Introduction
Colorectal cancer is a common type of cancer
worldwide; according to global cancer statistics, the number of new
cases of cancer originating from the colon and rectum reached
~1,800,000 in 2018(1). Despite
advances in our understanding of the molecular mechanism and the
development of new therapeutic methods, the prognosis of patients
with colorectal cancer remains poor, as the five-year overall
survival is near 60% (2). This is
due to the aggressive nature of colorectal cancer cells (3,4). To
overcome obstacles in the treatment of colorectal cancer, recent
studies have shifted their focus to the metabolism of tumor cells
that are pivotal for the survival and development of drug
resistance of colorectal cancer cells (5-7).
Cancer cells exhibit a strong requirement for energy
to sustain rapid proliferation and cell division (8). Unlike normal cells, most cancer cells
predominantly obtain energy via a high rate of glycolysis even in
aerobic conditions (9,10). During the pathogenesis of cancer,
reprogramming of metabolism leads to increased glucose consumption
and lactic acid fermentation, known as Warburg effect (9). Hexokinase 2 (HK2) is a member of the HK
family of enzymes that converts glucose to glucose-6-phosphate in
cells (11). Elevation of HK2
activity is commonly observed in cancer cells exhibiting Warburg
glycolysis (12). Glycolysis is also
required for colorectal cancer cells to activate signaling pathways
and maintain cell growth (13). In
colorectal cancer, studies found that HK2 was upregulated in
colorectal cancer cells, and its expression was associated with
worse overall survival of patients with colorectal cancer (14). Most recently, targeted therapy
against HK2 has shown beneficial effects on inhibiting the
proliferation of colorectal cancer cells (15).
MicroRNAs (miRNAs/miRs) are small non-coding
single-stranded molecules in cells (16). Mechanistically, miRNAs directly bind
to the 3'untranslated region (3'UTR) region in the mRNA of genes,
which leads to degradation of mRNA or inhibition of translation
(17). Through the tight control of
target gene expression, miRNAs are involved in almost all
physiological processes (18).
Dysregulation of miRNAs is associated with various types of cancer
(19). Several differentially
expressed miRNAs have been identified in colorectal cancer
(20,21). miRNAs such as miR-16, miR-17 and
miR-181a have been experimentally proven to act as oncogenes or
tumor suppressors in colorectal cancer cells (22-24).
miR-1, miR-133b, miR-124, miR-137 and miR-340 have been identified
as tumor suppressors via repressing the cellular metabolism of
colorectal cancer (6,25).
In the current study, the expression of miR-513a-3p
in colorectal cancer cells and tissues was investigated. Our data
suggested that miR-513a-5p inhibited glycolysis in colorectal
cancer cells via repressing HK2, indicating miR-513a-5p as a tumor
suppressor in colorectal cancer.
Materials and methods
Tissue collection
A total of 30 colorectal tumors and matched normal
tissues were collected from patients during surgery in the People's
Hospital of Boluo County (Huizhou, China) between February 2015 and
May 2017. All participants did not receive chemotherapy or
radiotherapy before surgery. Written informed consent was provided
by all patients, and the protocol of current study was approved by
the Ethics committee of the People's Hospital of Boluo County. The
samples were immediately stored in -80˚C before being subjected to
RNA extraction.
Cell culture
A normal colonic mucosal cell line, FHC, and
colorectal cancer cell lines, HCT116 and SW480, were purchased from
ATCC. Cells were cultured in DMEM medium (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) in an incubator at 37˚C with 5%
CO2.
RNA isolation and RT-qPCR
Total RNA was extracted from tissues and cells with
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
following manufacturer's protocol. The quantity and quality of RNA
was determined using a NanoDrop 2000 (Thermo Fisher Scientific,
Inc.). RNA was then reverse transcribed into first-stranded cDNA
with PrimeScript™ RT Reagent kit (Takara Bio, Inc.).
qPCR was performed on a Real-time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with a SYBR Green PCR kit (Takara
Bio, Inc.) following the manufacturer's protocol (26). The quantification of miRNA was
performed with the stem-loop RT-PCR as reported before (27). The RT-qPCR condition was: predenature
at 95˚C for 30 sec, followed by 35 cycles of denature at 95˚C for
10 sec and elongation at 55˚C for 20 sec. β-actin and U6 were used
as internal controls for mRNA and miRNA, respectively. The primer
sequences were as follows: β-actin forward,
5'-CATGTACGTTGCTATCCAGGC-3' and reverse,
5'-CTCCTTAATGTCACGCACGAT-3'; HK2 forward,
5'-GAGCCACCACTCACCCTACT-3' and reverse, 5'-CCAGGCATTCGGCAATGTG-3';
enolase 2 (ENO2) forward, 5'-AGCCTCTACGGGCATCTATGA-3' and reverse,
5'-TTCTCAGTCCCATCCAACTCC-3'; phosphoglycerate kinase 1 (PGK1)
forward, 5'-TGGACGTTAAAGGGAAGCGG-3' and reverse,
5'-GCTCATAAGGACTACCGACTTGG-3'; phosphofructokinase platelet (PFKP)
forward, 5'-GCATGGGTATCTACGTGGGG-3' and reverse,
5'-CTCTGCGATGTTTGAGCCTC-3'; hexokinase 1 (HK1) forward,
5'-GCTCTCCGATGAAACTCTCATAG-3' and reverse,
5'-GGACCTTACGAATGTTGGCAA-3'; aldolase A (ALDOA) forward,
5'-ATGCCCTACCAATATCCAGCA-3' and reverse,
5'-GCTCCCAGTGGACTCATCTG-3'; miR-513a-3p forward,
5'-GCCGAGTTTTAATTTATATT-3' and reverse, 5'-CTCAACTGGTGTCGTGGA-3';
and U6 forward, 5'-GCGCGTCGTGAAGCGTTC-3' and reverse,
5'-GTGCAGGGTCCGAGGT-3'. The sequence for the stem-loop primer was
5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAAATT-3'.
