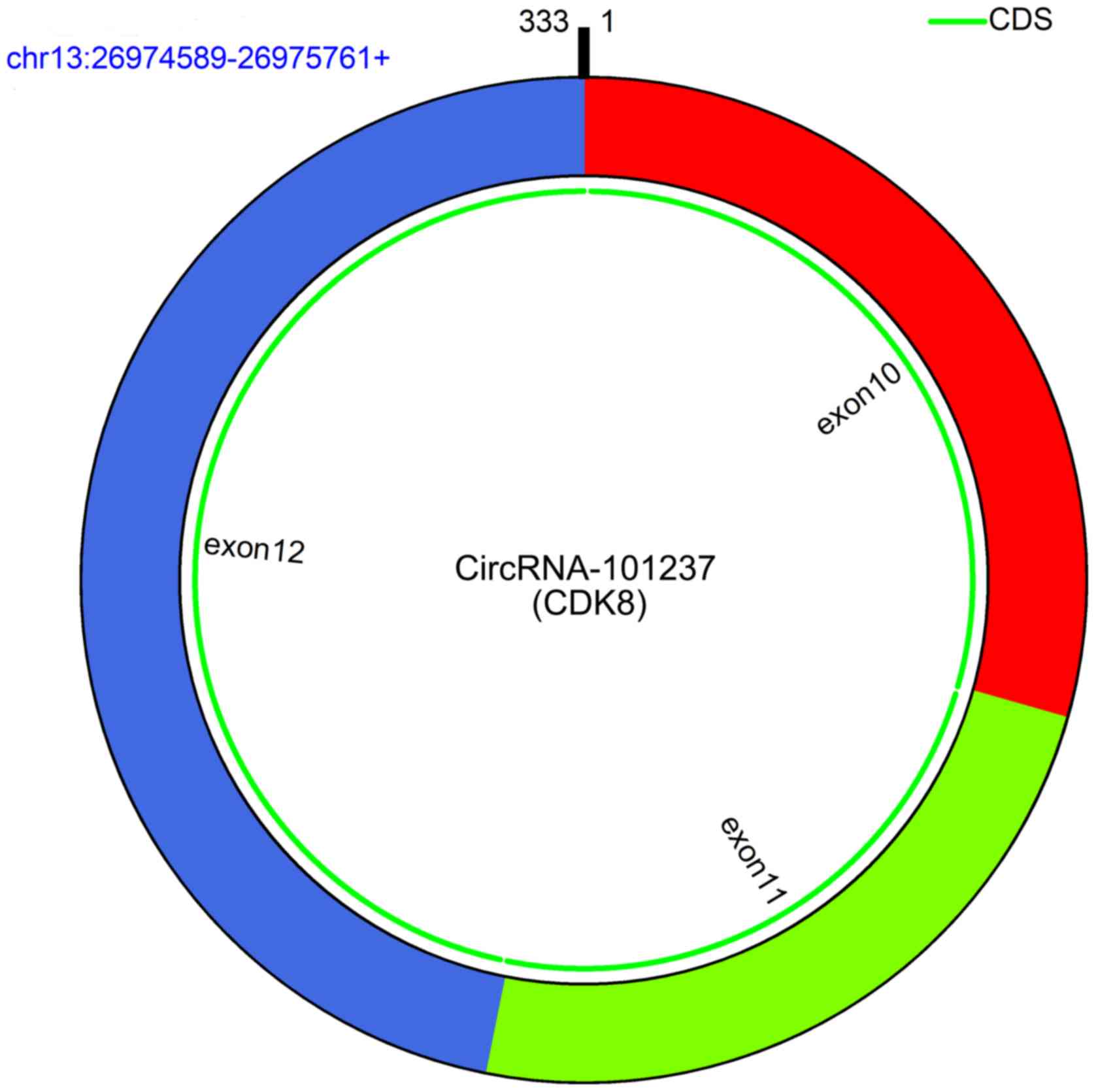
study then focused on the newly identified circRNA_101237, whose
encoding gene is located at chromosome (chr)13:26974589-26975761
and which is produced by backsplicing of exons 10, 11 and 12 of
cyclin-dependent kinase (CDK)8 (Fig.
1). CircRNA_101237 is upregulated in tumor tissues and serum of
patients with HCC compared with that in paracancerous tissues and
serum of healthy controls, respectively. In addition, the
association between serum circRNA_101237 levels and the clinical
outcome in patients with HCC was assessed.
Materials and methods
Patients and samples
A total of 100 HCC cancer tissues and matched
adjacent tissues were collected from Huainan First People's
Hospital and the First Affiliated Hospital of the Medical College
of Anhui University of Science and Technology (Huainan, China)
between September 2013 and September 2017. The serum samples from
another independent cohort, including 120 healthy individuals who
came for physical examination and 234 patients with HCC were also
collected from Huainan First People's Hospital and the First
Affiliated Hospital of the Medical College of Anhui University of
Science and Technology between September 2013 and September 2017.
