Introduction
Traumatic lung injury (TLI) is often caused by blunt chest trauma (1). In Europe, the incidence rate of TLI is 27% among patients with multiple traumatic injuries (2) and the mortality rate is 56% in polytrauma patients (3). Typical complications of TLI include pneumonia and acute respiratory distress syndrome (4), which are associated with high mortality and morbidity rates (5). The clinical pathophysiology of TLI includes hypercarbia, hypoxemia, decreased lung volume and compliance, increased respiratory effort and ventilation-perfusion mismatch (6,7). Currently, therapeutic options for TLI are limited to basic supportive measures, including mechanical ventilation (8), which results in considerable economic burden on the healthcare system (9). In addition, no reliable surveillance or predictive biomarkers are currently available for the clinical diagnosis or identification of patients with a high risk of severe complications. It is necessary to investigate credible and stable biomarkers to predict the outcomes and complications of TLI and study the molecular mechanisms of TLI, in order to develop novel therapeutic approaches.
Circular RNAs (circRNAs) are a type of endogenous non-coding RNA that are characterized by a covalently continuous closed-loop structure (10,11). circRNAs are more stable and resistant to RNase R than liner RNAs due to the lack of 5'caps and 3'tails (12,13), which exhibit clear advantages in acting as molecular biomarkers for disease diagnosis. Previous studies have reported that circRNAs can target microRNAs (miRNAs), which are known as competitive endogenous RNAs (ceRNAs), and regulate gene expression at the transcriptional level (14,15). Previous studies have indicated that circRNAs are associated with malignancies including non-small cell lung cancer and pancreatic ductal adenocarcinoma (16-20). To the best of our knowledge, no previous study has investigated the expression levels or role of circRNAs in the pathogenesis of TLI. Therefore, the present study aimed to identify the differentially expressed circRNAs in TLI and investigate the molecular mechanism of TLI.
The present study performed circRNA expression profiling based using a high-throughput microarray analysis to identify TLI-specific circRNAs. The results of the current study indicated that the expression of circRNA_101523 was significantly upregulated, whereas circRNA_102927 and circRNA_100562 were downregulated in plasma of patients with TLI compared with controls. Subsequently, a bioinformatics analysis was used to construct ceRNA networks of these three circRNAs. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis identified potential biological processes and pathways of these networks. The present study also investigated circRNA_102927 in an additional group of clinical samples. The results of the present study suggested that circRNA_102927 may serve a role in the development of TLI and may be a novel and valuable biomarker of TLI.
Materials and methods
Patient sample collection
All patients were admitted to the Department of Burns Surgery of Changhai Hospital, The First Affiliated Hospital of The Naval Medicine University in Shanghai, from February 2017 to February 2018. All analyses were completed prior to March 2019. All patients were admitted to the hospital for the first time within 24 h after injury and were diagnosed by two surgeons based on their injury history and CT result (21) upon admission. A total of 50 samples were selected for the current study, and these samples were divided into a ‘TLI group’ and a ‘control group’. In total, five pairs of samples were used in microarray analysis and an additional 20 pairs were used for verifying the expression of circRNAs chosen in the present study using reverse transcription-quantitative PCR (RT-qPCR). In the disease group, patients were included based on the following criteria: Patients who presented to the Burn Surgery Department with isolated thoracic trauma and aged >18 years. Patients with severe basic diseases, burns, sepsis, vasculitis, patients with a recent history of surgery or those who refused to participate were excluded. In the control group, healthy volunteers, who were staff within the hospital (Shanghai, China) without any underlying disease were recruited. Authors had access to all information that could identify individual participants during or after data collection. The characteristics of patients with TLI and controls in the extended cohort are presented in Table I. In total, ~3 ml blood sample was drawn per patient on the third day after admission. Evacuated tubes were used to collect venous blood samples and ethylenediaminetetraacetic acid dipotassium was used as an anticoagulant. All plasma samples were processed as previously described (22-24). The plasma samples were processed in a centrifuge at 3,000 x g for 10 min at 4˚C and separated into a RNase-free tube.
 |
Table I
Characteristics of patients in the present cohort study
|
Table I
Characteristics of patients in the present cohort study
| Variables |
Control (n=20) |
TLI (n=20) |
P-value |
| Age (years) |
48.25±12.15 |
49.15±16.51 |
0.130a |
| Sex (%) |
|
|
|
| Male |
12(60) |
14(70) |
0.507b |
| Female |
8(40) |
6(30) |
|
| BMI (kg/m2) |
24.03±1.18 |
23.94±1.44 |
0.408a |
The present study was approved by the Shanghai Changhai Hospital Ethics Committee (approval no. CHEC2018-139) and has been registered at ClinicalTrials.gov (clinical trial no. NCT03766204; https://clinicaltrials.gov/ct2/show/NCT03766204). All participants or their representatives provided signed informed consent.
RNA isolation and quality control
Total RNA was extracted from plasma samples using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) based on the manufacturer's protocol. RNA integrity was assessed using electrophoresis on a 1% denaturing agarose gel (25). RNA concentration was measured using a NanoDrop ND-1000 spectrophotometer (26).
RNA labeling and hybridization
Sample labeling and array hybridization were performed based on the manufacturer's protocol (Arraystar, Inc.). Total RNA was digested with RNase R (Epicentre; Illumina, Inc.) to remove linear RNA. The amplified circRNAs were then transcribed into fluorescent complementary RNA (cRNA) using a random priming method (Arraystar Super RNA Labeling kit; Arraystar, Inc.). The labeled cRNA were purified using the RNeasy Mini kit (Qiagen, Inc.) according to the manufacturer's protocol. The concentration of the labeled cRNA (pmol Cy3/µg cRNA) was measured using NanoDrop-1000 (Thermo Fisher Scientific, Inc.). For each sample, the labeled cRNA was processed as per a previously published protocol (27).