Protein isolation and western
blotting
Total protein was extracted from cells using RIPA
lysis buffer (Thermo Fisher Scientific, Inc.) according to the
manufacturer's guidelines. After preparation of lysates, the
concentration was determined with a bicinchoninic acid kit (Pierce;
Thermo Fisher Scientific, Inc.). A total of 20 µg of each sample
was electrophoresed on an 8% gel using SDS-PAGE, and proteins were
transferred to a PVDF membrane. The membrane was incubated in
primary antibodies and secondary antibodies for 1 h at room
temperature sequentially. The blots were detected and visualized by
a chemiluminescence detection system (Pierce; Thermo Fisher
Scientific, Inc.). Primary antibodies against HK2 (1:2,000; cat.
no. ab228819) and β-actin (1:10,000; cat. no. ab8226,) were
purchased from Abcam. HRP-conjugated secondary antibodies against
mouse (1:50,000; cat. no. SA00001-1) and rabbit (1:50,000; cat. no.
SA00001-2) were obtained from ProteinTech Group, Inc.
Overexpression of miR-513a-3p
The miR-513a-3p mimic
(5'-UAAAUUUCACCUUUCUGAGAAGG-3') and negative control (miR-NC) mimic
(5'-AUUGGAACGAUACAGAGAAGAUU-3') were synthesized and purchased from
Shanghai GenePharma Co., Ltd. 1x106 HCT116 or SW480
cells were transfected with 50 nM miR-513a-3p mimic or miR-NC mimic
using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. At 48 h after
transfection, cells were harvested for subsequent experiments.
Overexpression of HK2
Overexpression of HK2 was achieved via transfection
of the pcDNA3 plasmid containing full length cDNA of HK2. The cDNA
of HCT116 was prepared as aforementioned. The full length of HK2
was amplified from HCT116 cDNA and ligated into the pcDNA3 plasmid
(Addgene, Inc.). The primer sequences were: HK2-forward:
5'-AAGCTTATGATTGCCTCGCA-3'; HK2-reverse: 5'- TCTAGACTATCGCTGTC-3'.
A total of 2 µg pcDNA3-HK2 or pcDNA3 was co-transfected with 50 nM
miR-NC mimic or miR-513a-3p mimic into 1X106 HCT116 or
SW480 cells with Lipofectamine 3000. After 48 h, cells were
subjected to subsequent experiments.
Determination of cell
proliferation
The proliferation ability of cells was detected with
a Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular
Technologies, Inc.) according to the manufacturer's protocol. In
brief, 10,000 cells were plated into each well of 96-well plates.
At the timepoints of 0, 24, 48 and 72 h after transfection, 10 µl
CCK-8 solution was added to the wells and incubated for 2 h at
37˚C. The medium containing CCK-8 solution was then transferred to
another 96-well plate. The absorbance at 450 nm was detected, and
was considered to reflect cell number.
Colorimetric assay of glucose
uptake
The glucose uptake ability of HCT116 and SW480 cells
was determined with a Glucose Uptake Colorimetric Assay kit
(BioVision, Inc.) following manufacturers' protocol. A total of
2,000 cells were plated in each well of 96-well plates. Cells were
transfected with miR-NC mimic or miR-513a-3p mimic, as
aforementioned. After 24 h, the cells were starved for glucose by
preincubating with 100 µl Krebs-Ringer-Phosphate-HEPES buffer
containing 2% BSA for 40 min at 37˚C. Next, 10 µl 10 mM
2-deoxy-D-glucose was added and the cells were incubated for 20
min. The absorbance at 412 nm was detected every 5 mins until the
100 pM standard reached 1.5.
Detection of lactate levels
The lactate levels in the culture medium were
detected using a Lactate Colorimetric/Fluorometric Assay kit
(BioVision, Inc.) following the manufacturer's protocol. A total of
2,000 cells were seeded in each well of 96-well plates and
transfected with miR-513a-3p mimic or miR-NC mimic. After 48 h,
lactate assay buffer was added to each well, and after incubation
for 30 mins, absorbance at 570 nm was detected and was considered
to reflect lactate levels.
Bioinformatic analysis and dual
luciferase reporter assay
Using GEO database (https://www.ncbi.nlm.nih.gov/gds/), the differentially
expression miRNA in colon adenoma, colon carcinoma and normal
tissues were analyzed based on GSE115513 (including expression
profile of 381 normal tissues, 51 colon adenoma and 411 colon
carcinoma). The potential target genes and putative binding sites
for miR-513a-3p were analyzed with TargetScan software (www.targetscan.org/vert_72/). The 3'UTR
containing the putative binding sites for miR-513-3p was cloned
using HCT116 cDNA and ligated into the pGL3 plasmid (Promega
Corporation). The primer sequence: HK2-forward:
5'-CTCTAGAAACCCCTGAAATCG-3'; HK2-reverse:
5'-CTCTAGATTTGATTATTTTGGA-3'. A total of 1x105 cells
were seeded into each well in 24-well plates. Cells were
transfected with 2 µg pGL3-HK2-3'UTR-WT or pGL3-HK2-3'UTR-Mut and
miR-513a-3p mimic or miR-NC mimic using Lipofectamine 3000. At 24 h
after transfection, the relative luciferase activity was detected
using a Dual Luciferase Reporter Assay system (Promega
Corporation).
Statistical analysis
All data were calculated with GraphPad Prism 6
(GraphPad Software, Inc.) and presented as mean ± SD. Differences
between two groups were analyzed using a two-tailed Student's
t-test. Differences between multiple groups were assessed using
one-way ANOVA followed by the Dunnett's post hoc test. Pearson's
correlation analysis was used to analyze the correlation between
miR-513a-3p expression and HK2 mRNA expression levels. Each
experiment was repeated at least 3 times. P<0.05 was considered
to indicate a statistically significant difference.