The general demographic and clinicopathological characteristics of
234 patients with HCC are shown in Table SI. The medical records of HCC
patients with clinical TNM staging and survival information were
collected. Patients with cisplatin-resistant HCC were defined as
those with persistent disease at >6 weeks and those with
recurrent disease at >2 months after completion of
cisplatin-based chemotherapy. Patients with cisplatin-sensitive HCC
were defined as those without local residual lesions at 6 weeks or
no recurrence at 2 months after completion of cisplatin-based
chemotherapy. The cisplatin-based chemotherapy regimen consisted of
doxorubicin 60 mg/m2, followed by cisplatin 60
mg/m2 infused over 30 min on day 1. Chemotherapy cycles
were repeated every 21 days for 3 cycles (14).
Cell culture and treatment
The HCC cell lines HCCLM3, Hep3B and MHCC97-H were
obtained from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. All of the cells were grown routinely
in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) and cultured at 37˚C in a humidified atmosphere
with 5% CO2.
The HCC cell lines were exposed to cisplatin at 0,
0.5, 1 and 2 µg/ml for 48 h or at 1 µg/ml for 0, 12, 24 or 48 h,
and the expression of circRNA_101237 was then assessed by reverse
transcription-quantitative (RT-q) PCR.
Cisplatin-resistant cells
Parental Huh7 cells were obtained from The Cell Bank
of Type Culture Collection of the Chinese Academy of Sciences and
cultured in DMEM medium containing 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.). The cisplatin-resistant cells
(Huh7/DDP) were established as previously described (15). Briefly, Huh7 cells were treated with
a low concentration (10 ng/ml) of cisplatin (Sigma-Aldrich; Merck
KGaA) for 72 h. The medium containing 10 ng/ml of cisplatin was
refreshed every 3 days for a total of 5 times. Further resistance
was established by gradually raising the concentration of cisplatin
in the culture solution until a target resistance of 5 µg/ml
cisplatin was acquired.
CircRNA array
A total of 3 HCC tissues and the matched adjacent
tissue samples were randomly selected for circRNA microarray. The
circRNA microarray data were analyzed using Arraystar Human circRNA
Array V2 analysis (Arraystar) by Kangchen BioTech Inc. In brief,
total RNAs were digested with RNase R to remove linear RNAs and
enrich circular RNAs. The enriched circular RNAs were amplified and
transcribed into fluorescent circRNA utilizing a random priming
method (Arraystar Super RNA Labeling Kit; Arraystar). The labeled
circRNAs were hybridized onto the Arraystar Human circRNA Array V2
(8x15K; Arraystar). Following washing of the slides, the arrays
were scanned by the Agilent Scanner G2505C. Agilent Feature
Extraction software (version 11.0.1.1) was used to analyze the
acquired array images. Differentially expressed circRNAs were then
identified by analyzing the fold change, as well as the P-value.
The threshold for significantly up- and downregulated genes was set
as fold change >2.0 and P<0.05.
RT-qPCR analysis
Total RNA was extracted with TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) and reverse
transcribed to complementary DNA by using SuperScript™ IV VILO™
Master Mix with ezDNase™ Enzyme (cat. no. 11766050; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. PCR
amplification was then performed with TaqMan Fast Advanced Master
Mix (cat. no. 4444558; Thermo Fisher Scientific, Inc.) according to
the manufacturers' protocol. β-actin expression was assessed as an
endogenous control. qPCR was performed using the following
conditions: 50˚C for 2 min, 95˚C for 2 min and 40 circles of 95.0˚C
for 1 sec and 60˚C for 20 sec. The primers used were as follows:
circRNA_101237 forward, 5'-TGAGCTTGTGAGTGAGTGGT-3' and reverse,
5'-GCAAGGAGAATGGCGAGATG-3'; β-actin forward,
5'-TTGTTACAGGAAGTCCCTTGCC-3' and reverse,
5'-ATGCTATCACCTCCCCTGTGTG-3'. The 2-∆∆Cq method was used
to analyze the qPCR data (16).
Statistical analysis
Experiments were performed as three independent
replicates. Values are expressed as the mean ± standard deviation
and statistical analysis was performed using SPSS 17.0 statistical
software (SPSS, Inc.). Differences among the groups were estimated
by Student's t-test or one-way analysis of variance with Tukey's
post-hoc test. The cases of HCC were divided into a high
circRNA_101237 expression group (expression above the mean value)
or otherwise into a low circRNA_100053 expression group. Good
prognosis of patients with HCC was defined as a five-year overall
survival probability of ≥60%, while a lower probability was defined
as poor prognosis. The overall survival rate estimates over time
were calculated using the Kaplan-Meier method with log-rank tests.
The association between circRNA_101237 expression and
clinicopathological variables of HCC patients was evaluated using
the Chi-squared test. Univariate and multivariate logistic
regression analyses using the Cox proportional hazards model were
performed to analyze prognostic factors. P<0.05 was
considered to indicate statistical significance.