Microarray data analysis of circRNA expression
Agilent Feature Extraction software (version 11.0.1.1; Agilent Technologies, Inc.) was used for raw data extraction. R software limma package (version 3.6; Arraystar, Inc.) was then used to process the raw data. After the low intensity filtering, the circRNAs that were flagged in ‘P’ (Present) or ‘M’ (Marginal) under ‘All Targets Value’ and ≤5 samples were kept for further analyses. circRNAs with a fold change ≥2 and a significant value of P≤0.03 between the TLI and the controls were considered to have significantly different expression levels. In addition, hierarchical clustering analysis was conducted using Java Treeview (R package ggplot2; Stanford University School of Medicine) based on expression levels of differentially expressed circRNAs.
RT-qPCR
The expression profiles of five circRNAs were validated in five pairs of plasma samples using RT-qPCR. These five circRNAs were selected randomly among circRNAs identified by microarray analysis of which raw signal intensity >200. RT-qPCR was performed as previously described (20). In brief, total RNA was extracted with TRI Reagent®-BD (Molecular Research Center, Inc.). SuperScript™ III Reverse Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) was used to perform RT-qPCR in a GeneAmp™ PCR System 9700 (Applied Biosystems; Thermo Fisher Scientific, Inc.). The RT temperature protocol was set to 50˚C for 60 min followed by 70˚C for 15 min to inactivate the enzyme. The primer sequences of target circRNAs and β-actin are listed in Table SI. All the primers were synthesized by Invitrogen (Thermo Fisher Scientific, Inc.). RT-qPCR was performed in a 10 µl reaction volume, which included 2 µl cDNA, 5 µl of 2X Master Mix, 0.5 µl forward primer (10 µM), 0.5 µl reverse primer (10 µM) and 2 µl ddH2O. The thermocycling condition was set to 95˚C for 10 min for pre-denaturation, followed by 40 cycles of 95˚C for 10 sec and at 60˚C for 60 sec. The relative expression levels of circRNAs were calculated using the 2-ΔΔCq method (28). RT-qPCR product was detected using 1% agarose gel electrophoresis using a denaturing agarose gel stained using ethidium bromide (Biotium, Inc.) in 70˚C for 5 min.
Prediction of circRNA-miRNA-mRNA targeting network
circRNA and miRNA interaction was predicted using Arraystar's homemade miRNA target prediction software which was based on TargetScan (version 7.1) and miRanda (version V5). The differentially expressed circRNAs validated by RT-qPCR were annotated to include the circRNA, miRNA and mRNA co-expression information. GO (http://geneontology.org/) and KEGG (https://www.kegg.jp/kegg/pathway.html) analysis were conducted using R language with home-made script (Kangchen BioTech Co., Ltd.).
Statistical analysis
All experimental data were analyzed using SPSS software 23.0 (IBM Corp.) and the graphs were plotted in GraphPad Prism 7.0 (GraphPad Software, Inc.). Data are presented as mean ± standard deviation, median ± interquartile range or proportions as appropriate. Differences in circRNA expression levels between the two groups were calculated using a two-tailed Student's t-test or one-way ANOVA. P<0.05 was considered to indicate a statistically significant difference.
Results
Microarray identification of differentially expressed circRNAs
The present study identified the differences in circRNA expression profile patterns between five pairs of plasma samples from patients with TLI and matched controls using high-throughput human circRNA microarray. A box plot was drawn to visualize the intensities in all datasets after normalization, which showed a similar distribution of log2 ratios across all samples (Fig. 1A). The results of hierarchical clustering and scatter plot visualization presented different circRNA expression profiling among the ten samples (Fig. 1B and C). The results indicated that there was a total of 310 abnormally expressed circRNAs (fold change >2; P<0.03) in TLI plasma compared with controls. Among them, 250 circRNAs were downregulated and 60 were upregulated. A Volcano plot was used to visualize the significant differences (fold change >2; P<0.03) between TLI samples and controls (Fig. 1D). The results indicated that the expression profile of circRNAs in the plasma of patients with TLI was distinct compared with healthy controls.
