Introduction
Primary amyloidosis is a systematic disease
characterized by the pathological deposition of misfolded proteins
known as immunoglobulin light-chains (LCs) (1). Prognosis is closely associated with the
organs involved, especially the heart, which is affected in 50-80%
of patients with primary amyloidosis (2,3).
Although this disease was identified in the mid-19th century, the
mortality rate remains high, resulting from a difficult early
diagnosis and few effective treatment options (4). Patients with cardiac amyloidosis
usually present with symptoms of heart failure, angina and
conductive delays (3,5). Cardiac biomarkers, such as serum
cardiac troponin and brain natriuretic peptide (BNP), are sensitive
markers of cardiac dysfunction and the levels are elevated in
amyloidosis (3,5,6).
Treatment of cardiac amyloidosis depends on a combination of
therapies aimed at controlling hematologic disorders and relieving
cardiac damage (5). Heart failure in
cardiac amyloidosis faces specific therapeutic challenges in the
clinic. The routine therapeutic paradigm of heart failure cannot be
simply translated to cardiac amyloidosis (5-7).
Loop diuretics are central to the treatment for relieving
congestion in cardiac amyloidosis (3,5).
Angiotensin converting enzyme inhibitors, angiotensin receptor
blockers, beta blockers and mineralocorticoid receptor antagonists
can cause hypotension, renal insufficiency, or other
contraindications (3,5). They are often poorly tolerated and
require close patient monitoring (3,5).
Non-dihydropyridine calcium channel blockers and digoxin must be
avoided as they may worsen congestive heart failure or yield a risk
of toxicity (5). Patients usually
die from arrhythmia and congestive heart failure, and the disease
has a median survival time of <8 months and a 5-year survival
rate of <10% (3-5,8).
The disease progresses rapidly and the prognosis is poor. Thus, new
strategies for treatment are required to improve the survival
rate.
The pathological mechanism behind cardiac
amyloidosis remains to be elucidated. A previous study confirmed
that increased reactive oxygen species production in cardiomyocytes
induced by LC results in apoptosis and impaired heart function
(9). Moreover, altered metabolism,
mitochondrial dysfunction, impaired lysosomal/autophagy activity,
damaged calcium handling and altered p38 mitogen-activated protein
kinase pathway signaling have been reported to play important roles
in the process of cardiac dysfunction induced by cardiac
amyloidosis (4-10).
Recent studies revealed that impaired lysosomal and autophagy are
critical for mediating amyloidogenic LC toxicity (10-13).
The present study aimed to compare the direct toxic
effects of amyloidogenic LCs from various sources on cultured
cardiomyocytes, determine the changes in gene expression profiles
and identify common signaling pathway alterations in the
pathological process of amyloidosis.
Materials and methods
AL-09 synthesis
Amyloidogenic LC variable domain protein AL-09 is a
k LC that was first identified in a cardiac amyloidosis patient
(14,15). In the present study, recombinant
AL-09 protein was produced as described in previous studies
(14,16) by Nanjing Kingsray Biotechnology Co.,
Ltd with a 6xHis-tag added at the N-terminus. In brief, the target
gene was subcloned into the prokaryotic expression vector pET-12a
(GenScript) using NdeI/BamHI, and induced by 1 mM
isopropyl ß-D-1-thiogalactopyranoside in an Escherichia coli
BL21 (DE3) cell expression system (GenScript). The cells were
harvested and stored at -20˚C. Thawed cells were treated with
osmotic shock. AL-09 was extracted from inclusion bodies using 6 M
urea. The protein was purified using a Superdex 75 column (GE
Healthcare Life Sciences) on an AKTA fast protein liquid
chromatography system (GE Healthcare).
Western blotting to identify
AL-09
The molecular weight and purity of AL-09 were
analyzed by SDS-PAGE. Protein concentration was determined using a
Bradford protein assay (Beyotime Institute of Biotechnology).
Samples were stored at -80˚C in PBS (pH 7.4). A total of 2 µg AL-09
protein was electrophoretically separated by 4-20% gradient
SDS-PAGE (cat. no. M42012; GenScript) and then transferred onto
polyvinylidene difluoride membranes. Following blocking in 5%
non-fat milk at room temperature for 1 h, the membranes were
incubated overnight at 4˚C with mouse anti-His monoclonal antibody
(1:4,000; cat. no. A00186; GenScript). After washing, the membranes
were incubated with horseradish peroxidase (HRP)-conjugated goat
anti-mouse antibody (1:5,000; cat. no. A00160; Genscript) at room
temperature for 45 min. Protein signal was detected with a
LumiSensor™ Chemiluminescent HRP Substrate kit
(Genscript).
Human serum collection
Human serum was collected from a 61-year-old male
patient diagnosed with multiple myeloma with cardiac involvement
and from a healthy 28-year-old male donor without any known
disease. The biochemical test results of the healthy donor
presented with normal levels during the previous routine physical
examination. Formal written consents were obtained from the healthy
donor and the representative of the patient. The protocols used in
the present study were approved by the Medical Ethical Committee of
the First Affiliated Hospital of Nanjing Medical University
(Nanjing, China; approval no. 2015-SR-005).
Neonatal rat cardiomyocyte
culture
Cardiomyocytes were isolated from 1- to 3-day-old
male and female Sprague-Dawley rats (n=40-50 for each of the three
independent replicate experiments), which were provided by the
Experimental Animal Center of Nanjing Medical University. The
two-step procedure, consisting of enzyme digestion and
cardiomyocyte purification, was performed based on a previous study
(17). In brief, minced ventricles
were repeatedly digested in an enzyme solution, which included 0.4
mg/ml collagenase type II enzyme (Worthington Biochemical
Corporation) and 0.6 mg/ml trypsin (Gibco; Thermo Fisher
Scientific, Inc.). The pooled enzyme solution was centrifuged at
1,500 x g and 37˚C for 5 min, and the cardiomyocyte fraction was
resuspended in a solution containing horse serum (Gibco; Thermo
Fisher Scientific, Inc.). Further purification of cardiomyocytes
was performed using Percoll density gradient medium (Sigma-Aldrich;
Merck KGaA) and centrifugation at 1,500 x g and 21˚C for 30 min.
The enriched cardiomyocytes were resuspended in DMEM (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% horse serum and 5%
FBS (ScienCell Research Laboratories, Inc.) and 100 U/ml
penicillin/streptomycin (Beyotime Institute of Biotechnology), and
incubated at 37˚C with 5% CO2 for subsequent
experiments. The study protocol was approved by the Institutional
Animal Care and Use Committee of Nanjing Medical University
(approval no. IACUC-1708008).
CCK-8 assay
Cells were cultured in 96-well plates at a density
of 3x105 cells/well for 48 h, followed by starvation in
serum-free DMEM for 6-8 h. Cells were then randomly divided into
seven groups as follows: The control group, cultured in serum-free
DMEM; the P0.02 group, cultured with 0.02 mg/ml AL-09; the P0.1
group, cultured with 0.1 mg/ml AL-09; the P0.2 group, cultured with
0.2 mg/ml AL-09; the His group, cultured with 20 µg/ml 6xHis
peptide; the AL-10 group, cultured in 10% patient serum; and the
Z-10 group, cultured in 10% healthy serum. Cells were treated for a
further 48 h and the medium was replaced with serum-free DMEM at
100 µl/well. CCK-8 solution (Dojindo Molecular Technologies, Inc.)
was added (10 µl/well) to the medium and incubated at 37˚C for 1 h.
Subsequently, the absorbance was measured at a wavelength of 450 nm
using a microplate reader (BioTek Instruments, Inc.). Cell
viability was calculated as the absorbance ratio of treated groups
to the control group.
Apoptosis detection by flow
cytometry
Cardiomyocytes were seeded in 24-well plates at a
density of 3x105 cells/well for 48 h, starved for 6-8 h,
and cultured in serum-free DMEM, His peptide and AL-09 (0.2 mg/ml)
for 48 h. Cell apoptosis was determined using the Annexin V-Alexa
Fluor 647/propidium iodide (PI) Apoptosis Detection kit (FCMACS
Co., Ltd.) according to the manufacturer's instructions. Cells were
harvested and washed three times with ice-cold PBS, followed by
resuspension in 300 µl binding buffer. Subsequently, Annexin
V-Alexa Fluor 647 (5 µl) and PI (10 µl) were added to the samples
and incubated at room temperature for 15 min in the dark. The
percentages of Annexin V-Alexa Fluor 647-positive/PI-negative cells
(early apoptosis) and Annexin V-Alexa Fluor
647-positive/PI-positive cells (late apoptosis) were analyzed using
flow cytometry (BD FACStation™ 6.1 software; BD FACSCalibur; BD
Biosciences).