Results
CircRNA_101237 is upregulated in tumor
tissues and peripheral blood serum from patients with HCC
To investigate the role of circRNAs in HCC, circRNA
microarray was performed to screen differentially expressed
circRNAs. Upregulation was seen in 65 circRNAs and downregulation
was seen in 87 circRNAs in HCC tissues vs. adjacent controls. As
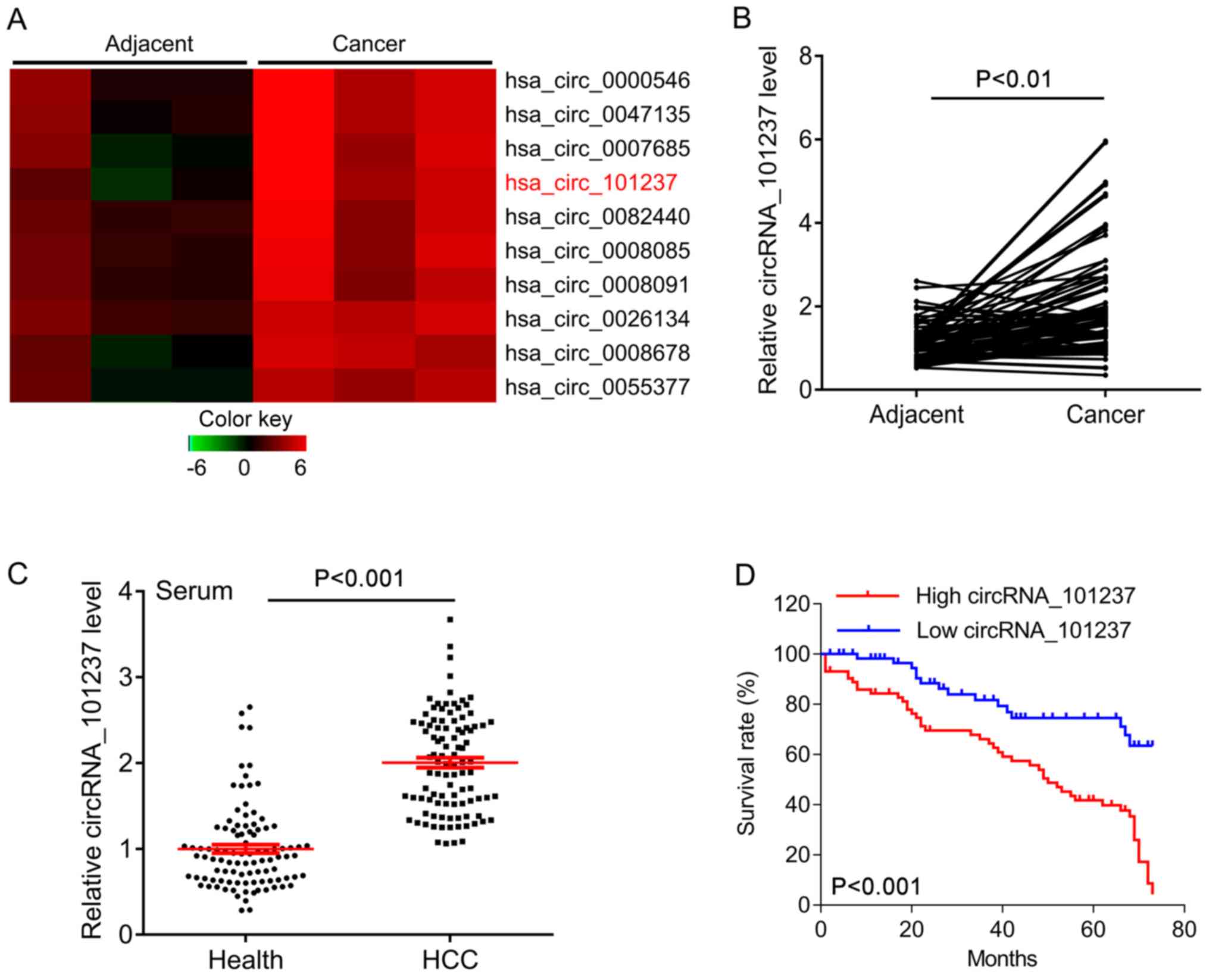
indicated in the heatmap, circRNA_101237 was significantly
upregulated in HCC tissues compared with that in the adjacent
controls (Fig. 2A). The expression
of circRNA_101237 was further confirmed in 100 HCC tissues and the
adjacent tissues. The quantitative results suggested that
circRNA_101237 was significantly increased in HCC tissues compared
with that in the adjacent tissues (Fig.
2B). To further investigate whether circRNA_101237 in the
peripheral blood may serve as a diagnostic biomarker for HCC,
circRNA_101237 expression was measured in an independent cohort,
including serum samples from 234 patients with HCC and 120 healthy
controls. CircRNA_101237 was also significantly upregulated in
serum samples from patients with HCC compared with those from
healthy controls (Fig. 2C).