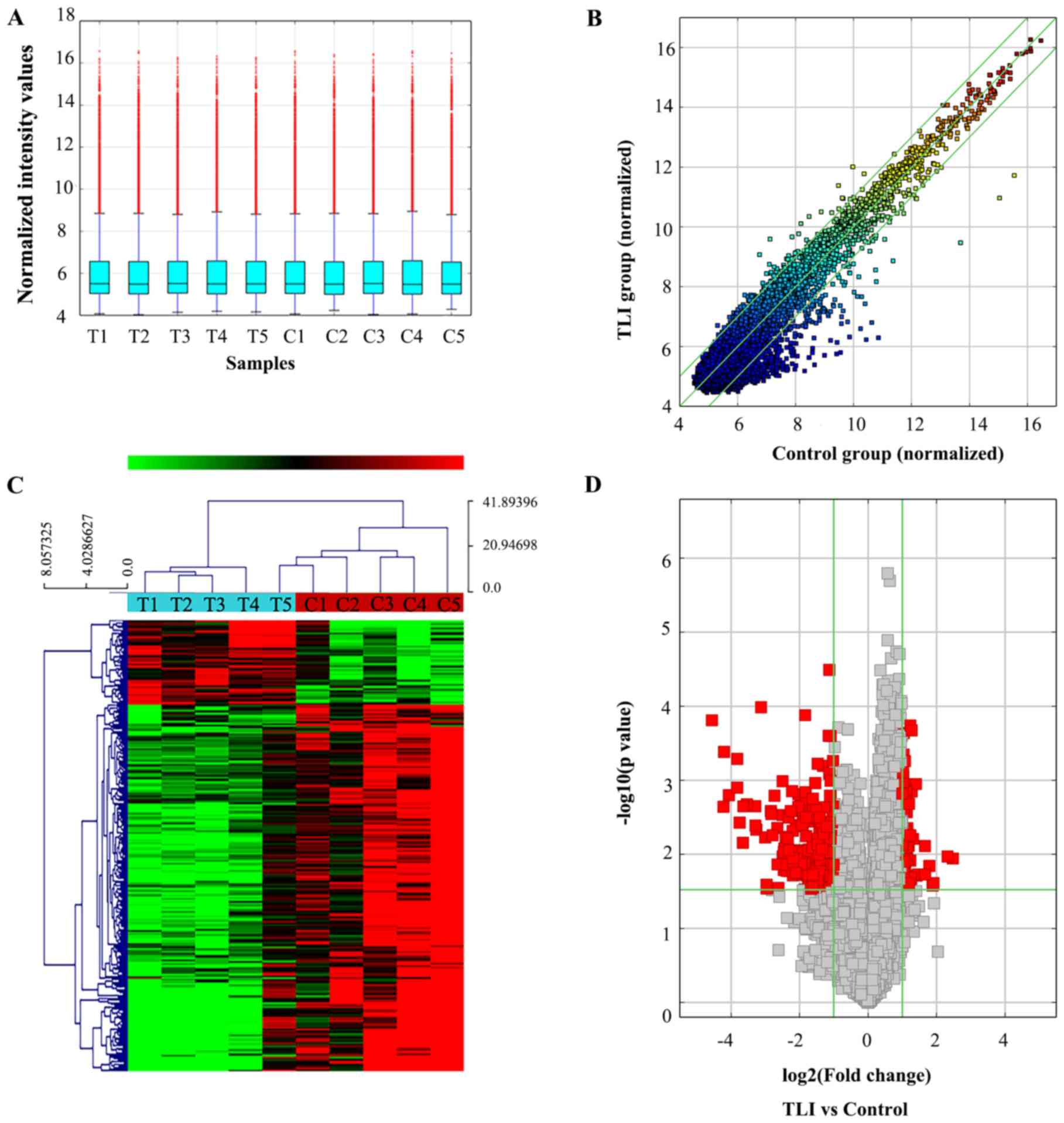 |
Figure 1
Differences and characterizations of circRNA expression profile between TLI plasma samples and paired normal samples. (A) Box plots of the distribution of circRNAs for the ten samples. The distributions were similar after normalization. (B) circRNAs in the scatter-plot above the top green line and below the bottom green line indicated >2-fold change of circRNAs between the two groups. (C) Unsupervised hierarchical clustering was built to show circRNA expression profiling among the all samples. Each row corresponds to one circRNA and each column represents the expression profile of one plasma sample. ‘Red’ indicates higher expression level and ‘green’ indicates lower expression level. (D) Volcano plots were used to visualize the differential circRNA expression. The vertical green lines corresponded to 2.0-fold up and down, and the horizontal green line represented a P<0.03. The red points in plot represent the differentially expressed circRNAs with statistical significance. TLI, traumatic lung injury; T, traumatic lung injury group; C, control; circRNAs, circular RNAs.
|
RT-qPCR validation of selected circRNAs
To identify the high-throughput microarray data, two upregulated circRNAs [homo sapiens circular RNA 101952 (hsa_circRNA_101952) and hsa_circRNA_101523] and three downregulated circRNAs (hsa_circRNA_102927, hsa_circRNA_100562 and hsa_circRNA_102034) were selected (Table II). The present study investigated the expression levels of these circRNAs in the plasma of patients with TLI compared with controls, using the same samples as in the microarray analysis, using RT-qPCR. Compared with control patients, the expression level of hsa_circRNA_101523 was significantly higher, whilst those of hsa_circRNA_102927 and hsa_circRNA_100562 was significantly lower; which a trend that is similar to the microarray results (Fig. 2).