RNA sequencing (RNA-Seq), Gene
Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analyses
To identify the common changes in the gene
transcription profiles in cardiomyocytes treated with amyloidogenic
LCs, two pairs of groups were designed to conduct a comparative
study. In one group, cardiomyocytes were cultured with 10% patient
serum (AL-10) or 10% healthy serum (Z-10, as a control). In another
group, cells were cultured with 0.2 mg/ml AL-09 or 20 µg/ml 6xHis
peptide (as a control). Cardiomyocytes were treated for 48 h. Total
RNA was extracted from cells using TRIzol® reagent (cat.
no. 15596-026; Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. RNA-seq was performed
at Guangzhou RiboBio Co., Ltd using a NEBNext Ultra RNA Library
Prep kit for Illumina (New England BioLabs, Inc.) according to the
manufacture's instructions. A HiSeq 2500 Sequencing system
(Illumina, Inc.) with SE50 sequencing strategy was employed.
Computational pipelines, including Tophat version 2.0.13
(https://ccb.jhu.edu/software/tophat/index.shtml) and
Gfold version 1.1.2 software (http://www.tongji.edu.cn/~zhanglab/GFOLD/index.html),
were employed to process the RNA-seq data. Genes were considered
significantly differentially expressed if they showed >1-fold
change and a Q-value of <0.001. The Q-value was obtained as a
correction of the P-value (18). GO
(http://www.geneontology.org/) and KEGG
pathway analysis (http://www.genome.ad.jp/kegg) were performed using the
Database for Annotation, Visualization and Integrated Discovery 6.8
(http://david.ncifcrf.gov). P<0.05 was used as
the significance threshold for the enrichment degree of GO
annotations and KEGG pathways.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was used to verify the RNA-seq results.
Genes that demonstrated a simultaneous change in expression levels
in the aforementioned two pairs of groups from the RNA-seq data
were used for subsequent analysis. The primers used in the present
study are listed in Table I.
Cardiomyocytes were treated following the same protocol as for
RNA-Seq. Total RNA was extracted from cells using
TRIzol® reagent (cat. no. 15596-026; Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. cDNA was synthesized using a RevertAid RT Reverse
Transcription kit (cat. no. K1691; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. RT-qPCR was performed
using the FastStart Universal SYBR Green Master (Rox) kit (cat. no.
4913850001; Roche; Merck KGaA) on a 7900 HT-Fast Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
following thermocycling conditions were used for the PCR: Initial
denaturation at 95˚C for 30 sec, followed by denaturation at 95˚C
for 5 sec and annealing and extension at 60˚C for 30 sec for 40
cycles. The data were analyzed according to the
2-ΔΔCq method (19). The internal reference gene GAPDH was
used for normalization.
 | Table IPrimers used for reverse
transcription-quantitative PCR. |
Table I
Primers used for reverse
transcription-quantitative PCR.
| mRNA | Forward primer | Reverse primer |
|---|
| Bmp6 |
5'-GAGCTTTACGTGAGCTTCCAGGAC-3' |
5'-GGCATTCATGTGTGCATTGAGAG-3' |
| Bmp4 |
5'-GAGGAGGAAGAAGAGCAGAGC-3' |
5'-GCGACGGCAGTTCTTATTCT-3' |
| Ptgs2
(Cox2) |
5'-CCACTTCAAGGGAGTCTGGAACAT-3' |
5'-TATCACACACTCTGTTGTGCTCCC-3' |
| Ptgs1
(Cox1) |
5'-CATTCTGCCCTCTGTACCCAAAGA-3' |
5'-GAGCTGGAGGAAATAGCCACTCAA-3' |
| Ereg |
5'-TCACAGCGACTTGCTTGACC-3' |
5'-GGCGCTGTTCAGACTTGTGG-3' |
| Tgfa |
5'-CGCACGTACACACACCAAAC-3' |
5'-TCAGTGCATCATATACAGAAGTCAGAA-3' |
| Plod2 |
5'-GGGGATCCATGCTCGCCCTGCTCTCC-3' |
5'-GGCTCGAGTTGTCTGCCAGCCGCTTATC-3' |
| Ccnb1 |
5'-CAACTGGAGGAAGAGCAGTCA-3' |
5'-CATCTGAACCTGTATTAGCCA-3' |
| Gapdh |
5'-ACCACAGTCCATGCCATCAC-3' |
5'-TCCACCACCATGTTGCTGTA-3' |
Statistical analysis
Normally distributed continuous variables are
presented as the mean ± SD. Statistical analysis was performed
using GraphPad Prism 6 (GraphPad Software, Inc). Comparisons of
continuous variables between groups were analyzed by unpaired
Student's t-test. One-way ANOVA with Dunnett's multiple comparison
post hoc test was used to compare means from different unrelated
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
Recombinant amyloidogenic LC
AL-09
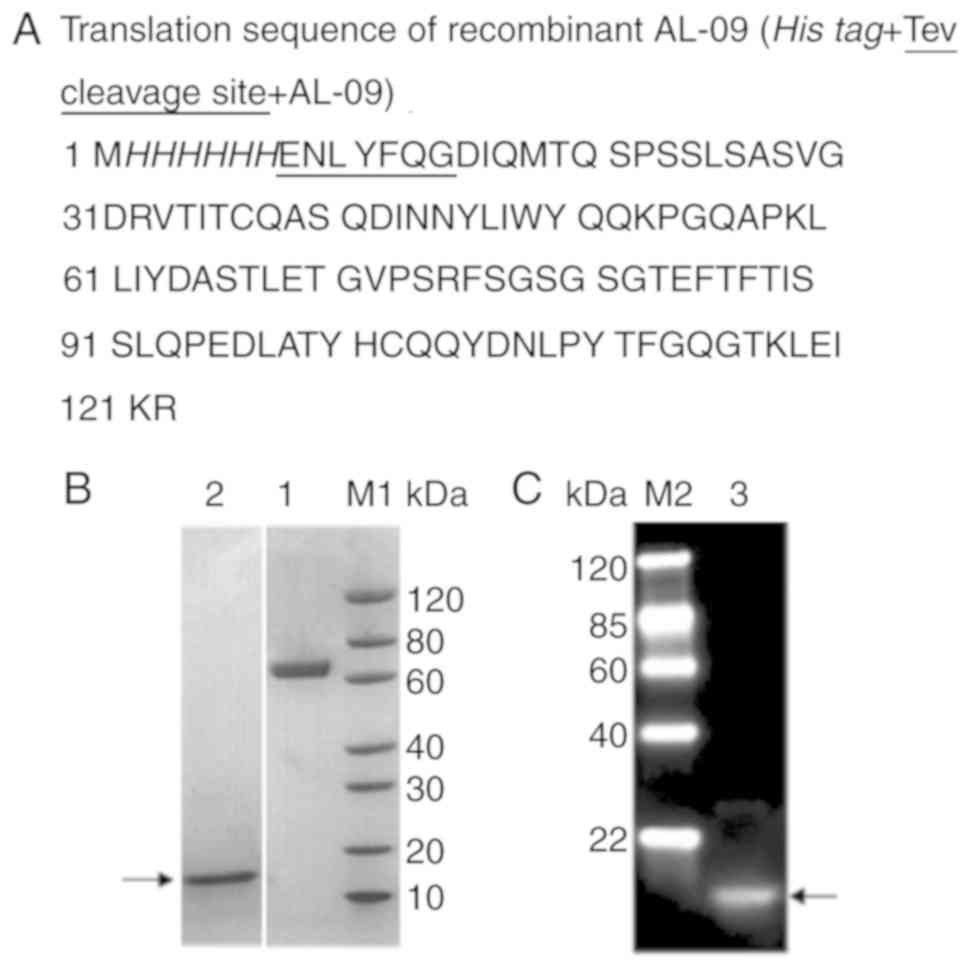
The translation sequence of recombinant AL-09 is
shown in Fig. 1A. The purity of the
protein was ~90%, and the molecular weight was ~13.7 kDa (Fig. 1B). The AL-09 with 6xHis-tag was
confirmed by western blotting (Fig.