High circRNA_101237 expression is
associated with poor outcome for patients with HCC
The association between circRNA_101237 expression
and clinicopathological features of HCC patients was then analyzed.
The HCC patients were stratified into a high circRNA_101237 group
and low circRNA_101237 group based on the mean level of
circRNA_101237. The results indicated that the expression of
circRNA_101237 was associated with tumor size (P<0.001), lymph
node metastasis (P=0.006), distant metastasis (P=0.0002), TNM stage
(P=0.0002) and Barcelona Clinic Liver Cancer (BCLC) stage
(P<0.001; Table I). In addition,
univariate analysis suggested that the serum levels of
circRNA_101237 (hazard ratio=3.29, P=0.01), as well as the tumor
size (hazard ratio=3.24, P=0.01), lymph node metastasis (hazard
ratio=2.76, P=0.03), distant metastasis (hazard ratio=5.72,
P=0.01), BCLC stage (hazard ratio=2.87, P=0.02) and TNM stage
(hazard ratio=4.15, P=0.03) were significantly associated with the
prognosis of patients with HCC (Table
II). Multivariate analysis revealed that the serum levels of
circRNA_101237 (hazard ratio=3.42, P=0.02), tumor size (hazard
ratio=3.14, P=0.03), lymph node metastasis (hazard ratio=3.76,
P=0.02), distant metastasis (hazard ratio=4.35, P=0.01), BCLC stage
(hazard ratio=3.25, P=0.03) and TNM stage (hazard ratio=3.93,
P=0.03) were independent prognostic factors for the survival of
patients with HCC (Table III). The
association between serum levels of circRNA_101237 in HCC patients
and overall survival was then further analyzed. The Kaplan-Meier
survival curves indicated that the patients with high
circRNA_101237 expression had a significantly poorer overall
survival than those with low circRNA_101237 expression (P<0.001;
Fig. 2D). Overall, the results
suggest that circRNA_101237 expression has a negative impact on the
prognosis of patients with HCC.
 | Table IClinical association between serum
circRNA_101237 levels and clinicopathological characteristics of
patients with hepatocellular carcinoma. |
Table I
Clinical association between serum
circRNA_101237 levels and clinicopathological characteristics of
patients with hepatocellular carcinoma.
| Serum
circRNA_101237 | |
|---|
| Variable | Low expression
(n=86) | High expression
(n=148) | χ2 test
P-value |
|---|
| Age (years) | | | 0.4070 |
|
<50 | 56 | 88 | |
|
≥50 | 30 | 60 | |
| Sex | | | 0.1460 |
|
Male | 64 | 96 | |
|
Female | 22 | 52 | |
| Tumor size (cm) | | | <0.001 |
|
<3 | 60 | 48 | |
|
≥3 | 26 | 100 | |
| Lymph node
metastasis | | | 0.0060 |
|
N0-1 | 52 | 61 | |
|
N2-4 | 34 | 87 | |
| Distant
metastasis | | | 0.0002 |
|
No | 57 | 60 | |
|
Yes | 29 | 88 | |
| TNM stage | | | 0.0002 |
|
I-II | 54 | 55 | |
|
III-IV | 32 | 93 | |
| BCLC stage | | | <0.001 |
|
0 or A | 32 | 13 | |
|
B | 26 | 34 | |
|
C | 20 | 56 | |
|
D | 8 | 45 | |
| ALBI grade | | | 0.089 |
|
1 | 34 | 42 | |
|
2 | 26 | 65 | |
|
3 | 26 | 41 | |
| Diabetes
mellitus | | | 0.147 |
|
No | 72 | 112 | |
|
Yes | 14 | 36 | |
| Body mass index
(kg/m2) | | | 0.48 |
|
<30 | 54 | 86 | |
|
≥30 | 32 | 62 | |
 | Table IIUnivariate analysis of prognostic
factors for patients with hepatocellular carcinoma. |
Table II
Univariate analysis of prognostic
factors for patients with hepatocellular carcinoma.
| Variable | Hazard ratio (95%
confidence interval) | P-value |
|---|
| Age (≥50/<50
years) | 1.06
(0.870-1.142) | 0.17 |
| Sex
(male/female) | 1.02
(0.943-1.165) | 0.34 |
| Tumor size
(≥3/<3 cm) | 3.24
(1.820-7.322) | 0.01 |
| Lymph node
metastasis (N2-4/N0-1) | 2.76
(1.346-5.276) | 0.03 |
| Distant metastasis
(yes/no) | 5.72 (2.