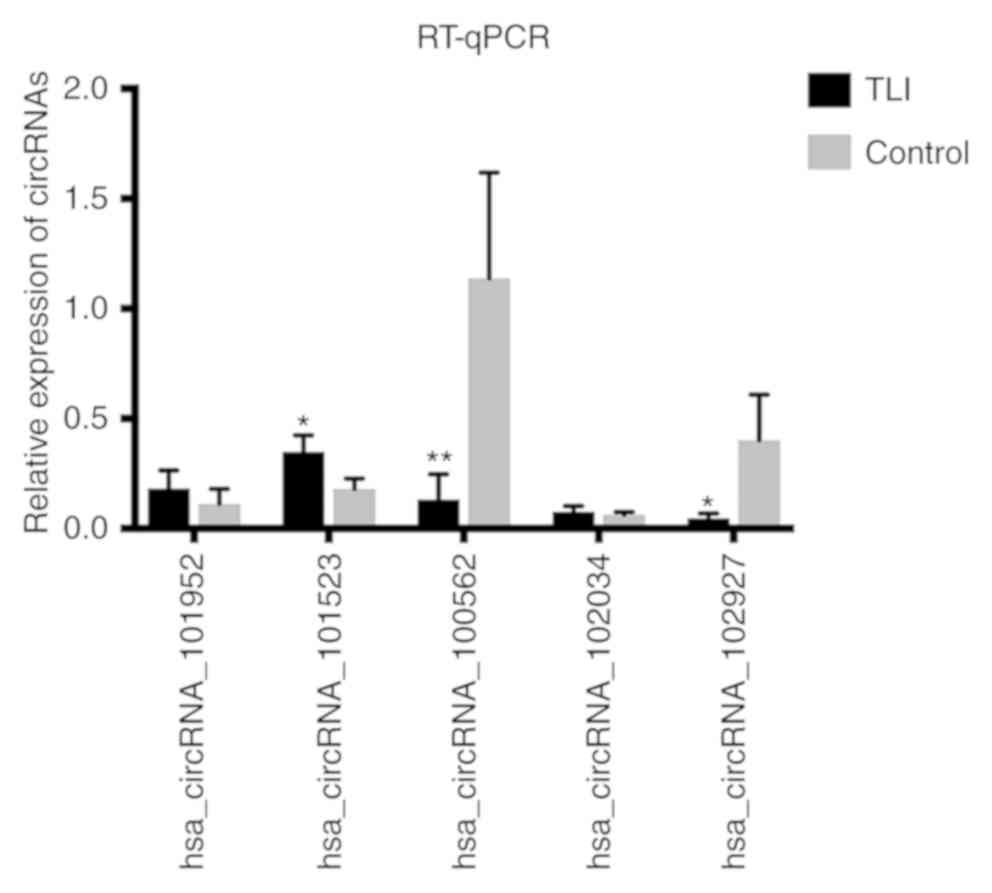 |
Figure 2
RT-qPCR identified five differentially expressed circRNAs in TLI. Expression levels of hsa_circRNA_102927, hsa_circRNA_101523 and hsa_circRNA_100562 matched the microarray results. *P<0.05 and **P<0.01 vs. Control. circRNAs, circular RNAs; hsa_circRNA, homo sapiens circular RNA; TLI, traumatic lung injury; RT-q, Reverse transcription-quantitative.
|
 |
Table II
Biological information of the selected five circRNAs.
|
Table II
Biological information of the selected five circRNAs.
| circRNA ID |
Fold change |
P-value |
circRNA type |
Chromosome |
Best transcripta |
Gene symbol |
| Upregulated |
|
|
|
|
|
|
| hsa_circRNA_101952 |
2.9682598 |
0.00125 |
Exonic |
chr17 |
NM_015113 |
ZZEF1 |
| hsa_circRNA_101523 |
2.801168 |
0.00062 |
Exonic |
chr15 |
NM_014547 |
TMOD3 |
| Downregulated |
|
|
|
|
|
|
| hsa_circRNA_102927 |
2.6298996 |
0.01442 |
Exonic |
chr2 |
NM_032276 |
RHBDD1 |
| hsa_circRNA_100562 |
7.8189314 |
0.02890 |
Exonic |
chr10 |
NM_012425 |
RSU1 |
| hsa_circRNA_102034 |
7.8189314 |
0.00681 |
Exonic |
chr17 |
NM_018307 |
RHOT1 |
Bioinformatics prediction of circRNA-miRNA-mRNA interaction
To investigate the potential molecular functions of the selected circRNAs, circRNA and miRNA interactions were predicted using Arraystar's homemade miRNA target prediction software which was based on TargetScan and miRanda. A total of 31 miRNAs were targeted by the three circRNAs. To further investigate how these three circRNAs could regulate gene expression as ceRNAs, the present study predicted the related target genes according to the predicted miRNAs targeted by these three circRNAs using MirdbV5 and TargetScan7.1. Using the overlapping results of the two databases (Fig. S1), which comprised 568 target genes for the identified circRNAs, a prediction network was built using Cytoscape (version 3.7.2; https://cytoscape.org/) (Fig. 3). All circRNAs, miRNAs and genes were listed in Table SII.