1C), and the protein concentration was 0.4 mg/ml.
Case clinic information
A 61-year-old male patient, diagnosed as suffering
from multiple myeloma, restrictive cardiomyopathy, hypertension and
New York Heart Association class III (7) heart failure on December 7, 2016, at the
First Affiliated Hospital of Nanjing Medical University, was
recruited to the present study. Bone marrow smears showed that the
percentage of immature plasma cells was 10% and the presence of
bone marrow hyperplasia. Immunohistochemical analysis showed that
the bone marrow was positive CD38(+), CD138(+), κ LC(-), λ LC(+),
CD56 (-) and multiple myeloma oncogene 1(+). Serum immunofixation
electrophoresis was positive for immunoglobulin G LCs. The serum
and urine λ LC levels were 1.71 g/l (normal range, 0.9-2.1 g/l) and
200 mg/l (normal range, 0-3.9 mg/l), respectively. The serum
N-terminal (NT)-pro BNP level was 8,016 ng/l (normal range, 0-125
ng/l). Echocardiography showed left ventricular hypertrophy, mild
pericardial effusion and a left ventricular ejection fraction
(LVEF) of 40.5%. Myocardial perfusion imaging with 99mTc sestamibi
at rest showed spotted sparse distribution in the left ventricle, a
dilated left ventricular cavity, a diffuse decrease in wall motion
and thickening of the left ventricular wall, with the LVEF at 35%.
Cardiac MRI 3T enhanced scanning presented with the following
results: LVEF, 38.3%; end systolic volume, 54.1 ml; end diastolic
volume, 87.7 ml; stroke volume, 33.6 ml; cardiac output, 2.99
l/min; cardiac index, 1.79 l/min/m2 and end diastolic
myocardial mass, 204.6 g. Ring-shaped high signal shadows were
observed under the endocardium of the left and right ventricles.
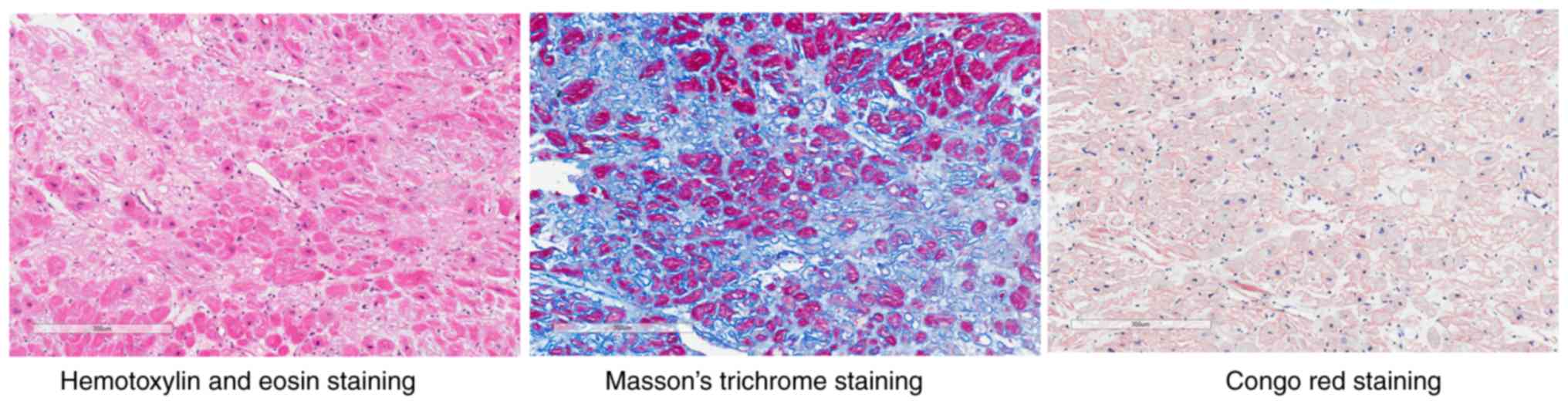
Hematoxylin and eosin staining and Masson's trichrome staining of
myocardial biopsy specimens showed myocardial degeneration,
disordered arrangement of myocardial fibers, adipose tissue
infiltration, dilation of interstitial small vessels, fibrous
proliferation and hyalinization. Congo red staining showed the
presence of pink dye in the myocardium. The histological slides
were scanned and photographed using an Aperio CS2 image capture
device with Aperio ImageScope v12.3.2.7001 software (Leica
Biosystems) (Fig. 2). Serum (4 ml)
was collected from this patient and stored at -80˚C until further
use. Control serum was obtained from a 28-year old healthy male
without any distinct disease at a routine physical examination.
Amyloidogenic LCs decrease cellular
viability and increase apoptosis
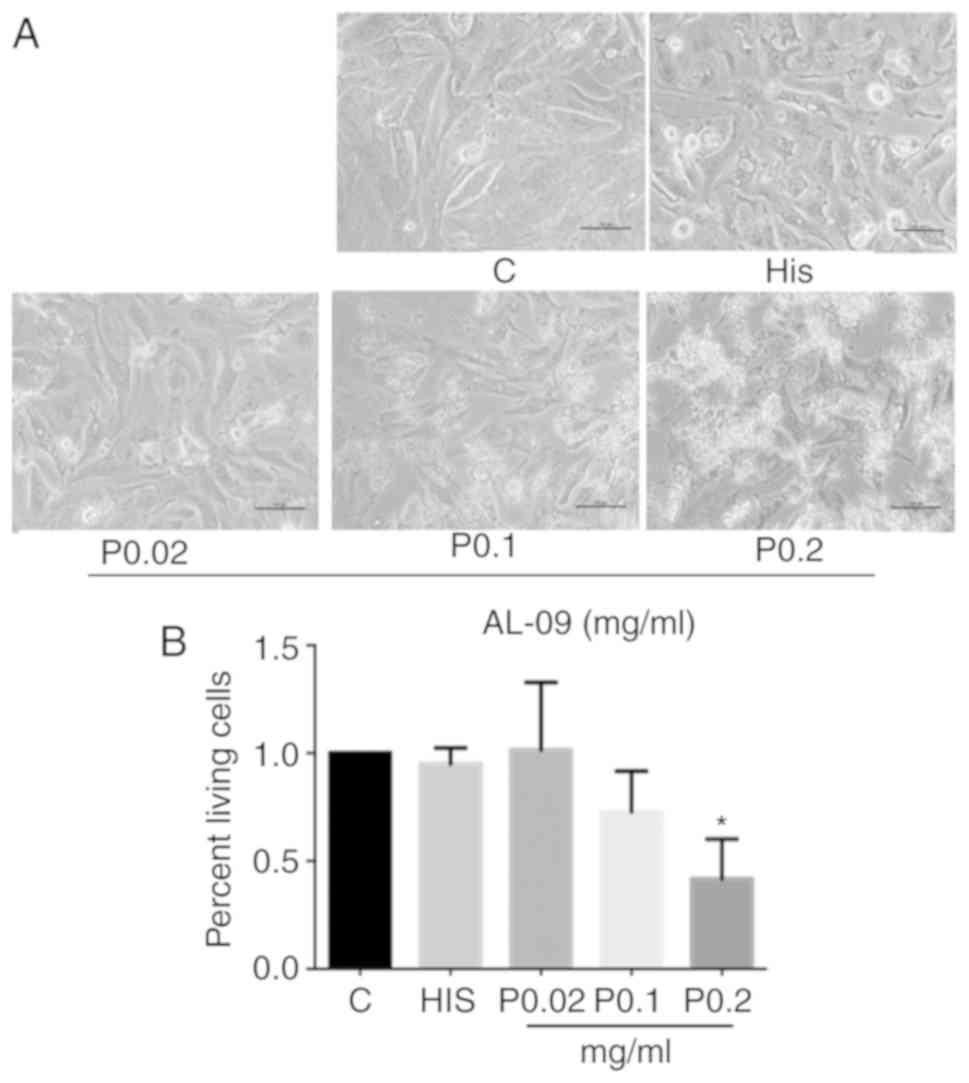
Cultured cardiomyocytes in each group showed normal