Results
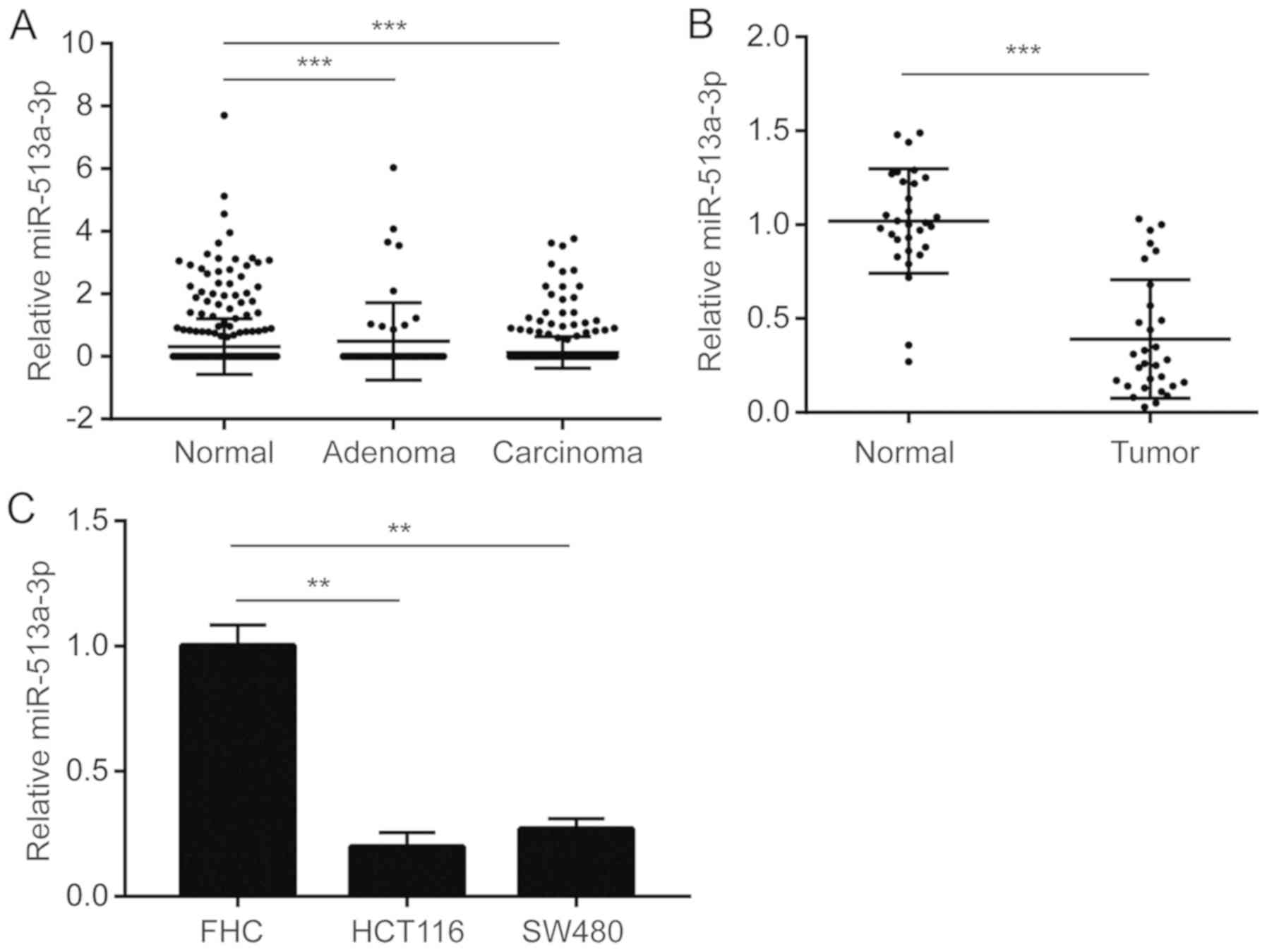
Downregulation of miR-513a-3p in
colorectal cancer
miRNA expression data of colorectal cancer was
retrieved from published datasets. miR-513a-3p was found to be
significantly downregulated in colon adenoma and colon carcinoma
compared with normal colon tissues (Fig.
1A). For validation, 30 pairs of tumor and normal tissues were
collected from patients with colorectal cancer. RT-qPCR results
suggested that miR-513a-3p was downregulated in colorectal tumors
(Fig. 1B). In addition, miR-513a-3p
levels were also lower in the colorectal cancer cell lines HCT116
and SW480 compared with the normal colonic mucosal cell line, FHC
(Fig. 1C).
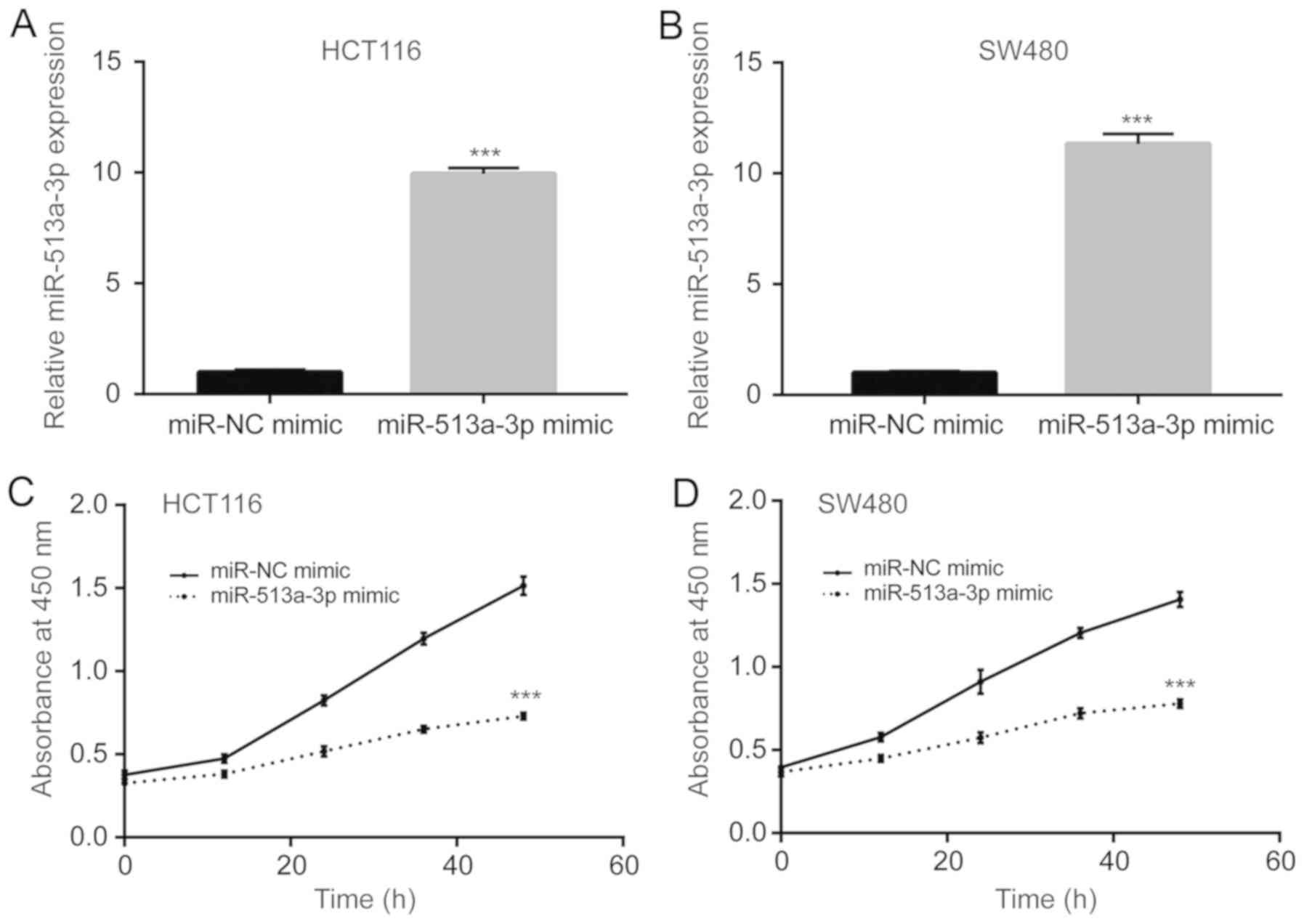
Overexpression of miR-513a-3p
suppresses colorectal cancer cell proliferation and metabolism
To investigate the role of miR-513a-3p in colorectal
cancer cells, miR-513a-3p mimic was transfected into HCT116 and
SW480 cells. Transfection of miR-513a-3p mimic induced a 10-fold