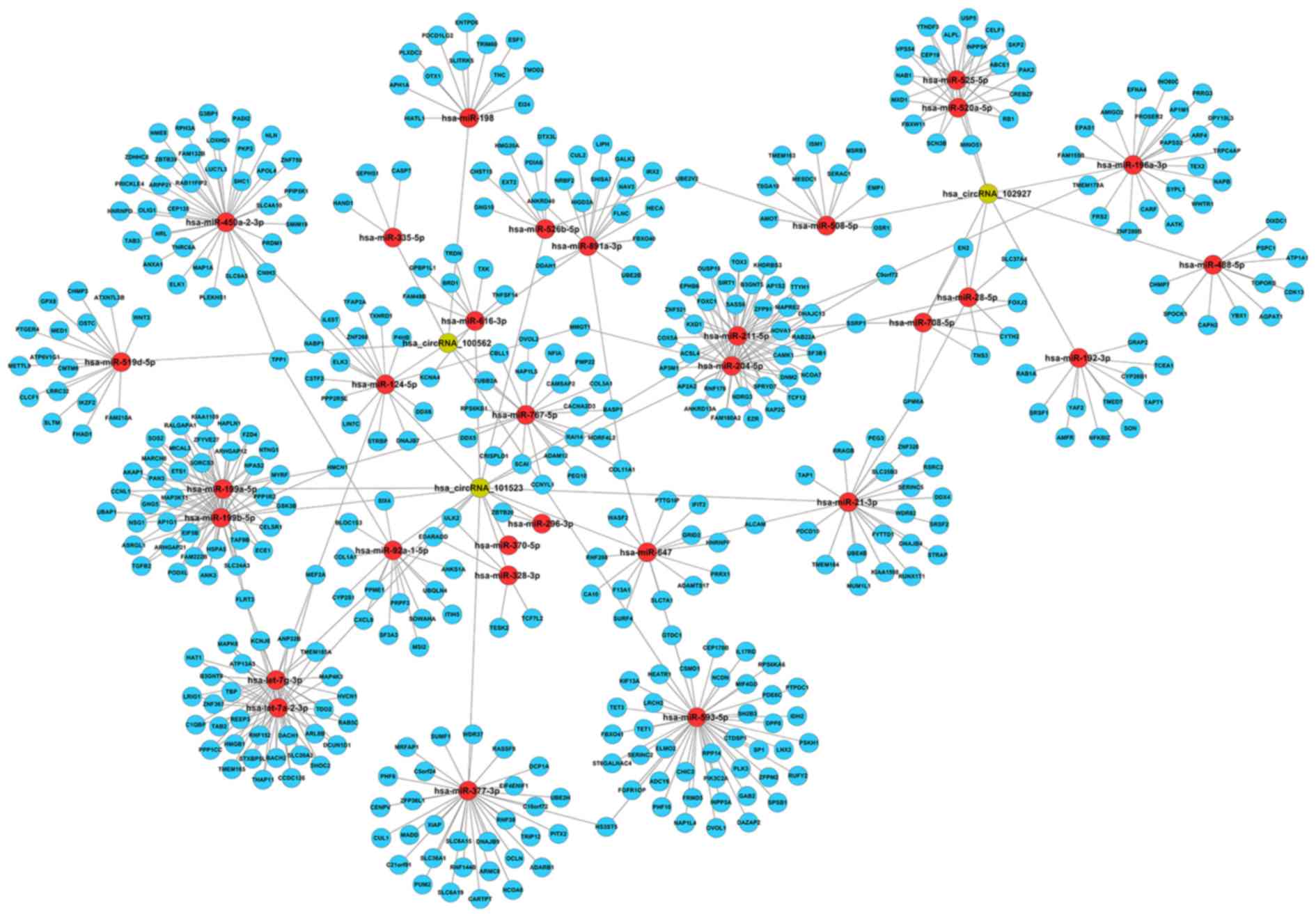 |
Figure 3
Targeted miRNAs and mRNAs network prediction. Prediction circRNAs-miRNAs-mRNAs network constructed by Cytoscape after sequence-pairing prediction. circRNAs, circular RNAs; miRNA, microRNA. All circRNAs, miRNAs and mRNAs included in network were listed in Table SII.
|
Functional analysis of predicted co-expression genes for the circRNAs
The present study performed GO analysis on the ten most significantly enriched genes to identify their functional roles in terms of molecular functions, cellular components and biological processes (BP; Fig. 4A). The results suggested the predicted genes were closely associated with biological processes, including cellular differentiation, positive regulation of transcription, positive regulation of macromolecule metabolic processing and DNA-templated transcription. Mostly, these genes were indicated to serve roles in protein binding in intracellular membrane-bound organelles or the nucleoplasm. KEGG analysis for these genes showed that the top ten significantly enriched pathways included hepatocellular carcinoma, mTOR signaling pathway and herpes simplex virus infection (Fig. 4B). The results suggested these processes and pathways may be associated with the pathophysiology process of TLI.
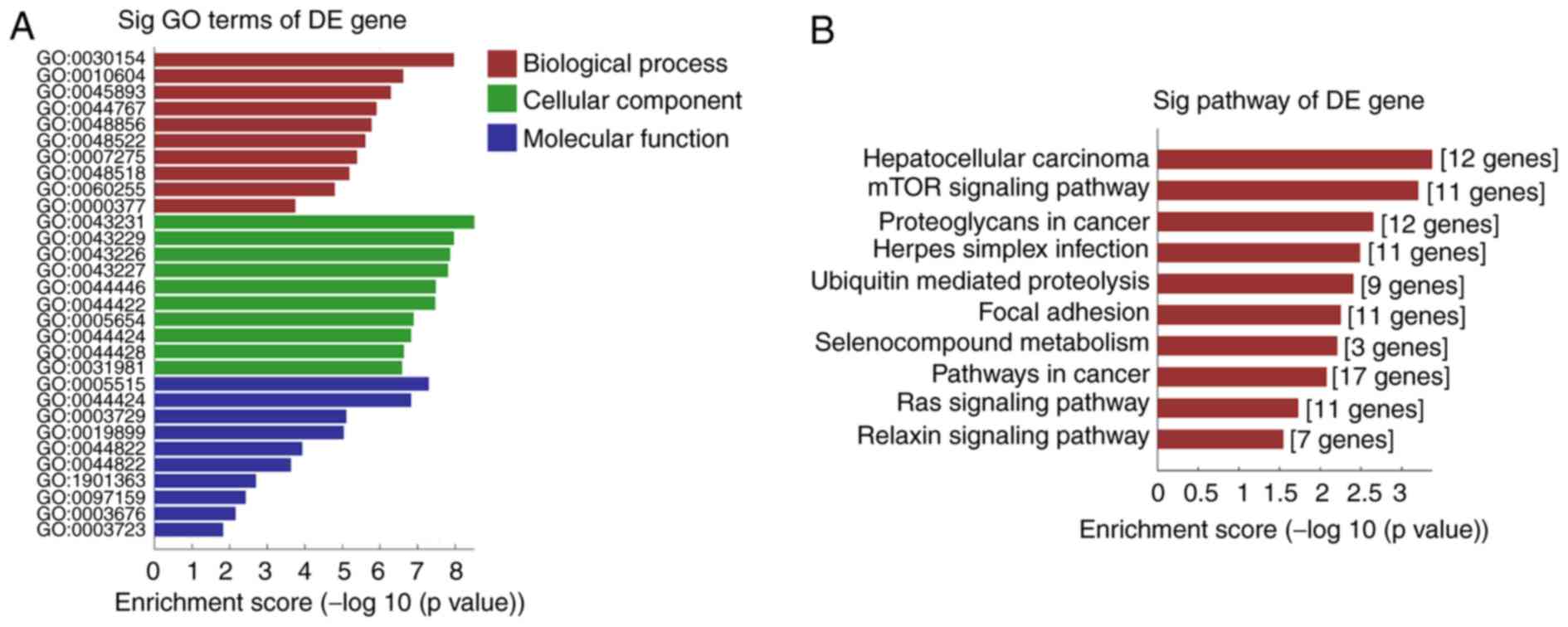 |
Figure 4
Enriched GO terms and KEGG pathways of host genes of differentially expressed circRNAs in TLI. (A) Significantly enriched GO biological processes, cellular components and molecular functions of differentially expressed circular RNAs in TLI. (B) Significantly enriched top ten KEGG pathways of differentially expressed circular RNAs in TLI. circRNAs, circular RNAs; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; TLI, traumatic lung injury; DE, differentially expressed.