cell bodies, intact pseudopodia and high pulsation under light
microscopy. Cells treated with a higher concentration of AL-09
peptide (0.2 mg/ml) showed more cellular debris compared with the
control group (Fig. 3A). The results
of CCK-8 assays demonstrated that the viability of cardiomyocytes
cultured with AL-09 (0.2 mg/ml) decreased to 42% of the control
group (P<0.05; Fig. 3B). The
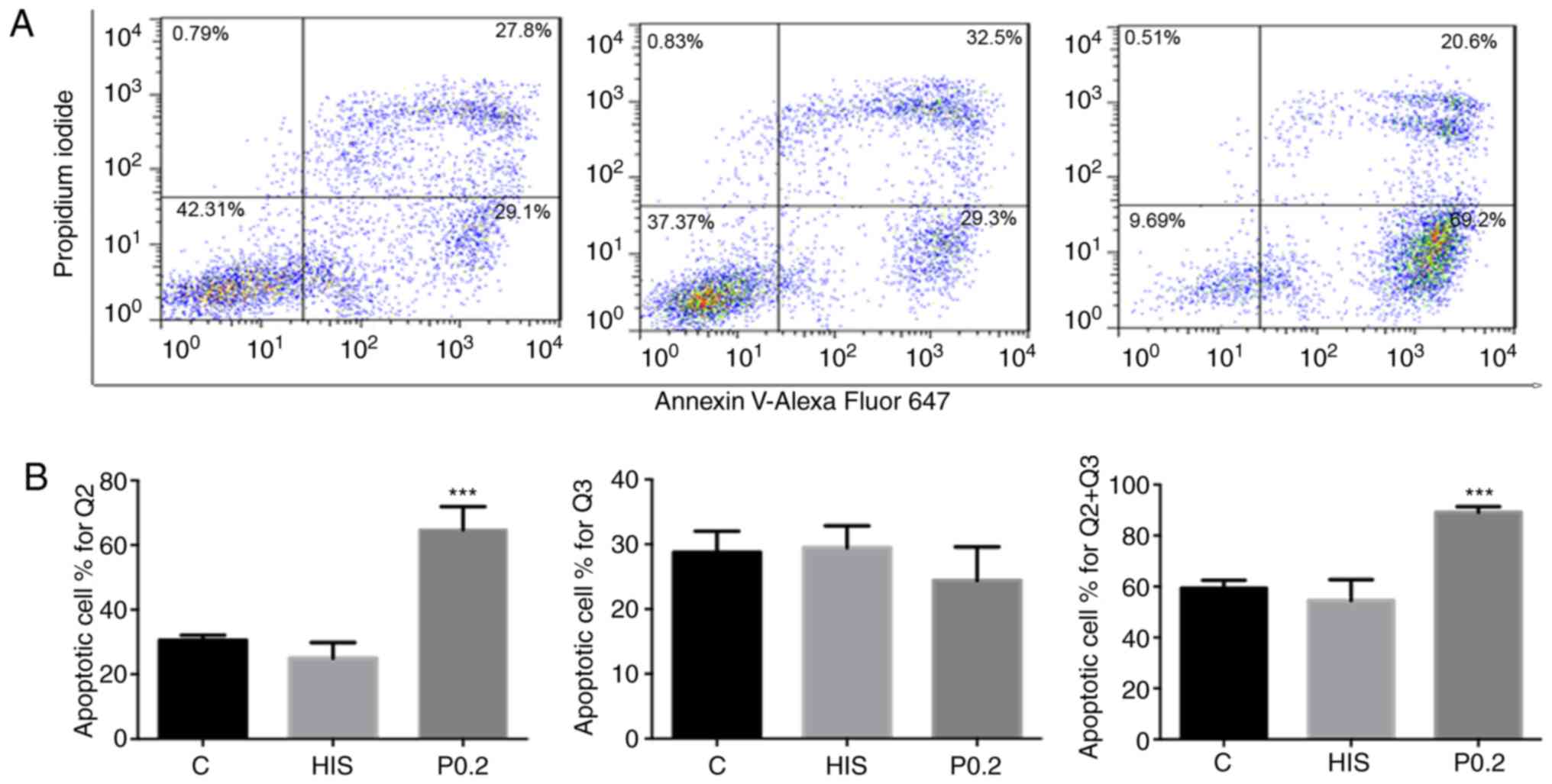
results of flow cytometry detection were consistent with the CCK-8
assay. Cells in AL-09-treated groups presented with higher
percentages of late apoptosis and total apoptotic cells compared
with those in the control group (P<0.05; Fig. 4). No significant difference was found
in the percentages of early apoptotic cells between the
AL-09-treated and control groups (Fig.
4).
No obvious morphological changes were observed in
groups treated with human serum (data not shown). The viability of
the cardiomyocytes cultured with 10% AL patient serum decreased to
72% of that of the control cells cultured with 10% healthy serum
(0.72±0.07 vs. 0.98±0.10; P<0.05; data not shown).
Identification of differentially
expressed genes (DEGs)
A total of 1,376 genes were differentially expressed
in the peptide-paired group [Ph (6xHis peptide group); Pa (AL-09
peptide group)], and 869 genes were differentially expressed in the
serum-paired group [Ms (normal serum group); Mc (AL patient serum
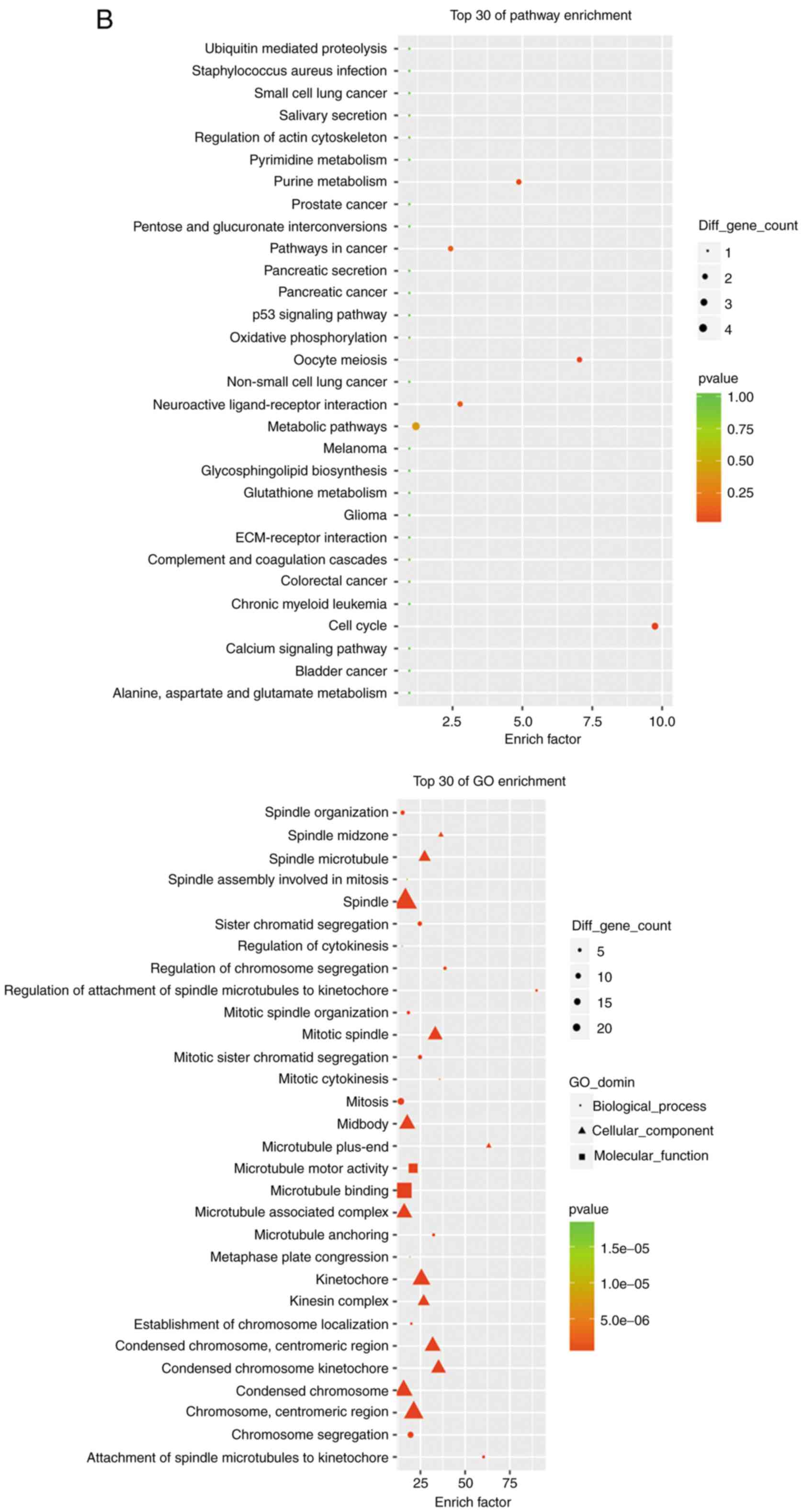
group)]. Fig. 5A and B shows GO and KEGG analyses of upregulated
and downregulated DEGs co-existing in the two paired groups,
respectively. Fig. 5C shows a Venn
diagram of DEGs in both paired groups. A total of 256 DEGs
co-existed in both paired groups by comprehensive comparison. It
was found that 127 genes were upregulated and 88 genes were
simultaneously downregulated in both the Ms-Mc and the Ph-Pa
groups. There were 41 genes that presented different changes in the
Ms-Mc and Ph-Pa groups. Among the upregulated DEGs, the main
high-enrichment GO terms regarding cardiomyocytes included
‘regulation of calcium ion transport into cytosol’, ‘protein
activation cascade’, ‘growth factor activity’, ‘dioxygenase
activity’ and ‘anchored component of membrane’. Upregulated DEGs
were involved in signal transduction, metabolism, apoptosis,
adhesion and heart physiology. Upregulated KEGG pathways included
‘amino sugar and nucleotide sugar metabolism’, ‘lysine
degradation’, ‘TGF-β signaling pathway’ and ‘ErbB signaling
pathway’ (Table II). The
downregulated DEGs are involved in proliferation and heart
functions. KEGG pathways included ‘p53 signaling pathway’, ‘cell
cycle’, ‘oocyte meiosis’ and ‘purine metabolism’ (Table III).