703-9.352) | 0.01 |
| TNM stage
(III-IV/I-II) | 4.15
(1.492-7.626) | 0.03 |
| Serum
circRNA_101237 levels (high/low) | 3.29
(2.632-8.544) | 0.01 |
| BCLC stage
(C-D/0-B) | 2.87
(1.375-6.432) | 0.02 |
| ALBI grade
(III/I-II) | 1.2
(0.978-3.651) | 0.06 |
| Diabetes mellitus
(yes/no) | 1.08
(0.774-1.692) | 0.17 |
| Body mass index
(≥30/<30 kg/m2) | 1.03
(0.641-1.863) | 0.36 |
 | Table IIIMultivariate analysis of independent
prognostic factors for patients with hepatocellular carcinoma. |
Table III
Multivariate analysis of independent
prognostic factors for patients with hepatocellular carcinoma.
| Variable | Hazard ratio (95%
confidence interval) | P-value |
|---|
| Tumor size
(≥3/<3 cm) | 3.14
(2.522-5.541) | 0.03 |
| Lymph node
metastasis (N2-4/N0-1) | 3.76
(1.765-8.547) | 0.02 |
| Distant metastasis
(yes/no) | 4.35
(3.431-8.651) | 0.01 |
| TNM stage
(III-IV/I-II) | 3.93
(2.086-6.322) | 0.03 |
| BCLC stage
(C-D/0-B) | 3.25
(2.268-8.634) | 0.03 |
| Serum
circRNA_101237 levels (high/low) | 3.42
(2.215-6.532) | 0.02 |
Circ_101237 expression is associated
with cisplatin resistance in patients with HCC
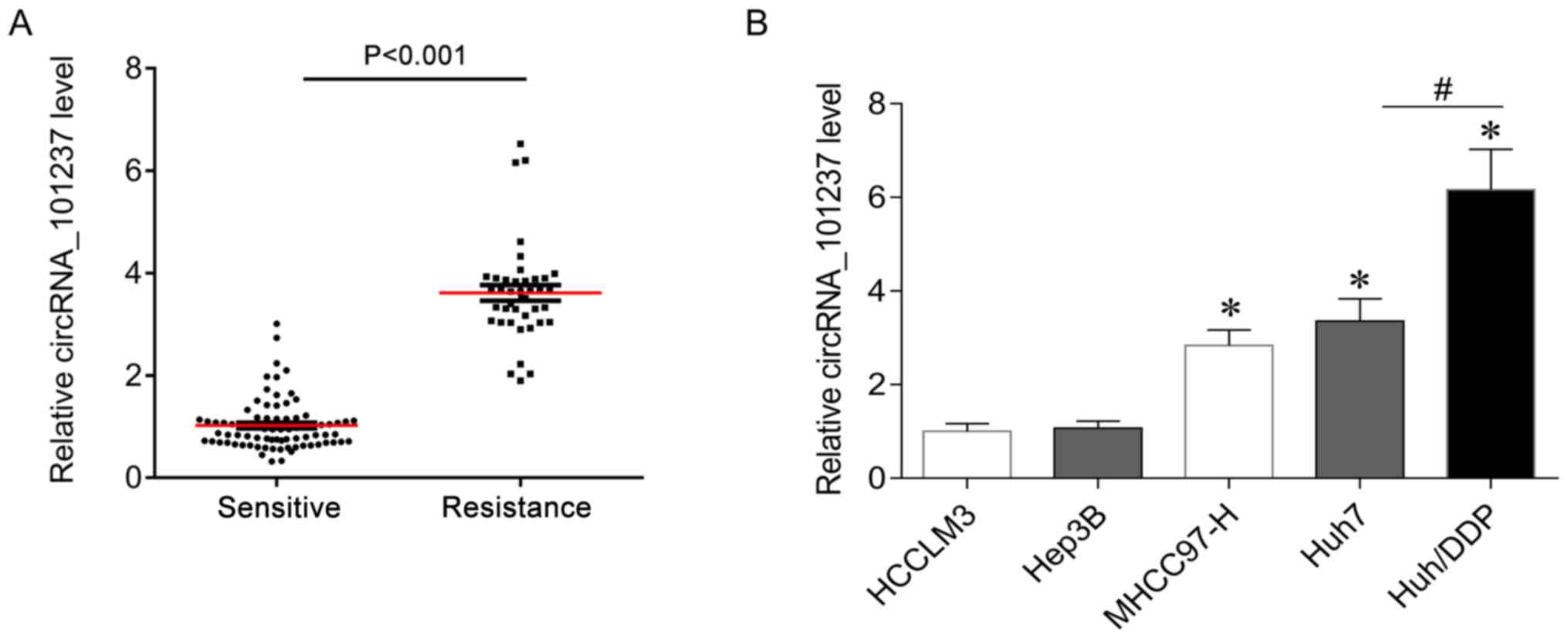
Of note, circRNA_101237 was increased by ~3-fold in
the serum of cisplatin-resistant HCC patients (n=50) compared with
that in cisplatin-sensitive patients (n=62) (Fig. 3A). In addition, the expression of
circRNA_101237 in liver cancer cell lines was determined, and the
results indicated that circRNA_101237 was significantly upregulated
in the MHCC97-H and Huh7 cell lines as compared with that in HCCLM3
cells (Fig. 3B). Of note,
circRNA_101237 levels in cisplatin-resistant Huh7/DDP cells was
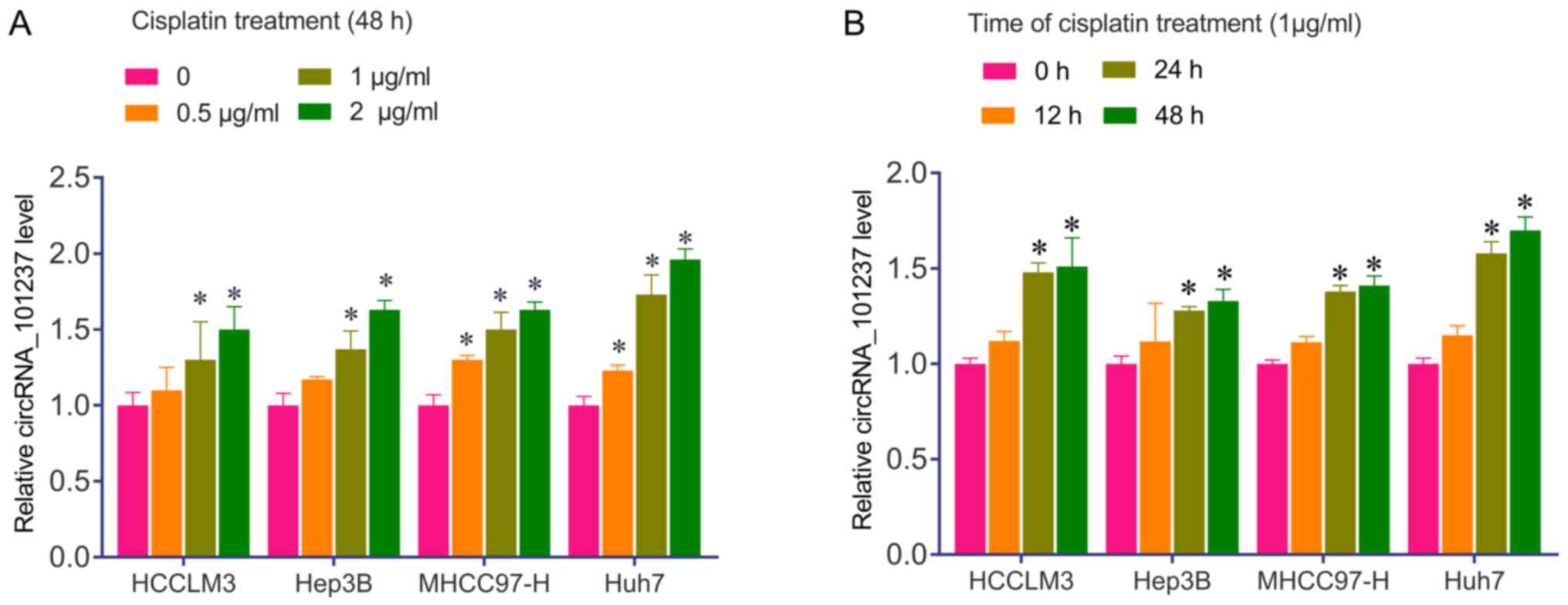
higher than that in the parental cells (Fig. 3B). In addition, HCCLM3, Hep3B,
MHCC97-H and Huh7 cells were confirmed to respond to cisplatin by
upregulating circRNA_101237 in a time- and cisplatin dose-dependent