|
Expanded identification of circRNAs expression in TLI
An expanded sample size was used to investigate the results of the present study, with an additional 20 matched pairs of clinical plasma samples used to identify the expression levels of hsa_circ_102927 by RT-qPCR. The present results identified a significant downregulation of hsa_circ_102927 in the plasma of patients with TLI (0.34-fold; P<0.001; Fig. 5A). Subsequently, a receiver-operating characteristic (ROC) curve was drawn to measure the ability of hsa_circ_102927 to distinguish TLI plasma from normal controls. The area under the ROC curve (AUC) was 0.978 (Fig. 5B). The specificity and sensitivity were both 95.0%. The optimal cutoff value of hsa_circ_102927 was 0.01595.
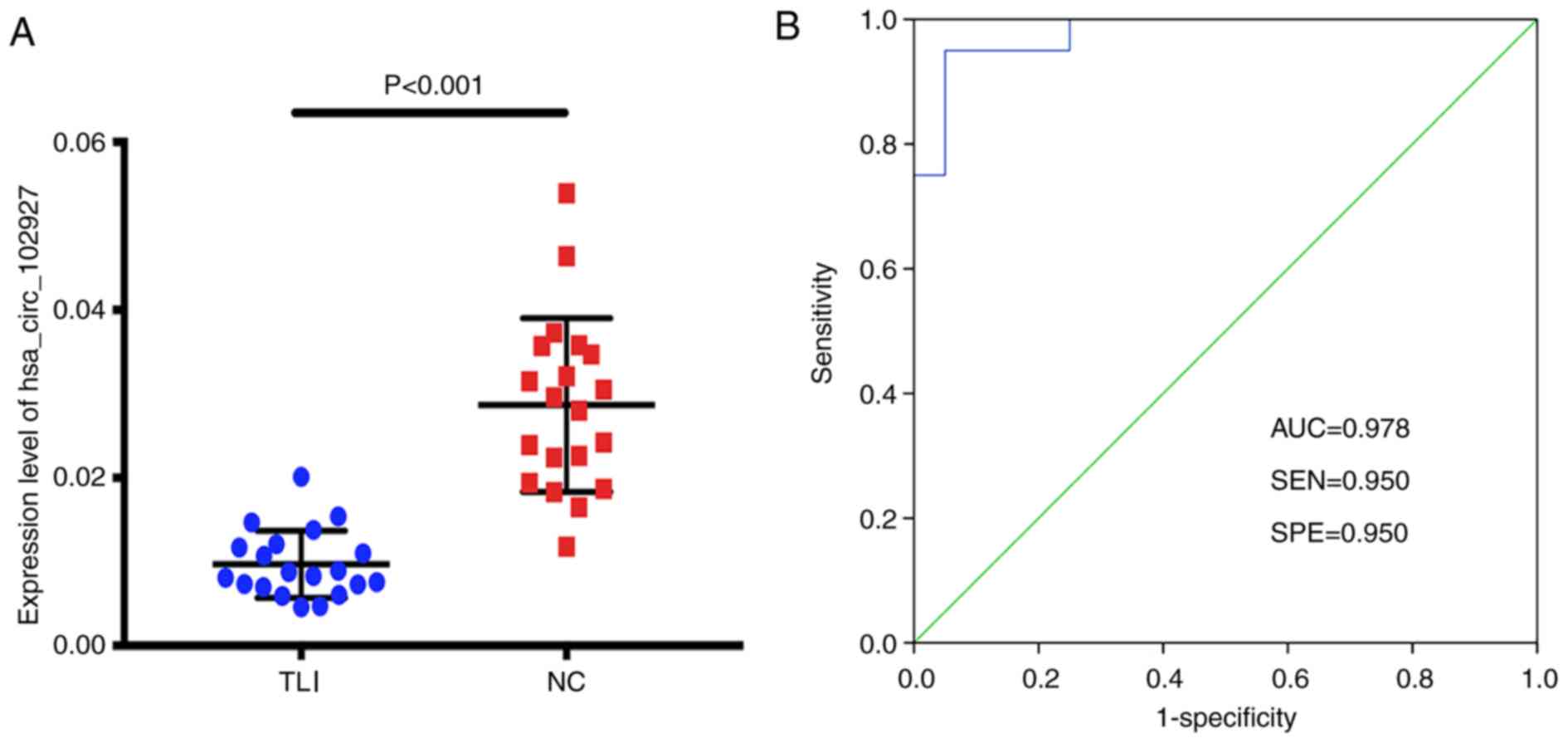 |
Figure 5
Hsa_circ_102927 is significantly downregulated in patients with TLI. (A) Expression levels of hsa_circ_102927 in the TLI group were significantly lower compared with the control group. P<0.001; n=20. (B) A ROC curve of hsa_circ_102927 was drawn to differentiate TLI from controls. circRNA, circular RNA; ROC, receiver-operating characteristic; AUC, area under the ROC curve; hsa_circRNA, homo sapiens circular RNA; TLI, traumatic lung injury; NC, negative control; SPE, specificity; SEN, sensitivity.