increase in the expression of miR-513a-3p in HCT116 cells (Fig. 2A). Similarly, miR-513a-3p expression
also increased in SW480 cells after transfection with miR-513a-3p
mimic (Fig. 2B). The proliferation
of colorectal cancer cells was detected with a CCK-8 kit. It was
found that miR-513a-3p overexpression significantly reduced
proliferation of HCT116 cells (Fig.
2C). In SW480 cells, overexpression of miR-513a-3p also
inhibited proliferation (Fig. 2D).
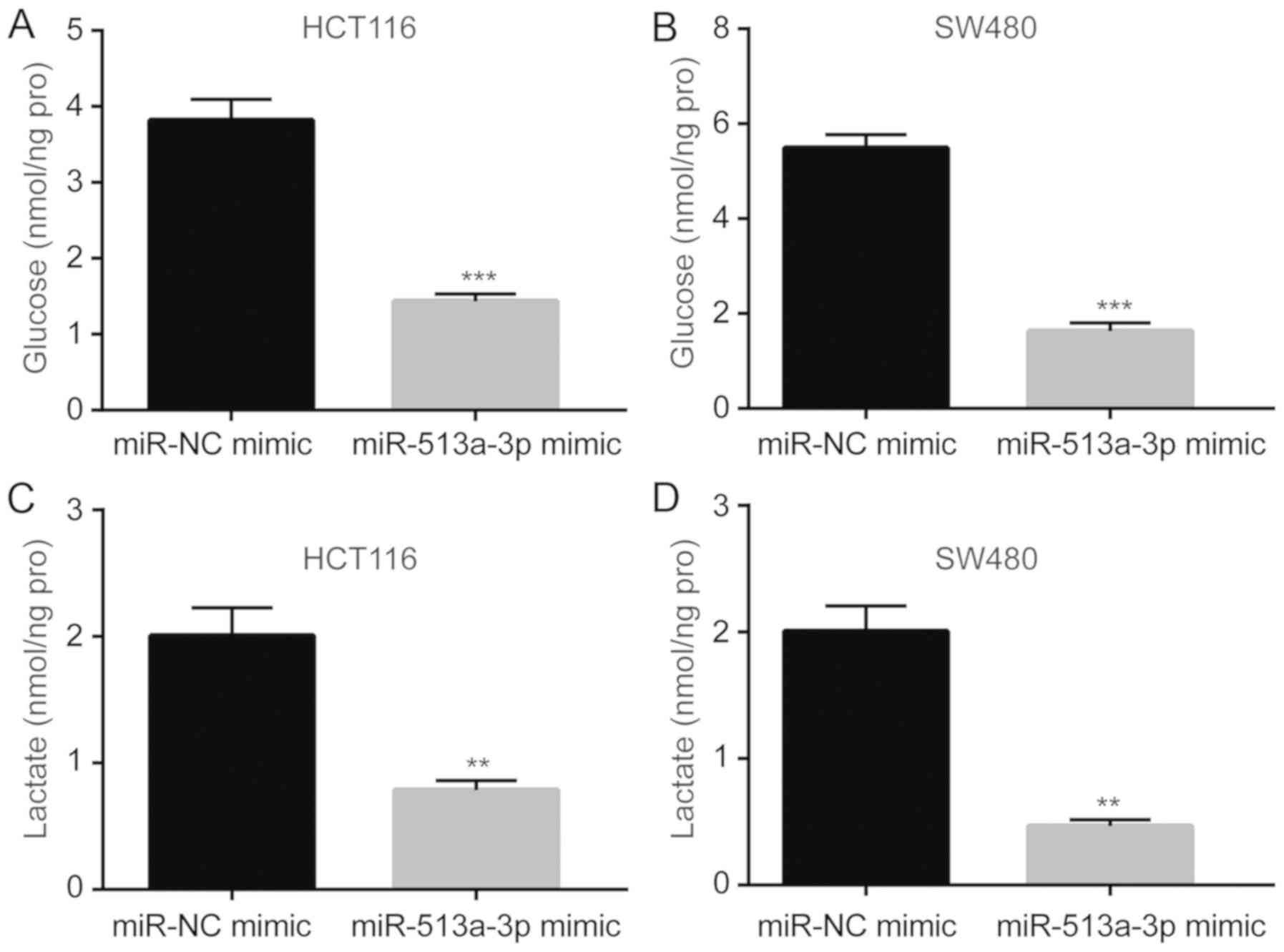
Increased proliferation of colorectal cancer cells is associated
with aberrant cell metabolism, including an abnormally high rate of
glycolysis. miR-513a-3p overexpression significantly reduced the
glucose uptake rate in HCT116 and SW480 cells (Fig. 3A and B). Additionally, lactate levels in the
supernatant of cultured cells were significantly decreased after
miR-513a-3p overexpression (Fig. 3C
and D). These results suggested that
miR-513a-3p negatively regulated the glycolytic process of
colorectal cancer cells to inhibit proliferation.