manner (Fig. 4). These results
suggest that circRNA_101237 may be used as a biomarker for HCC
diagnosis and cisplatin resistance in patients with HCC.
Discussion
In the present study, a novel circRNA,
circRNA_101237, was identified as a diagnostic biomarker for HCC
and a potential therapeutic target. CircRNA_101237 was upregulated
in HCC tumor tissues compared with that in adjacent tissues. It was
further confirmed that circRNA_101237 is upregulated in serum
samples from patients with HCC. The upregulation of circRNA_101237
was positively associated with tumor size, lymph node metastasis,
distant metastasis and TNM stage of HCC patients. In addition,
univariate and multivariate analysis suggested that circRNA_101237
is an independent predictor of prognosis in patients with HCC.
These results indicate that circRNA_101237 has a key role in the
development of HCC.
If cancer is diagnosed at an early stage, the 5-year
survival rate of HCC patients is better (probably >70%)
(17). Early diagnosis of HCC is
difficult due to inflammation and cirrhosis. Therefore, there is an
urgent requirement to develop novel biomarkers for early diagnosis
of HCC (11). Recently, serum
alpha-fetoprotein, phosphatidylinositol-3, osteopontin, Golgi
protein-73 and various miRNAs have been reported as promising early
biomarkers of HCC, providing insight into the mechanisms that drive
tumorigenesis, which may lead to the development of more effective
treatment strategies (18). In the
present study, circRNA_101237 was revealed as a novel biomarker for
HCC. However, in the present cohort of HCC patients, no detailed
information was available regarding HBV, HCV and non-alcoholic
fatty liver disease. As the influence of hepatitis/NAFLD was
unknown, it was not reasonable to analyze the association between
circRNA_101237 and cirrhosis in patients with liver cancer.