|
Discussion
There has been increasing development in high-throughput RNA-sequencing microarray and bioinformatics analysis tools in recent decades (16). In addition, there has been greater research interest in circRNAs. However, to the best of our knowledge, there are no previous studies focused on circRNAs in TLI, therefore, the present study may be the first to study circRNA expression profile in patients with TLI. The present study exhibits advantages to ensure accuracy of results. Firstly, the present study collected valuable clinical samples from patients with TLI, which highlights the validity and reliability of the analyzed results. Secondly, the screening conditions were set as fold change >2 and P<0.03 to select differentially expressed circRNAs. Thirdly, published research relating to miRNAs was used to filter the results of the bioinformatics analysis (29). Finally, a group of expanded size of samples were used to investigate the expression level of hsa_circ_102927. The present results indicated that hsa_circ_102927 was significantly downregulated, with an average fold change of 0.34 compared with controls. The present ROC analysis suggested that hsa_circ_102927 was highly specific (95%) and sensitive (95%), with an AUC of 0.978. The present results suggested a potential direction for further research in TLI.
As an accidental injury, TLI is accompanied with multiple trauma (30). The main pathophysiological changes peak at 72 h after injury (31). Therefore, the present study performed a microarray on plasma collected from the patients on the third day after admission. The microarray analysis identified circRNAs that were significantly dysregulated in patients with TLI compared with controls; including 250 that were downregulated and 60 that were upregulated. The present results indicated the importance of microarray analysis in the validation of differentially expressed circRNAs. Next, five highly expressed circRNAs identified from the microarray data were selected for further verification. After RT-qPCR analysis, the selected circRNAs hsa_circRNA_101523, hsa_circRNA_102927 and hsa_circRNA_100562 were identified to be significantly differently expressed in patients with TLI. The present results indicated that these circRNAs may be differentially regulated and may have potential functions in TLI.
miRNAs regulate the translation of genes by mRNA degradation or inhibiting mRNA translation by binding to the 3' untranslated region of their target genes (32). Based on two databases, 568 target genes of the predicted miRNAs were selected, and GO and KEGG analyses were subsequently performed. The ceRNA network constructed in the present study had the same mRNAs, along with involvement of the top ten significant enrichment BPs. The results of the present study suggested the three circRNAs, as well as their co-expression ceRNA networks, may be potential molecular factors involved in TLI pathogenesis.
However, there are some limitations to the present study, despite analyzing the microarray data and RT-qPCR validation. TLI occurs after severe accidents, and whilst the present study established criteria to select patients, it was difficult to maintain the homogeneity of all samples. The present study detected five samples from patients with high-throughput microarray, but the baseline severity of TLI was not the same across all patients. Therefore, the RT-qPCR results are not identical with the microarray analysis. The present RT-qPCR validation results indicated that hsa_circRNA_101952 and hsa_circRNA_102034 did not significantly change. As a result, more patients with TLI of similar severity are required to further validate circRNAs expression. The present study selected miRNAs based on databases and literature to construct the ceRNA network, and whether these microRNAs are expressed differently in TLI requires further investigation.
Future studies should investigate the expression levels of miRNAs, as well as mRNAs, in the ceRNA network. Furthermore, future studies should investigate how the ceRNA pathways affect the development of TLI in in vitro and in vivo experiments.
In summary, the present study identified the circRNA expression profile of the plasma from patients with TLI compared with controls. Moreover, three highly expressed circRNAs were investigated using RT-qPCR. Bioinformatics analysis indicated the potential ceRNA networks and the biological processes and pathways involved in TLI. The present results suggested that hsa_circ_102927 may serve as a potential biomarker for TLI diagnosis and may be involved in the pathology of TLI. In addition, the present results identified an important research basis for exploring other lung diseases with similar pathological mechanisms to TLI.
Supplementary Material
Figure S1. A total of 568 microRNA.targeted genes were predicted using the overlapping results of the two databases MirdbV5 and TargetScan7.1.
Table SI. Sequences of the primers for the detection of selected circRNAs targets and β-actin.
Table SII. The list of ceRNA network prediction.
Acknowledgements
Not applicable.