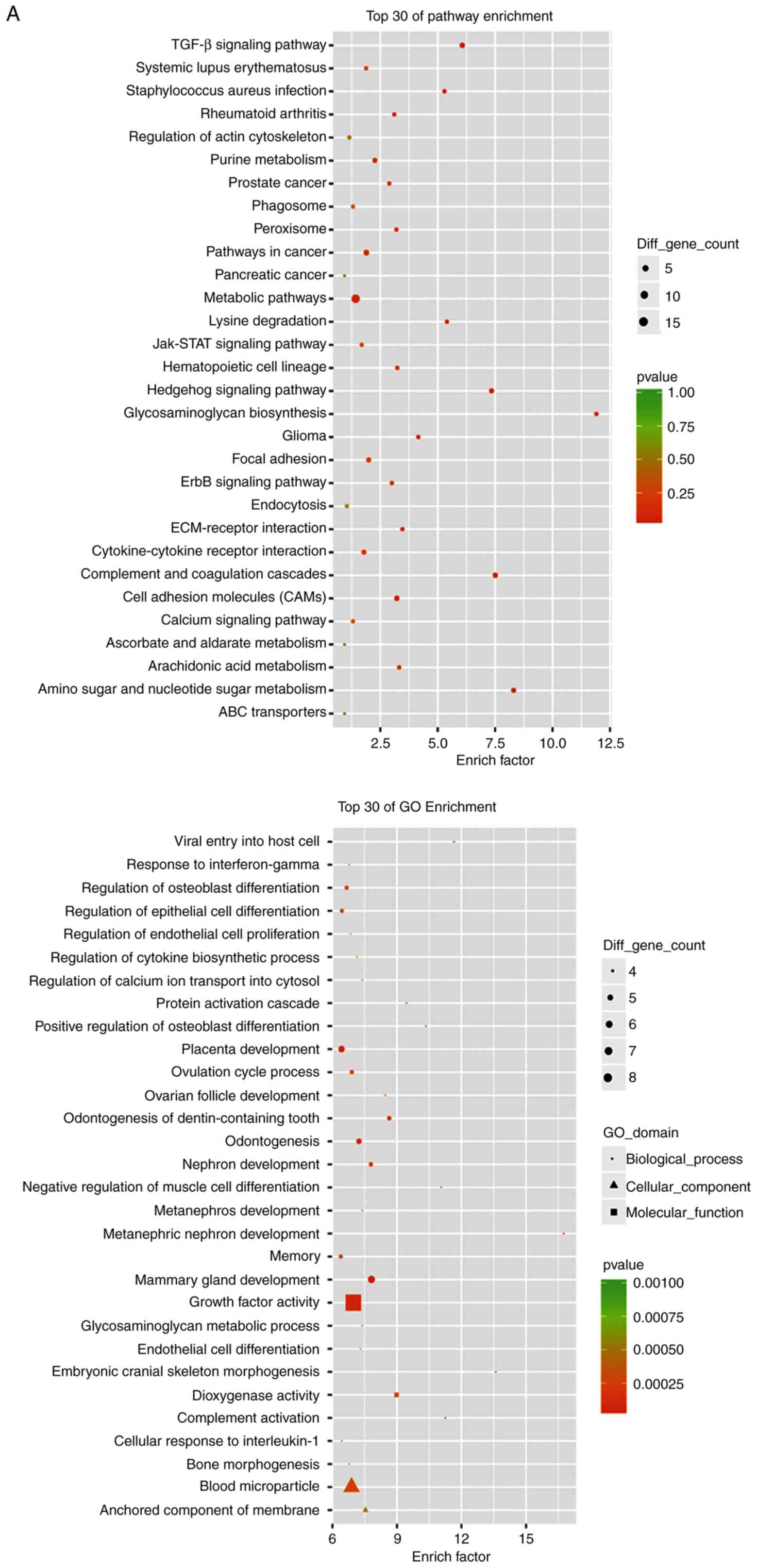 | Figure 5RNA sequencing analysis results. (A)
GO and KEGG analyses of upregulated DEGs in both Ph-Pa and Ms-Mc
paired groups. RNA sequencing analysis results. (B) GO and KEGG
analyses of downregulated DEGs in both Ph-Pa and Ms-Mc paired
groups. (C) Venn diagram of DEGs of both paired groups. The 256
genes, including 127 upregulated and 88 downregulated genes, were
analyzed using gene enrichment pathway analysis. GO, Gene Ontology;
KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially
expressed genes; AL-09, recombinant amyloidogenic light-chain
protein; Ph, 6xHis peptide group; Pa, AL-09 peptide group; Ms,
healthy serum group; Mc, AL patient serum group. |
 | Table IIKyoto Encyclopedia of Genes and
Genomes pathway analysis of upregulated differentially expressed
genes in both paired groups. |
Table II
Kyoto Encyclopedia of Genes and
Genomes pathway analysis of upregulated differentially expressed
genes in both paired groups.
| Term | P-value | Genes |
|---|
| Amino sugar and
nucleotide sugar metabolism | 0.0008 | Uap1, Gfpt2,
Ugdh |
| Hedgehog signaling
pathway | 0.0013 | Bmp4, Bmp6,
Gas1 |
| Lysine
degradation | 0.0101 | Plod2,
Lbp |
| Staphylococcus
aureus infection | 0.0107 | C1r,
C1s |
| TGF-β signaling
pathway | 0.0070 | Bmp4, Bmp6,
Thbs2 |
| Glioma | 0.0203 | Pdgfra,
Tgfa |
| Arachidonic acid
metabolism | 0.0359 | Ptgs2,
Ptgs1 |
| Hematopoietic cell
lineage | 0.0383 | Cd55,
Csf1 |
| Peroxisome | 0.0396 | Xdh,
Lbp |
| Rheumatoid
arthritis | 0.0422 | Csf1,
Angpt1 |
| ErbB signaling
pathway | 0.0463 | Ereg,
Tgfa |
 | Table IIIKyoto Encyclopedia of Genes and
Genomes pathway analysis of downregulated differentially expressed
genes in both paired groups. |
Table III
Kyoto Encyclopedia of Genes and
Genomes pathway analysis of downregulated differentially expressed
genes in both paired groups.
| Term | P-value | Genes |
|---|
| p53 signaling
pathway | 0.0017 | Ccnb1,
Rrm2 |
| Cell cycle | 0.0006 | Ccnb1, Pttg1,
Cdkn2c |
| Oocyte meiosis | 0.0057 | Ccnb1,
Pttg1 |
| Purine
metabolism | 0.0152 | Adssl1,
Rrm2 |
Validation analysis by RT-qPCR
KEGG pathway analysis showed that among the
upregulated DEGs, bone morphogenetic protein 4 (BMP4) and 6
(BMP6) were involved the ‘TGF-β signaling pathway’ and the
‘Hedgehog signaling pathway’. Prostaglandin G/H synthase 1
(PTGS1/COX1) and 2 (PTGS2/COX2) were associated with
‘arachidonic acid metabolism’. Epiregulin (EREG) and
TGFA were associated with the ‘ErbB signaling pathway’, and
procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 (PLOD2)
was associated with ‘lysine degradation’. In the KEGG pathway
analysis of downregulated DEGs, it was shown that E3
ubiquitin-protein ligase CCNB1IP1 (CCNB1) was involved in
the ‘p53 signaling pathway’, the ‘cell cycle’ and ‘oocyte meiosis’.