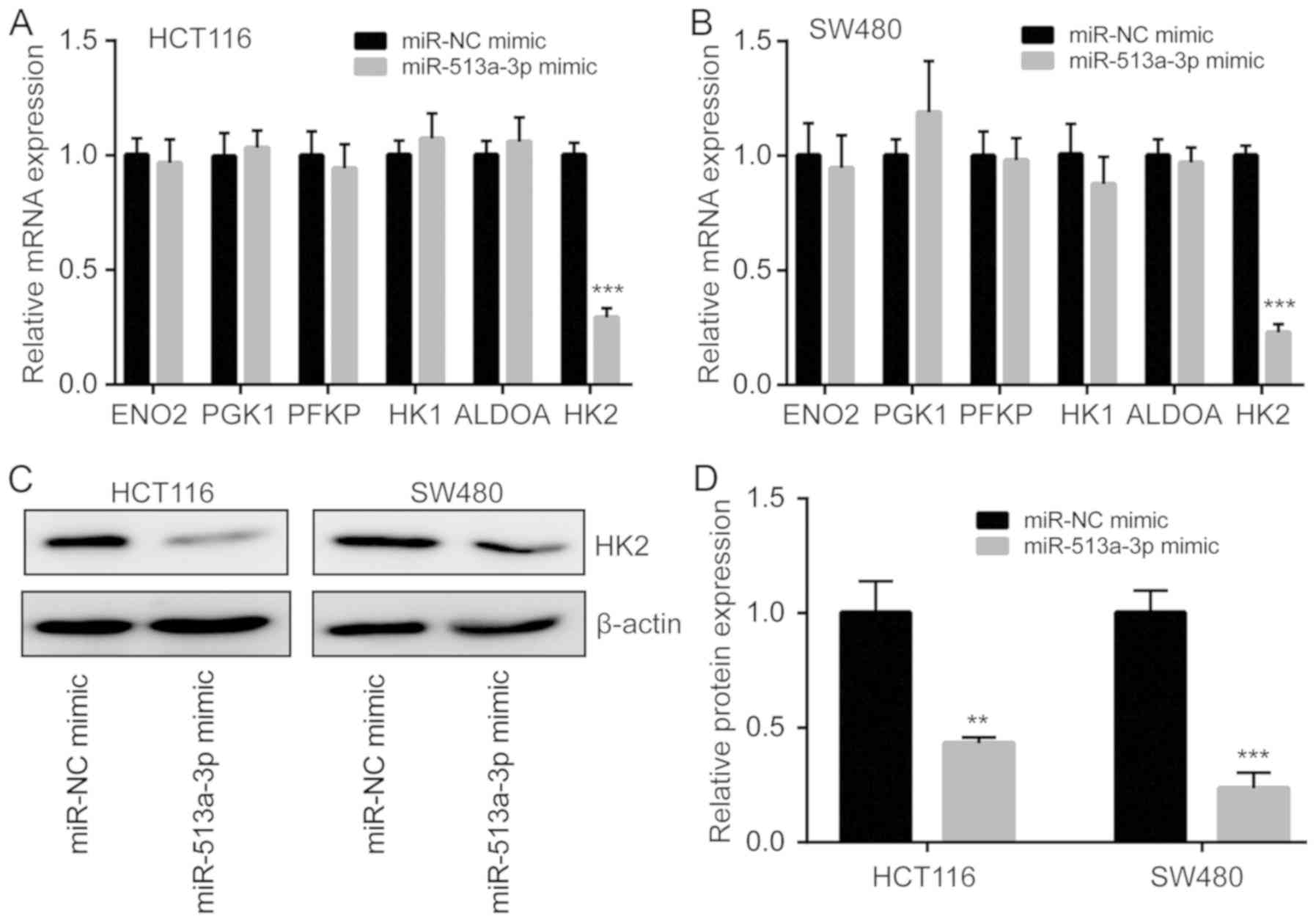
miR-513a-3p inhibits HK2 expression in
colorectal cancer cells
To investigate how miR-513a-3p regulates glycolysis
in colorectal cancer cells, RT-qPCR was performed to detect the
expression of multiple key genes involved in glycolysis, including
ENO2, PGK1, PFKP, HK1, ALDOA and HK2 (28,29). The
results showed that mRNA expression of HK2 was significantly
reduced while the expression of ENO2, PGK1, PFKP, HK1 and ALDOA was
not changed in HCT116 cells transfected with miR-513a-3p mimic
(Fig. 4A). In SW480 cells, the
decrease of HK2 mRNA expression was also observed following
miR-513a-3p overexpression (Fig.
4B). Additionally, western blotting showed that miR-513a-3p
overexpression decreased HK2 protein expression in both HCT116 and
SW480 cells (Fig. 4C and D).