Furthermore, in the future, analysis of the correlation between
circRNA_101237 and recently used biomarkers may provide meaningful
information for early diagnosis of HCC. These points will be
further investigated in future studies by our group.
The present results indicated that circRNA_101237
levels were increased in cisplatin-resistant HCC tumor tissues and
cisplatin-resistant Huh7 cells, and cisplatin exposure induced an
increase in circRNA_101237 expression, suggesting that
circRNA_101237 may be a biomarker of cisplatin resistance in
patients with HCC. Cisplatin has been widely used to treat a
variety of cancer types, including HCC (19). Patients with HCC initially respond to
cisplatin therapy but resistance frequently occurs, which is
associated with increased DNA repair, altered cell accumulation and
increased drug efflux mediated by multidrug resistance proteins
(20). It has been reported that
cisplatin resistance in HCC may occur through the loss of
Runt-associated transcription factor 3 and upregulation of
cyclophilin B (21,22). Recently, certain key biomarkers of
cisplatin resistance have revealed novel molecular mechanisms of
resistance (23). The role of
circRNAs in the development of chemotherapeutic drug resistance has
also been highlighted (24-26).
For instance, hsa_circ_0004674 is increased in chemoresistant
osteosarcoma cells and tissues and is associated with poor
prognosis (27). Upregulation of
circRNA-MTO1 promotes monastrol-induced cytotoxicity and reverses
monastrol resistance by inhibiting Eg5(28). CircRNA-PVT1 (plasmocytoma variant
translocation) is significantly upregulated in osteosarcoma, serum
and chemoresistant cell lines, including those with doxorubicin and
cisplatin resistance. CircRNA-PVT1 knockdown overcomes the
resistance of osteosarcoma cells to doxorubicin and cisplatin
(29). The present study indicated
that patients with cisplatin-resistant HCC and cisplatin-resistant
Huh7/DDP cells had increased levels of circRNA_101237, but it
remains elusive whether circRNA_101237 knockdown is able to inhibit
HCC cell proliferation and sensitize HCC cells to cisplatin. These
biological roles of circRNA_101237 in HCC cells will be
demonstrated in the future and the underlying mechanisms will be
revealed.
In conclusion, the present study indicated that
circRNA_101237 is upregulated in tissues and serum of patients with
HCC and may serve as a diagnostic and prognostic biomarker. In
addition, the levels of circRNA_101237 were increased in the serum
of cisplatin-resistant HCC patients and in cisplatin-resistant Huh7
cells. The present results provide evidence that circRNA_101237 may
be used as a diagnostic biomarker for HCC and a potential
therapeutic target for overcoming cisplatin resistance.
Supplementary Material
Table SI. General demographic and
clinicopathological characteristics of patients with HCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL made substantial contributions to the design of
the study. SZ, JW and YW analyzed and interpreted the patient data.
SZ and JW performed the cell biological experiments. All authors
contributed to the writing of the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Huainan First People's Hospital and the First
Affiliated Hospital of the Medical College of Anhui University of
Science and Technology (Huainan, China). All subjects provided
written informed consent to participate in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sia D, Villanueva A, Friedman SL and
Llovet JM: Liver cancer cell of origin, molecular class, and
effects on patient prognosis. Gastroenterology. 152:745–761.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ikeda K: Recent advances in medical
management of hepatocellular carcinoma. Hepatol Res. 49:14–32.
2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kawaoka T, Aikata H, Kobayashi T, Uchikawa
S, Ohya K, Kodama K, Nishida Y, Daijo K, Osawa M, Teraoka Y, et al:
Comparison of hepatic arterial infusion chemotherapy between
5-fluorouracil-based continuous infusion chemotherapy and low-dose
cisplatin monotherapy for advanced hepatocellular carcinoma.
Hepatol Res. 48:1118–1130. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q
and Wang C: Comprehensive circular RNA profiling reveals that
hsa_circ_0005075, a new circular RNA biomarker, is involved in
hepatocellular carcinoma development. Medicine (Baltimore).
95(e3811)2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sun X and Malhotra A: Noncoding RNAs
(ncRNA) in hepato cancer: A review. J Environ Pathol Toxicol Oncol.
37:15–25. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lanzafame M, Bianco G, Terracciano LM, Ng
C and Piscuoglio S: The role of long non-coding RNAs in
hepatocarcinogenesis. Int J Mol Sci. 19(E682)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Qiu LP, Wu YH, Yu XF, Tang Q, Chen L and
Chen KP: The emerging role of circular RNAs in hepatocellular
carcinoma. J Cancer. 9:1548–1559. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu BH, Zhang BB, Liu XQ, Zheng S, Dong KR
and Dong R: Expression profiling identifies circular RNA signature
in hepatoblastoma. Cell Physiol Biochem. 45:706–719.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang M, Yang Y, Xu J, Bai W, Ren X and Wu
H: CircRNAs as biomarkers of cancer: A meta-analysis. BMC Cancer.