The mRNA expression of eight genes (Bmp4, Bmp6,
Ptgs1, Ptgs2, Ereg, Tgfa, Plod2 and Ccnb1) was
further confirmed by RT-qPCR in both paired groups. The results of
RT-qPCR were consistent with the RNA-seq results (Fig. 6). The mRNA expression levels of
Bmp4, Bmp6, Ptgs1, Ptgs2, Ereg, Tgfa and Plod2
were significantly upregulated in both the amyloidosis patient
serum and AL-09 peptide groups compared with the levels in their
respective controls. Bmp6 showed the most significant
increase in the Ph-Pa group, and Ptgs2 showed the most
significant increase in the Ms-Mc group. The mRNA expression levels
of Ccnb1 showed significant downregulation in the AL-09
peptide group, but no significant difference was observed in the AL
patient serum group compared with their respective controls.
 | Figure 6mRNA expression levels of eight genes
(Bmp4, Bmp6, Ptgs1, Ptgs2, Ereg, Tgfa, Plod2 and
Ccnb1) were determined using reverse
transcription-quantitative PCR. **P<0.01,
***P<0.001 and ****P<0.0001. vs. Ph or
Ms groups. Ph, 6xHis peptide group; Pa, AL-09 peptide group; Ms,
healthy serum group; Mc, AL patient serum group; Bmp, bone
morphogenetic protein; Ptgs, prostaglandin G/H synthase;
Ereg, epiregulin; Tgfa, transforming growth factor-α;
Plod2, procollagen-lysine,2-oxoglutarate 5-dioxygenase 2;
Ccnb1, E3 ubiquitin-protein ligase CCNB1IP1. |
Discussion
Cardiac amyloidosis often occurs due to myocardial
infiltration by immunoglobulin proteins, such as in LC amyloidosis,
which is the most common form of systemic amyloidosis (3). Cardiac amyloidosis often confers a poor
prognosis, particularly in patients affected by systemic
amyloidosis (3). In the present
study, the patient diagnosed with multiple myeloma was
characterized by the presence of bone marrow hyperplasia, with a
higher percentage of immature plasma cells. The plasma cells
produced immunoglobulin G LCs, predominantly the λ variant. The
patient presented with notably higher urine λ LC levels. The
amyloidogenic LC underwent extracellular misfolding, aggregated
into soluble oligomers and amyloid fibrils, and was deposited
within the heart. Histology of the myocardial biopsy confirmed
cardiac amyloidosis. The patient experienced heart failure, as
demonstrated by echocardiography, radionuclide myocardial perfusion
imaging and cardiac MR 3T enhanced scanning. The level of serum
NT-pro BNP, a biomarker for heart failure, was increased to 8,016
ng/l, which predicted a poor prognosis for the patient (3).
AL amyloidosis is lethal due to the presence of the
toxic LC and not as a result of the malignant behavior of the
plasma cell clones (3). The present
study was designed to investigate the toxic effects of LCs from
various sources on cardiomyocytes. The serum of the aforementioned
patient was isolated, added to the culture medium of cardiomyocytes
and compared with serum from a healthy subject. Cells treated with
patient serum presented with decreased cellular viability. AL-09 is
a variant of the κ LC protein, which has proven to be a major
contributing protein to the development of myocardial amyloid
plaques (11). Recombinant AL-09 was
prepared with a 6xHis tag. Cultured cardiomyocytes were treated
with recombinant AL-09 peptide and 6xHis peptide. Cells treated
with recombinant AL-09 peptide presented with decreased cellular
viability and a higher percentage of apoptotic cells. The results
indicated that the λ and κ amyloidogenic LCs were directly harmful
to the cultured cells.
To identify common signaling pathway alterations
from the toxic λ/κ LCs on cardiomyocytes, the results of RNA-seq of
two paired groups (recombinant AL-09 peptide versus 6xHis peptide;
serum from the AL patient versus serum from the healthy subject)
were analyzed. The present study focused on the DEGs that co-exist
in both paired groups, which were involved in signal transduction,
metabolism, cell proliferation, cell apoptosis, cellular adhesion
and heart physiology. The KEGG pathways for the upregulated genes
included the ‘TGF-β signaling pathway’, ‘lysine degradation’, the
‘Hedgehog signaling pathway’ and the ‘ErbB signaling pathway’. The
KEGG pathways for downregulated genes included the ‘p53 signaling
pathway’ and the ‘cell cycle’. No concordant change on genes
involved in autophagy/lysosomal was found in either paired
group.
The TGF-β-BMP signaling pathway plays an important
role in cell proliferation, apoptosis and fibrosis (20). TGF-β can induce endothelial cell
inflammation, proliferation and migration, and the immune response,
as well as enhancing the synthesis of extracellular matrix proteins
(20,21). TGF-β and/or BMP6 can promote
hepatocyte apoptosis and fibrosis (22). Activation of the TGF-β-BMP signaling
pathway promotes neuronal apoptosis in Alzheimer's disease (AD)
(23). The dysfunction of cerebral
blood vessels and cognitive deficits were shown to be reversed in a
study inhibiting the TGF-β signaling pathway (24). In the cardiovascular system, the
TGF-β signaling pathway is involved in the development of
myocardial hypertrophy and extracellular matrix
deposition/remodeling, and in the promotion of apoptosis and heart
failure (25). Therefore, inhibition
of the TGF-β-BMP pathway reduces the progression of cardiac
remodeling and aids in reducing the risk of heart failure (26,27).
Activation of the TGF-β-BMP signaling pathway plays a role in the
development of amyloid cardiomyopathy. The present study confirmed
that Bmp4 and Bmp6 gene expression levels were both
increased in AL-09 peptide-treated and patient serum-treated
cardiomyocytes compared with the levels in their respective
controls, suggesting the activation of the TGF-β-BMP signaling
pathway. BMP4 and BMP6 are members of the TGF-β superfamily; they
bind with BMP receptors on the surface of the cell membrane, and
are involved in a variety of signal transduction pathways such as
maintaining homeostasis, regulating hormone levels and controlling
tissue growth and development (20,27).
BMP4 induces cardiomyocyte hypertrophy and apoptosis by activating
the JNK signaling pathway and altering the in vivo redox
state (28,29). BMP6 is involved in the
pathophysiology of chronic heart failure (30). Increasing BMP6 expression, induced by
TGF activation, promotes tissue fibrosis and new vascular
development, as well as inhibiting cell proliferation (22,31,32). The
decreased cell viability and increased apoptosis observed in the
present study may in part be due to the activation of the TGF-β-BMP
signaling pathway, which may be a target for treating cardiac
amyloidosis.
Prostaglandins are involved in AD, another disease
characterized by extracellular deposition of the amyloid β-peptide
(33). The disease is also
characterized by the appearance of extensive neuronal loss,
intracellular neurofibrillary tangles and changes in synapses of
the hippocampus and cerebral cortex (33). The inflammatory pathways are
activated by these processes, and the production of inflammatory
mediators, including cytokines and prostaglandins, is induced by
microglia, astrocytes and infiltrating leukocytes (33). Prostaglandins are small arachidonic
acid-derived lipids from multi-enzymatic pathways in which COXs and
phospholipases act in a rate-limiting capacity (33). COXs, also known as PTGSs, include two
isozymes: A constitutive PTGS1 (COX1) and an inducible PTGS2 (COX2)
(33). The pharmacological blockade
of PTGS has been demonstrated to reverse the progression of AD
(34). To the best of our knowledge,
no study has proven that PTGSs are also activated in cardiac
amyloidosis. Prostaglandins and PTGSs might exhibit similar effects
in both the central nervous and cardiovascular systems. In the
present study, Ptgs1 and Ptgs2 gene expression levels
were both increased in the AL-09 peptide-treated and patient
serum-treated cardiomyocytes, which suggested that there was an
increased production of prostaglandins. Further studies on the role
of PTGSs and prostaglandins in amyloid cardiomyopathy may
contribute to development of successful therapeutic strategies.