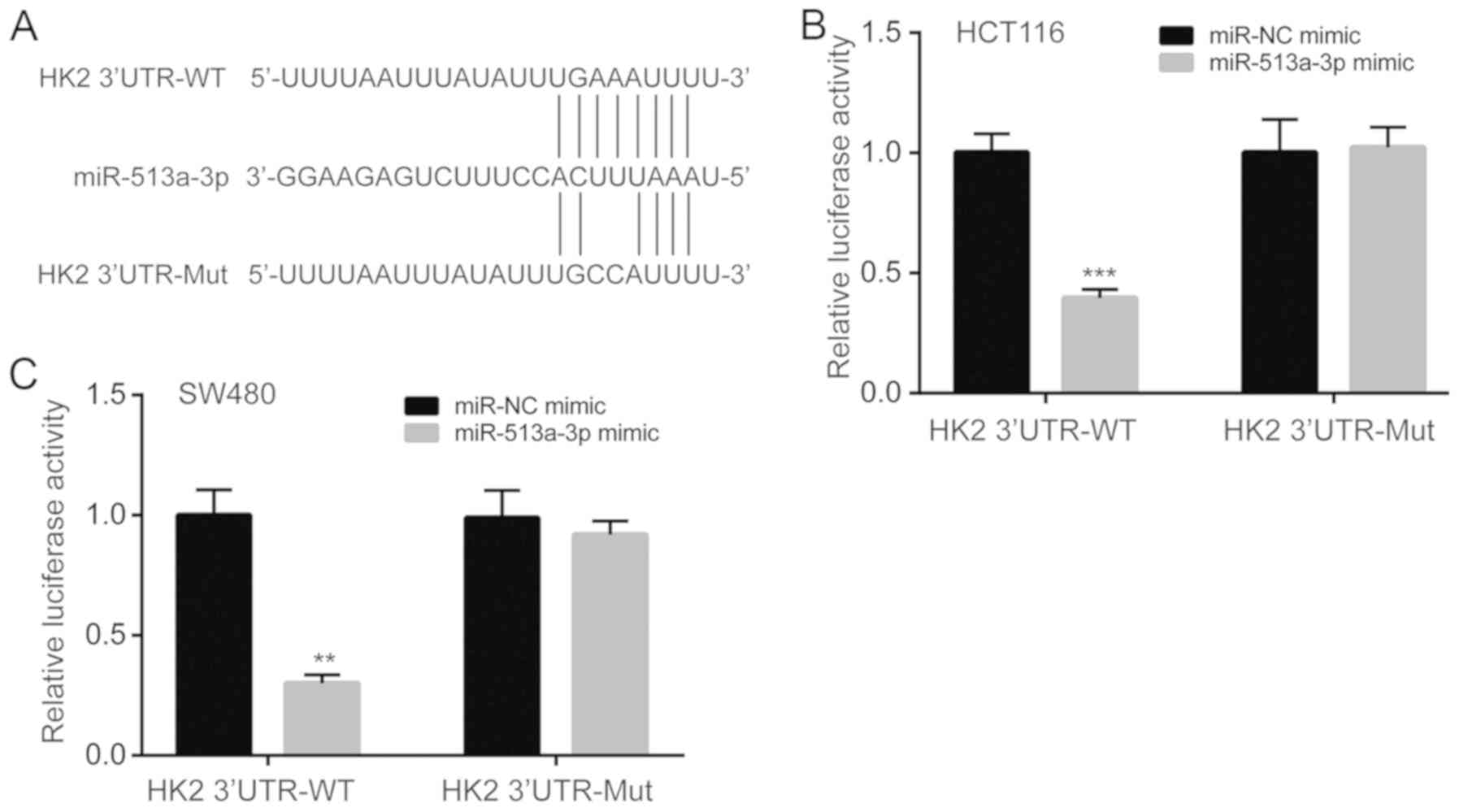
miR-513a-3p directly represses HK2
expression
To explore whether miR-513a-3p directly regulated
HK2 expression, bioinformatic analysis was performed. It was found
that there was a putative binding site for miR-513a-3p in the 3'UTR
of HK2 mRNA (Fig. 5A). A luciferase
reporter plasmid containing wild-type (HK2 3'UTR-WT) or mutant (HK2
3'UTR-Mut) 3'UTR of the HK2 mRNA was constructed (Fig. 5A). The dual luciferase reporter assay
showed that miR-513a-3p overexpression reduced relative luciferase
activity in HCT116 cells transfected with HK2 3'UTR-WT, but not in
cells transfected with HK2 3'UTR-Mut (Fig. 5B). The results in HCT116 cells were
consistent with results in SW480 cells, as the miR-513a-3p mimic
also reduced relative luciferase activity in SW480 cells
transfected with HK2 3'UTR-WT only (Fig.
5C). These results indicated that miR-513a-3p directly bound to
HK2 mRNA to repress its expression in colorectal cancer cells.
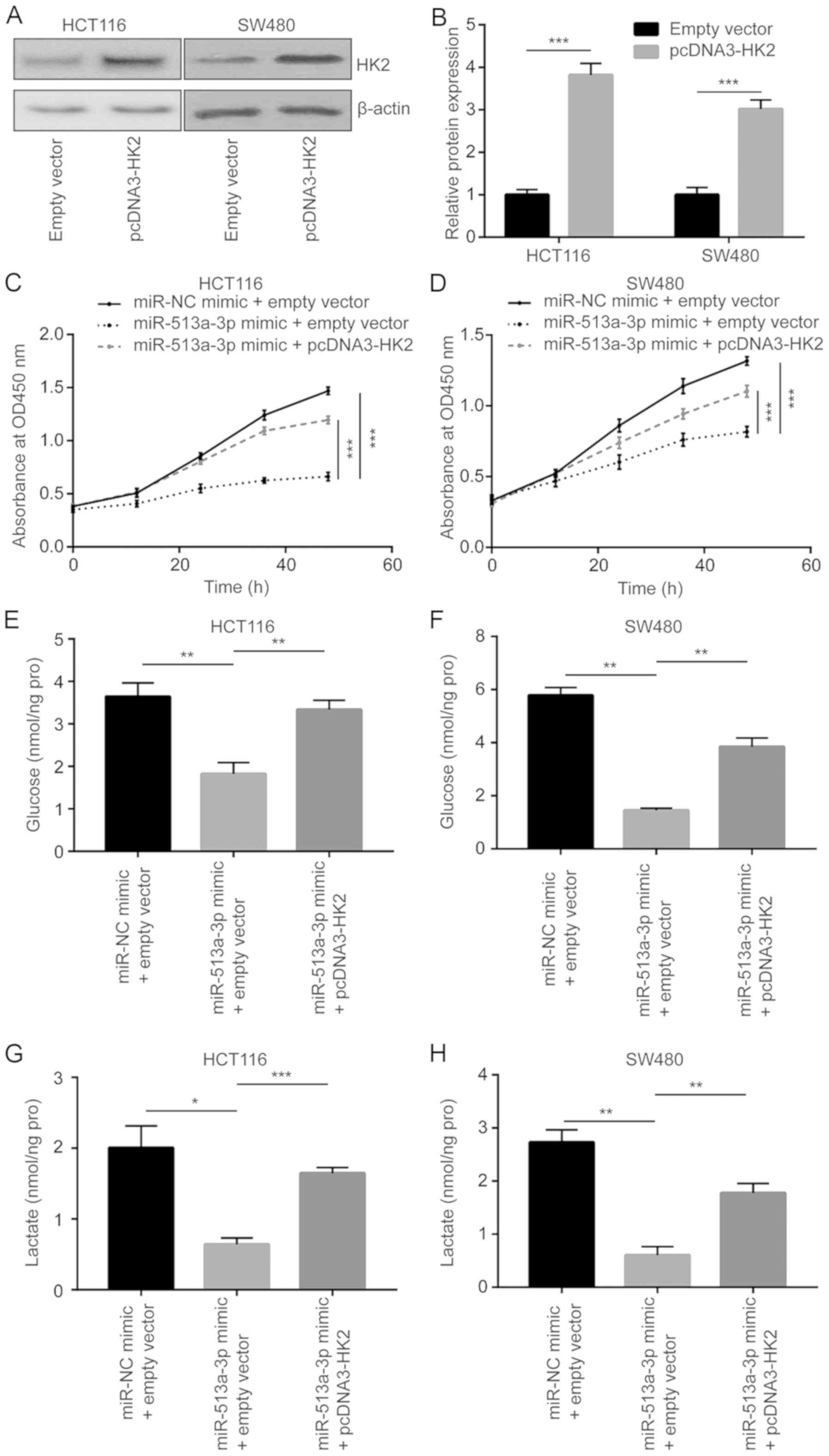
miR-513a-3p inhibits proliferation and
glycolysis of colorectal cancer cells via targeting HK2
A plasmid containing the HK2 cDNA sequence was
constructed. Transfection of pcDNA3-HK2 upregulated HK2 protein
expression in HCT116 and SW480 cells (Fig. 6A and B). A proliferation assay revealed that
overexpression of HK2 reversed the reduction of proliferation
induced by miR-513a-3p mimic in HCT116 and SW480 cells (Fig. 6C and D). Additionally, the decrease of glucose