18(303)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D
and Ni Y: CircRNA circ_0067934 promotes tumor growth and metastasis
in hepatocellular carcinoma through regulation of
miR-1324/FZD5/Wnt/β-catenin axis. Biochem Biophys Res Commun.
497:626–632. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shi L, Yan P, Liang Y, Sun Y, Shen J, Zhou
S, Lin H, Liang X and Cai X: Circular RNA expression is suppressed
by androgen receptor (AR)-regulated adenosine deaminase that acts
on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis.
8(e3171)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang X, Qiu S, Luo P, Zhou H, Jing W,
Liang C and Tu J: Down-regulation of hsa_circ_0001649 in
hepatocellular carcinoma predicts a poor prognosis. Cancer Biomark.
22:135–142. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y,
Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as
the sponge of microRNA-9 to suppress hepatocellular carcinoma
progression. Hepatology. 66:1151–1164. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Shao YY, Liang PC, Wu YM, Huang CC, Huang
KW, Cheng JC, Hsu CH, Hsu C, Cheng AL and Lin ZZ: A pilot study of
hepatic arterial infusion of chemotherapy for patients with
advanced hepatocellular carcinoma who have failed anti-angiogenic
therapy. Liver Int. 33:1413–1419. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wakamatsu T, Nakahashi Y, Hachimine D,
Seki T and Okazaki K: The combination of glycyrrhizin and
lamivudine can reverse the cisplatin resistance in hepatocellular
carcinoma cells through inhibition of multidrug
resistance-associated proteins. Int J Oncol. 31:1465–1472.
2007.PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fu J and Wang H: Precision diagnosis and
treatment of liver cancer in China. Cancer Lett. 412:283–288.
2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Tsuchiya N, Sawada Y, Endo I, Saito K,
Uemura Y and Nakatsura T: Biomarkers for the early diagnosis of
hepatocellular carcinoma. World J Gastroenterol. 21:10573–10583.
2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Amable L: Cisplatin resistance and
opportunities for precision medicine. Pharmacol Res. 106:27–36.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hu Y, Zhu QN, Deng JL, Li ZX, Wang G and
Zhu YS: Emerging role of long non-coding RNAs in cisplatin
resistance. Onco Targets Ther. 11:3185–3194. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kataoka J, Shiraha H, Horiguchi S,
Sawahara H, Uchida D, Nagahara T, Iwamuro M, Morimoto H, Takeuchi
Y, Kuwaki K, et al: Loss of Runt-related transcription factor 3
induces resistance to 5-fluorouracil and cisplatin in
hepatocellular carcinoma. Oncol Rep. 35:2576–2582. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kim Y, Jang M, Lim S, Won H, Yoon KS, Park
JH, Kim HJ, Kim BH, Park WS, Ha J and Kim SS: Role of cyclophilin B
in tumorigenesis and cisplatin resistance in hepatocellular
carcinoma in humans. Hepatology. 54:1661–1678. 2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Achkar IW, Abdulrahman N, Al-Sulaiti H,
Joseph JM, Uddin S and Mraiche F: Cisplatin based therapy: The role
of the mitogen activated protein kinase signaling pathway. J Transl
Med. 16(96)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Xu N, Chen S, Liu Y, Li W, Liu Z, Bian X,
Ling C and Jiang M: Profiles and bioinformatics analysis of
differentially expressed Circrnas in taxol-resistant non-small cell
lung cancer cells. Cell Physiol Biochem. 48:2046–2060.
2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ding B, Lou W, Xu L and Fan W: Non-coding
RNA in drug resistance of hepatocellular carcinoma. Biosci Rep.
38(BSR20180915)2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Shao F, Huang M, Meng F and Huang Q:
Circular RNA signature predicts gemcitabine resistance of
pancreatic ductal adenocarcinoma. Front Pharmacol.
9(584)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kun-Peng Z, Xiao-Long M, Lei Z, Chun-Lin
Z, Jian-Ping H and Tai-Cheng Z: Screening circular RNA related to
chemotherapeutic resistance in osteosarcoma by RNA sequencing.
Epigenomics. 10:1327–1346. 2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Liu Y, Dong Y, Zhao L, Su L and Luo J:
Circular RNAMTO1 suppresses breast cancer cell viability and
reverses monastrol resistance through regulating the TRAF4/Eg5
axis. Int J Oncol. 53:1752–1762. 2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kun-Peng Z, Xiao-Long M and Chun-Lin Z:
Overexpressed circPVT1, a potential new circular RNA biomarker,
contributes to doxorubicin and cisplatin resistance of osteosarcoma
cells by regulating ABCB1. Int J Biol Sci. 14:321–330.
2018.PubMed/NCBI View Article : Google Scholar
|