EREG is a secreted peptide hormone that belongs to
the epidermal growth factor (EGF) family (35). EREG binds to the EGF receptor (EGFR)
and ErbB4, and can stimulate signaling of ErbB2 and ErbB3 through
ligand-induced heterodimerization with a cognate receptor (35). EREG has functions in a variety of
biological processes, including wound healing, inflammation, oocyte
maturation and tissue repair, by regulating angiogenesis and
vascular remodeling, as well as by stimulating cell proliferation
(35). TGF-α is another ligand for
EGFR (36). TGF-α is responsible for
activating a cell proliferation, differentiation and development
signaling pathway. A study on AD confirmed that accumulated amyloid
β interacts with the ErbB signaling pathway (37). To the best of our knowledge, no study
has demonstrated that the ErbB signaling pathway is involved in
cardiac amyloidosis. In the present study, abnormal ErbB signaling
was elicited both in the AL-09 peptide-treated and patient
serum-treated cardiomyocytes. The upregulation of Ereg and
Tgfa gene expression levels were confirmed using RT-qPCR.
However, the underlying mechanisms remain to be elucidated.
The myocardial extracellular space is a complicated
and dynamic environment that is essential for the normal structure
and function of the heart (38). The
physiological pathways that regulate normal collagen turnover
control and the pathological development of fibrosis are slowly
being understood (38). The
myocardial biopsy with Masson’s trichrome stain from the patient in
the present study showed abundant collagen accumulation. Lysyl
hydroxylases 2 (encoded by the Plod2 gene) is a vital enzyme
that mediates stabilized collagen crosslink formation; it is
localized to the cisternae of the rough endoplasmic reticulum and
catalyzes the lysyl residues hydroxylation in collagen-like
peptides (39). The consequent
hydroxylysyl groups become sites of carbohydrate adherence in
collagen and are therefore essential for intermolecular crosslink
stability (39). The effect of PLOD2
on cancer has been previously reviewed (39). In the present study, Plod2
mRNA levels were upregulated both in AL-09 peptide-treated and
patient serum-treated cardiomyocytes, which in turn may contribute
to abnormal collagen accumulation in cardiac amyloidosis.
Based on KEGG pathway analysis of downregulated
DEGs, the ‘p53 signaling pathway’, the ‘cell cycle’, ‘oocyte
meiosis’ and ‘purine metabolism’ were listed as associated terms
for both the AL-09 peptide-treated and patient serum-treated
cardiomyocytes. CCNB1 was shown to be involved in the ‘p53
signaling pathway’, the ‘cell cycle’ and ‘oocyte meiosis’. CCNB1 is
a regulatory protein that is involved in mitosis and is required
for the correct control of the G2/M transition in the
cell cycle. The p53 tumor suppressor prevents this transition by
decreasing the intracellular CCNB1 protein levels and attenuating
the CCNB1 promoter activity (40). A
previous study showed that amyloid-β directly binds to cyclin B1 in
AD (41). In amyloidosis, cyclin D1
was identified as a prominent driver of the disease (42). In the present study, RNA-seq results
shown that Ccnb1 gene expression decreased both in the AL-09
peptide and the AL patient serum groups compared with their
respective controls. RT-qPCR results validated that the mRNA
expression levels of Ccnb1 statistically decreased in
peptide-treated group, but no significant difference was observed
in the AL patient serum group compared with their respective
controls. Further investigation is needed to explain the
discrepancy. The underlying mechanism of action behind the link
between cyclins and amyloidosis needs to be further elucidated.
In conclusion, the present study confirmed that
amyloidogenic LCs (λ and κ) from different sources may directly
damage the viability of cultured cardiomyocytes and promote cell
apoptosis. A total of 256 DEGs co-existed both in the λ and κ
LC-treated cardiomyocytes, of which 127 genes were upregulated and
88 genes were downregulated. The TGF-β-BMP signaling pathway, the
ErbB signaling pathway, prostaglandins and collagen production were
activated in LC-treated cardiomyocytes, with higher mRNA expression
levels of Bmp4, Bmp6, Ptgs1, Ptgs2, Ereg, Tgfa and
Plod2 observed. The p53 signaling pathway and the cell cycle
were hindered, characterized by decreasing Ccnb1 gene
expression levels. A limitation of the present study was that the
recombinant AL-09 protein was only confirmed using SDS-PAGE and
western blotting. No mass spectrometry or other biochemical
analysis was performed. Meanwhile, in vivo experiments were
not performed and protein expression changes were not investigated,
which would be beneficial to confirm the reported changes. There
were also no functional experiments for the associated pathways.
Further studies are needed to demonstrate the underlying mechanism
of action behind the gene alterations and cardiac amyloidosis, and
thus to provide biomarkers for diagnosis and to help develop
improved treatment strategies.
Acknowledgements
Not applicable.
Funding
This study was supported by the Open Project of
Jiangsu Commission of Health (grant no. XK05200903).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FX, XC and DX designed the study. FX, YY, PL and ZB
performed the experiments. FX, FW, WS and DX collected clinical
data. FX, HW and XC analyzed the results. FX, XC, HW and DX wrote
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The clinical study was approved by Medical Ethical
Committee of the First Affiliated Hospital of Nanjing Medical
University (approval no. 2015-SR-005). Formal written consent was
obtained from the healthy donor and the representative of the
patient. The research on experimental animals was approved by the
Institutional Animal Care and Use Committee of Nanjing Medical
University (approval no. IACUC-1708008).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liao R, Jain M, Teller P, Connors LH, Ngoy
S, Skinner M, Falk RH and Apstein CS: Infusion of light chains from
patients with cardiac amyloidosis causes diastolic dysfunction in
isolated mouse hearts. Circulation. 104:1594–1597. 2001.PubMed/NCBI
|
|
2
|
Guan J, Mishra S, Shi J, Plovie E, Qiu Y,
Cao X, Gianni D, Jiang B, Del Monte F, Connors LH, et al:
Stanniocalcin1 is a key mediator of amyloidogenic light chain
induced cardiotoxicity. Basic Res Cardiol. 108(378)2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Aimo A, Buda G, Fontana M, Barison A,
Vergaro G, Emdin M and Merlini G: Therapies for cardiac light chain
amyloidosis: An update. Int J Cardiol. 271:152–160. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Shi J, Guan J, Jiang B, Brenner DA, Del
Monte F, Ward JE, Connors LH, Sawyer DB, Semigran MJ, Macgillivray
TE, et al: Amyloidogenic light chains induce cardiomyocyte
contractile dysfunction and apoptosis via a non-canonical p38alpha
MAPK pathway. Proc Natl Acad Sci USA. 107:4188–4193.
2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Tuzovic M, Yang EH, Baas AS, Depasquale
EC, Deng MC, Cruz D and Vorobiof G: Cardiac amyloidosis: Diagnosis
and treatment strategies. Curr Oncol Rep. 19(46)2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tanaka K, Essick EE, Doros G, Tanriverdi
K, Connors LH, Seldin DC and Sam F: Circulating matrix
metalloproteinases and tissue inhibitors of metalloproteinases in
cardiac amyloidosis. J Am Heart Assoc. 2(e005868)2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
WRITING COMMITTEE MEMBERS. Yancy CW,
Jessup M, Bozkurt B, Butler J, Casey DE Jr, Drazner MH, Fonarow GC,
Geraci SA, Horwich T, et al: 2013 ACCF/AHA guideline for the
management of heart failure: A report of the American College of
Cardiology Foundation/American Heart Association Task Force on
practice guidelines. Circulation. 128:e240–e327. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Lavatelli F, Imperlini E, Orru S, Rognoni
P, Sarnataro D, Palladini G, Malpasso G, Soriano ME, Di Fonzo A,
Valentini V, et al: Novel mitochondrial protein interactors of
immunoglobulin light chains causing heart amyloidosis. FASEB J.