uptake induced by miR-513a-3p mimic was also reversed after HK2
overexpression in HCT116 and SW480 cells (Fig. 6E and F). The lactate levels recovered after HK2
overexpression in HCT116 and SW480 cells treated with miR-513a-3p
mimic (Fig. 6G and H).
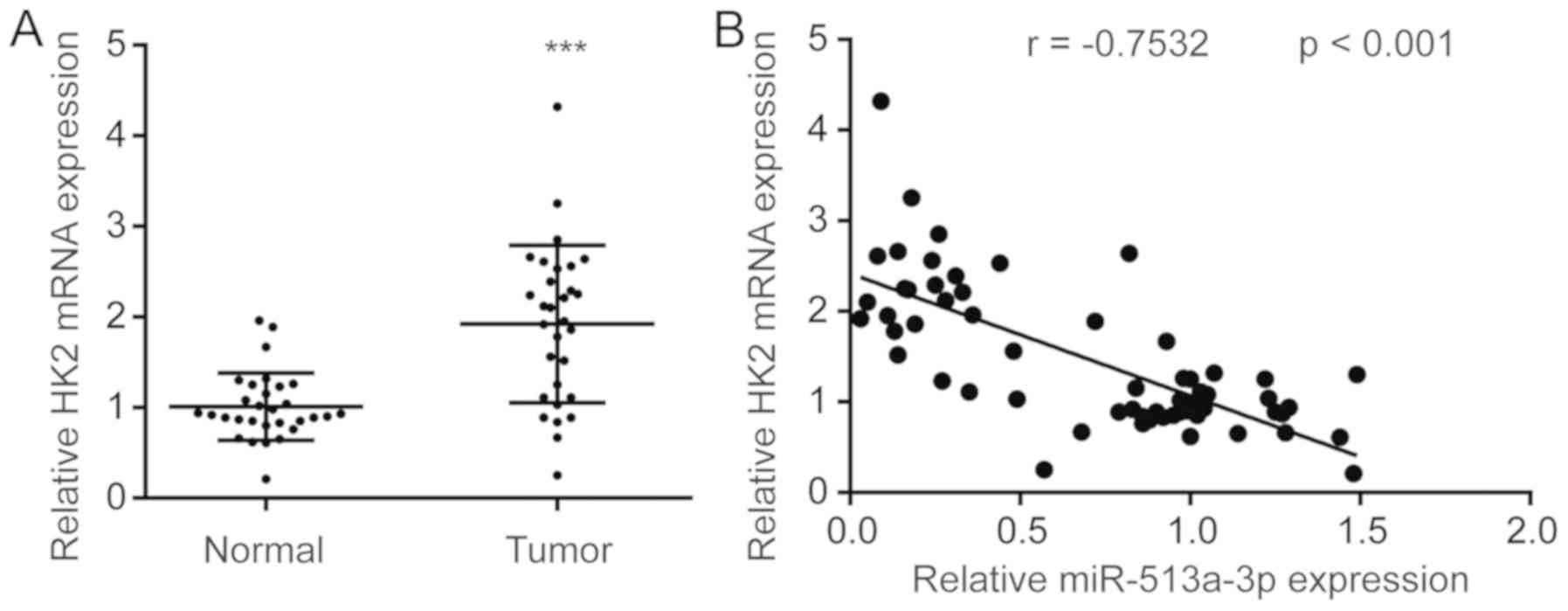
Expression of miR-513a-3p is
negatively correlated with HK2 mRNA expression in colorectal tumors
and normal tissues
To examine the clinical association between
miR-513a-3p and HK2, HK2 mRNA expression was detected in colorectal
tumors and normal tissues. HK2 mRNA levels were significantly
upregulated in colorectal tumors compared with normal tissues
(Fig. 7A). Pearson correlation
analysis suggested that miR-513a-3p levels were negatively
correlated with HK2 mRNA expression in tumors and normal tissues
from patients with colorectal cancer (Fig. 7B).
Discussion
Numerous studies have demonstrated that miRNAs are
pivotal for colorectal cancer progression and may be used as
therapeutic targets and biomarkers for patients with colorectal
cancer (23,24,30).
Several studies indicated that miR-513a-3p was involved in
carcinogenesis (31,32). Zhang et al (31) found that miR-513a-3p was upregulated
in cisplatin-resistant lung cancer cells, and that it sensitized
lung cancer cells to cisplatin treatment via targeting GSTP1.
Another study showed that miR-513a-3p regulated the inflammatory
process and the migration of lung cancer cells through direct
repression of Integrin Beta-8(32).