Funding
The present study was funded by National Natural Science Foundation of China (grant nos. 81772125, 81670015 and 81801937).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YJ, FZ, YS and ZFX conceived and designed the experiments. QY and CW performed the experiments. GSW, YJ and KAW analyzed the data. BHZ and FZ collected the patient samples. BHZ analyzed the raw data of microarray. YJ and GSW wrote the manuscript. ZFX, YS and FZ reviewed the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by The Medical Ethical Board of Changhai Hospital (approval no. CHEC2018-139) and has been registered at ClinicalTrials.gov (clinical trial no. NCT03766204; https://clinicaltrials.gov/ct2/show/NCT03766204). All participants or their representatives provided signed informed consent. Patient information was anonymized and de-identified before analysis.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Wu XJ, Xia ZY, Wang LL, Luo T, Zhan LY, Meng QT and Song XM: Effects of penehyclidine hydrochloride on pulmonary contusion from blunt chest trauma in rats. Injury. 43:232–236. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Balci AE, Kazez A, Eren S, Ayan E, Ozalp K and Eren MN: Blunt thoracic trauma in children: Review of 137 cases. Eur J Cardiothorac Surg. 26:387–392. 2004.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Gaillard M, Herve C, Mandin L and Raynaud P: Mortality prognostic factors in chest injury. J Trauma. 30:93–96. 1990.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Calfee CS, Eisner MD, Ware LB, Thompson BT, Parsons PE, Wheeler AP, Korpak A and Matthay MA: Acute Respiratory Distress Syndrome Network National Heart Lung and Blood Institute: Trauma-associated lung injury differs clinically and biologically from acute lung injury due to other clinical disorders. Crit Care Med. 35:2243–2250. 2007.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Suresh MV, Yu B, Lakshminrusimha S, Machado-Aranda D, Talarico N, Zeng L, Davidson BA, Pennathur S and Raghavendran K: The protective role of MnTBAP in oxidant-mediated injury and inflammation in a rat model of lung contusion. Surgery. 154:980–990. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Van Eeden SF, Klopper JF, Alheit B and Bardin PG: Ventilation-perfusion imaging in evaluating regional lung function in nonpenetrating injury to the chest. Chest. 95:632–638. 1989.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Cohn SM: Pulmonary contusion: Review of the clinical entity. J Trauma. 42:973–979. 1997.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Klein Y, Cohn SM and Proctor KG: Lung contusion: Pathophysiology and management. Curr Opin Anaesthesiol. 15:65–68. 2002.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Suresh MV, Thomas B, Machado-Aranda D, Dolgachev VA, Kumar Ramakrishnan S, Talarico N, Cavassani K, Sherman MA, Hemmila MR, Kunkel SL, et al: Double-stranded RNA interacts with toll-like receptor 3 in driving the acute inflammatory response following lung contusion. Crit Care Med. 44:e1054–e1066. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ebbesen KK, Hansen TB and Kjems J: Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ledford H: Circular RNAs throw genetics for a loop. Nature. 494(415)2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hsiao KY, Sun HS and Tsai SJ: Circular RNA-New member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 242:1136–1141. 2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 19:141–157. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al: Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 495:333–338. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kulcheski FR, Christoff AP and Margis R: Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 238:42–51. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S and Yuan H: Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 8(2878)2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Huang WJ, Wang Y, Liu S, Yang J, Guo SX, Wang L, Wang H and Fan YF: Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting PLK1 expression. Cancer Lett. 422:70–80. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, et al: Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 66:1151–1164. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Huang ZK, Yao FY, Xu JQ, Deng Z, Su RG, Peng YP, Luo Q and Li JM: Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from active tuberculosis patients. Cell Physiol Biochem. 45:1230–1240. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Guo W, Zhang B, Mu K, Feng SQ, Dong ZY, Ning GZ, Li HR, Liu S, Zhao L, Li Y, et al: Circular RNA GRB10 as a competitive endogenous RNA regulating nucleus pulposus cells death in degenerative intervertebral disk. Cell Death Dis. 9(319)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wicky S, Wintermark M, Schnyder P, Capasso P and Denys A: Imaging of blunt chest trauma. Eur Radiol. 10:1524–1538. 2000.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jiang Y, Brynskikh AM, S-Manickam D and Kabanov AV: SOD1 nanozyme salvages ischemic brain by locally protecting cerebral vasculature. J Control Release. 213:36–44. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Jiang Y, Arounleut P, Rheiner S, Bae Y, Kabanov AV, Milligan C and Manickam DS: SOD1 nanozyme with reduced toxicity and MPS accumulation. J Control Release. 231:38–49. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Jiang Y, Fay JM, Poon CD, Vinod N, Zhao Y, Bullock K, Qin S, Manickam DS, Yi X, Banks WA and Kabanov AV: Nanoformulation of brain-derived neurotrophic factor with target receptor-triggered-release in the central nervous system. Adv Funct Mater. 28(1703982)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Natarajan G, Perriotte-Olson C, Bhinderwala F, Powers R, Desouza CV, Talmon GA, Yuhang J, Zimmerman MC, Kabanov AV and Saraswathi V: Nanoformulated copper/zinc superoxide dismutase exerts differential effects on glucose vs lipid homeostasis depending on the diet composition possibly via altered AMPK signaling. Transl Res. 188:10–26. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jiang Y, Gaudin A, Zhang J, Agarwal T, Song E, Kauffman AC, Tietjen GT, Wang Y, Jiang Z, Cheng CJ and Saltzman WM: A ‘top-down’ approach to actuate poly(amine-co-ester) terpolymers for potent and safe mRNA delivery. Biomaterials. 176:122–130. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Singh M, George AK, Homme RP, Majumder A, Laha A, Sandhu HS and Tyagi SC: Expression analysis of the circular RNA molecules in the human retinal cells treated with homocysteine. Curr Eye Res. 44:287–293. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ferruelo A, Penuelas O and Lorente JA: MicroRNAs as biomarkers of acute lung injury. Ann Transl Med. 6(34)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Suresh MV, Thomas B, Dolgachev VA, Sherman MA, Goldberg R, Johnson M, Chowdhury A, Machado-Aranda D and Raghavendran K: Toll-like receptor-9 (TLR9) is requisite for acute inflammatory response and injury following lung contusion. Shock. 46:412–419. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Cohn SM and Dubose JJ: Pulmonary contusion: An update on recent advances in clinical management. World J Surg. 34:1959–1970. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Xia B, Lu J, Wang R, Yang Z, Zhou X and Huang P: miR-21-3p Regulates influenza A virus replication by targeting histone deacetylase-8. Front Cell Infect Microbiol. 8(175)2018.PubMed/NCBI View Article : Google Scholar
|



