29:4614–4628. 2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Brenner DA, Jain M, Pimentel DR, Wang B,
Connors LH, Skinner M, Apstein CS and Liao R: Human amyloidogenic
light chains directly impair cardiomyocyte function through an
increase in cellular oxidant stress. Circ Res. 94:1008–1010.
2004.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Imperlini E, Gnecchi M, Rognoni P, Sabidò
E, Ciuffreda MC, Palladini G, Espadas G, Mancuso FM, Bozzola M,
Malpasso G, et al: Proteotoxicity in cardiac amyloidosis:
Amyloidogenic light chains affect the levels of intracellular
proteins in human heart cells. Sci Rep. 7(15661)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Guan J, Mishra S, Qiu Y, Shi J, Trudeau K,
Las G, Liesa M, Shirihai OS, Connors LH, Seldin DC, et al:
Lysosomal dysfunction and impaired autophagy underlie the
pathogenesis of amyloidogenic light chain-mediated cardiotoxicity.
EMBO Mol Med. 6:1493–1507. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Pattison JS and Robbins J: Protein
misfolding and cardiac disease: Establishing cause and effect.
Autophagy. 4:821–823. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Edwards HV, Cameron RT and Baillie GS: The
emerging role of HSP20 as a multifunctional protective agent. Cell
Signal. 23:1447–1454. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Sikkink LA and Ramirez-Alvarado M:
Cytotoxicity of amyloidogenic immunoglobulin light chains in cell
culture. Cell Death Dis. 1(e98)2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
McLaughlin RW, De Stigter JK, Sikkink LA,
Baden EM and Ramirez-Alvarado M: The effects of sodium sulfate,
glycosaminoglycans, and Congo red on the structure, stability, and
amyloid formation of an immunoglobulin light-chain protein. Protein
Sci. 15:1710–1722. 2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Baden EM, Owen BA, Peterson FC, Volkman
BF, Ramirez-Alvarado M and Thompson JR: Altered dimer interface
decreases stability in an amyloidogenic protein. J Biol Chem.
283:15853–15860. 2008.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Louch WE, Sheehan KA and Wolska BM:
Methods in cardiomyocyte isolation, culture, and gene transfer. J
Mol Cell Cardiol. 51:288–298. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Audic S and Claverie JM: The significance
of digital gene expression profiles. Genome Res. 7:986–995.
1997.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Danilewicz M and Wagrowska-Danilewicz M:
Immunohistochemical analysis of transforming growth factor beta-1
in AA and AL renal amyloidosis. Pol J Pathol. 57:193–198.
2006.PubMed/NCBI
|
|
21
|
Weiss R, Lifshitz V and Frenkel D: TGF-β1
affects endothelial cell interaction with macrophages and T cells
leading to the development of cerebrovascular amyloidosis. Brain
Behav Immun. 25:1017–1024. 2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lam HB, Yeh CH, Cheng KC, Hsu CT and Cheng
JT: Effect of cholinergic denervation on hepatic fibrosis induced
by carbon tetrachloride in rats. Neurosci Lett. 438:90–95.
2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Adams SL, Benayoun L, Tilton K, Mellott
TJ, Seshadri S, Blusztajn JK and Delalle I: Immunohistochemical
analysis of activin receptor-like kinase 1 (ACVRL1/ALK1) expression
in the rat and human hippocampus: Decline in CA3 during progression
of Alzheimer’s disease. J Alzheimers Dis. 63:1433–1443.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Papadopoulos P, Tong XK, Imboden H and
Hamel E: Losartan improves cerebrovascular function in a mouse
model of Alzheimer’s disease with combined overproduction of
amyloid-beta and transforming growth factor-β1. J Cereb Blood Flow
Metab. 37:1959–1970. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Dobaczewski M, Chen W and Frangogiannis
NG: Transforming growth factor (TGF)-β signaling in cardiac
remodeling. J Mol Cell Cardiol. 51:600–606. 2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Villar AV, García R, Llano M, Cobo M,
Merino D, Lantero A, Tramullas M, Hurlé JM, Hurlé MA and Nistal JF:
BAMBI (BMP and activin membrane-bound inhibitor) protects the
murine heart from pressure-overload biomechanical stress by
restraining TGF-β signaling. Biochim Biophys Acta. 1832:323–335.
2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wu F, Yao H, Bader A, Dong F, Zhu F, Wu N,
Wang B, Li H, Brockmeyer NH and Altmeyer P: Decorin gene transfer
inhibited the expression of TGFbeta1 and ECM in rat mesangial
cells. Eur J Med Res. 12:360–368. 2007.PubMed/NCBI
|
|
28
|
Lu J, Sun B, Huo R, Wang YC, Yang D, Xing
Y, Xiao XL, Xie X and Dong DL: Bone morphogenetic protein-2
antagonizes bone morphogenetic protein-4 induced cardiomyocyte
hypertrophy and apoptosis. J Cell Physiol. 229:1503–1510.
2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wu X, Sagave J, Rutkovskiy A, Haugen F,
Baysa A, Nygård S, Czibik G, Dahl CP, Gullestad L, Vaage J and
Valen G: Expression of bone morphogenetic protein 4 and its
receptors in the remodeling heart. Life Sci. 97:145–154.
2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Banach J, Gilewski W, Slomka A, Buszko K,
Błażejewski J, Karasek D, Rogowicz D, Żekanowska E and Sinkiewicz
W: Bone morphogenetic protein 6-a possible new player in
pathophysiology of heart failure. Clin Exp Pharmacol Physiol.
43:1247–1250. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Knittel T, Fellmer P, Muller L and
Ramadori G: Bone morphogenetic protein-6 is expressed in
nonparenchymal liver cells and upregulated by transforming growth
factor-beta 1. Exp Cell Res. 232:263–269. 1997.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yano R, Golbar HM, Izawa T, Sawamoto O,
Kuwamura M and Yamate J: Participation of bone morphogenetic
protein (BMP)-6 and osteopontin in cisplatin (CDDP)-induced rat
renal fibrosis. Exp Toxicol Pathol. 67:99–107. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Fattahi MJ and Mirshafiey A: Positive and
negative effects of prostaglandins in Alzheimer’s disease.
Psychiatry Clin Neurosci. 68:50–60. 2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Bitto A, Giuliani D, Pallio G, Irrera N,
Vandini E, Canalini F, Zaffe D, Ottani A, Minutoli L, Rinaldi M, et
al: Effects of COX1-2/5-LOX blockade in Alzheimer transgenic
3xTg-AD mice. Inflamm Res. 66:389–398. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Riese DJ II and Cullum RL: Epiregul in:
Roles in normal physiology and cancer. Semin Cell Dev Biol.
28:49–56. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Singh B, Carpenter G and Coffey RJ: EGF
receptor ligands: Recent advances. F1000Res.
5(F1000)2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhang H, Zhang L, Zhou D, He X, Wang D,
Pan H, Zhang X, Mei Y, Qian Q, Zheng T, et al: Ablating ErbB4 in PV
neurons attenuates synaptic and cognitive deficits in an animal
model of Alzheimer’s disease. Neurobiol Dis. 106:171–180.
2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
White SK, Sado DM, Flett AS and Moon JC:
Characterising the myocardial interstitial space: The clinical
relevance of non-invasive imaging. Heart. 98:773–779.
2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Du H, Pang M, Hou X, Yuan S and Sun L:
PLOD2 in cancer research. Biomed Pharmacother. 90:670–676.
2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Innocente SA, Abrahamson JL, Cogswell JP
and Lee JM: p53 regulates a G2 checkpoint through cyclin B1. Proc
Natl Acad Sci USA. 96:2147–2152. 1999.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Milton NG: The amyloid-beta peptide binds
to cyclin B1 and increases human cyclin-dependent kinase-1
activity. Neurosci Lett. 322:131–133. 2002.PubMed/NCBI View Article : Google Scholar
|
|
42
|
da Silva Filho MI, Försti A, Weinhold N,
Meziane I, Campo C, Huhn S, Nickel J, Hoffmann P, Nöthen MM, Jöckel
KH, et al: Genome-wide association study of immunoglobulin light
chain amyloidosis in three patient cohorts: Comparison with
myeloma. Leukemia. 31:1735–1742. 2017.PubMed/NCBI View Article : Google Scholar
|