The results of the present study suggest an antitumor role for
miR-513a-3p in colorectal cancer. miR-513a-3p was found to be
downregulated in colorectal cancer via bioinformatic analysis and
RT-qPCR validation. In two colorectal cancer cell lines,
overexpression of miR-513a-3p inhibited proliferation. As abnormal
metabolism is essential for sustained proliferative signaling in
colorectal cancer cells (6),
glycolysis was examined in colorectal cancer cells transfected with
miR-513a-3p mimic. Interestingly, the glucose uptake rate was
suppressed following miR-513a-3p overexpression. Additionally, the
lactate level in the culture medium of cells with miR-513a-3p
overexpression was decreased. The present study demonstrated that
miR-513a-3p influenced the metabolism and proliferation of
colorectal cancer cells.
The aberrant activation of metabolism in cancer
cells is the consequence of promoter methylation, gene mutation and
altered expression of non-coding RNA (10,33). In
hepatocellular carcinoma, the promoter of HK2 is hypomethylated,
resulting in enhanced HK2 expression (34). HK2 is essential for initiation of
KRAS mutation-driven tumors in mouse (35). In KRAS mutant colorectal cancer cells
(HCT116 and DLD-1), HK2 expression was significantly higher
compared with colorectal cancer cells with wild type KRAS (36). In colon and hepatocellular cancer
cells, miRNAs such as miR-98 and miR-125 were found to be negative
regulators of glycolysis via targeting HK2 (37,38). In
the current study, several key regulators of glycolysis were
screened in cells overexpressing miR-513a-3p. HK2 was downregulated
after miR-513a-3p overexpression in HCT116 and SW480 cells, two
KRAS-mutant colorectal cancer cell lines. miR-513a-3p was predicted
and validated to be a novel direct regulator of HK2 in cancer
cells. HK2 is pivotal for cell proliferation, stemness, metabolism
and drug resistance of colorectal cancer cells (39,40). In
the current study, HK2 overexpression reversed the inhibition of
cell proliferation caused by miR-513a-3p overexpression.
Furthermore, HK2 overexpression enhanced glucose uptake and
elevated lactate levels, which were inhibited by miR-513a-3p
overexpression. The results of this study suggest that
miR-513a-3p/HK2 interaction is a driver of glycolysis and cell
proliferation in colorectal cancer.
In summary, the present study explored the role of
miR-513a-3p in colorectal cancer. Downregulation of miR-513a-3p
directly increased HK2 expression and promoted proliferation and
metabolism in colorectal cancer cells. These findings may serve as
rationale for targeting miR-513a-3p and HK2 as a novel therapeutic
approach for patients with colorectal cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JY conceived and designed the present study. CL, ZY
and JY performed the experiments. CL and ZY coordinated the
research and analyzed the data. JY wrote the manuscript and
supervised the study. All of the authors received the final version
of manuscript and approved for publication.
Ethics approval and consent to
participate
The current study was approved by the Ethical
Committee of the People's Hospital of Boluo County (Huizhou,
China). Each patient provided written informed consent prior to the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Moghimi-Dehkordi B and Safaee A: An
overview of colorectal cancer survival rates and prognosis in Asia.
World J Gastro Oncol. 4:71–75. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Hayat MJ, Howlader N, Reichman ME and
Edwards BK: Cancer statistics, trends, and multiple primary cancer
analyses from the surveillance, epidemiology, and end results
(SEER) Program. Oncologist. 12:20–37. 2007.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang M, Liu T, Sun H, Weng W, Zhang Q,
Liu C, Han Y and Sheng W: Pim1 supports human colorectal cancer
growth during glucose deprivation by enhancing the Warburg effect.
Cancer Sci. 109:1468–1479. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Taniguchi K, Sakai M, Sugito N, Kumazaki
M, Shinohara H, Yamada N, Nakayama T, Ueda H, Nakagawa Y, Ito Y, et
al: PTBP1-associated microRNA-1 and -133b suppress the Warburg
effect in colorectal tumors. Oncotarget. 7:18940–18952.
2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Nijhuis A, Thompson H, Adam J, Parker A,
Gammon L, Lewis A, Bundy JG, Soga T, Jalaly A, Propper D, et al:
Remodelling of microRNAs in colorectal cancer by hypoxia alters
metabolism profiles and 5-fluorouracil resistance. Hum Mol Genet.
26:1552–1564. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hsu PP and Sabatini DM: Cancer cell
metabolism: Warburg and beyond. Cell. 134:703–707. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Vartanian A, Agnihotri S, Wilson MR,
Burrell KE, Tonge PD, Alamsahebpour A, Jalali S, Taccone MS,
Mansouri S, Golbourn B, et al: Targeting hexokinase 2 enhances
response to radio-chemotherapy in glioblastoma. Oncotarget.
7:69518–69535. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Jin Z, Gu J, Xin X, Li Y and Wang H:
Expression of hexokinase 2 in epithelial ovarian tumors and its
clinical significance in serous ovarian cancer. Eur J Gynaecol
Oncol. 35:519–524. 2014.PubMed/NCBI
|
|
13
|
Graziano F, Ruzzo A, Giacomini E,
Ricciardi T, Aprile G, Loupakis F, Lorenzini P, Ongaro E, Zoratto
F, Catalano V, et al: Glycolysis gene expression analysis and
selective metabolic advantage in the clinical progression of
colorectal cancer. Pharmacogenomics J. 17:258–264. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Katagiri M, Karasawa H, Takagi K, Nakayama
S, Yabuuchi S, Fujishima F, Naitoh T, Watanabe M, Suzuki T, Unno M
and Sasano H: Hexokinase 2 in colorectal cancer: A potent
prognostic factor associated with glycolysis, proliferation and
migration. Histol Histopathol. 32:351–360. 2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kudryavtseva AV, Fedorova MS, Zhavoronkov
A, Moskalev AA, Zasedatelev AS, Dmitriev AA, Sadritdinova AF,
Karpova IY, Nyushko KM, et al: Effect of lentivirus-mediated shRNA
inactivation of HK1, HK2, and HK3 genes in colorectal cancer and
melanoma cells. BMC Genet. 17 (Suppl 3)(S156)2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Treiber T, Treiber N and Meister G:
Regulation of microRNA biogenesis and function. Thromb Haemost.
107:605–610. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhao Y and Srivastava D: A developmental
view of microRNA function. Trends Biochem Sci. 32:189–197.
2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Mo YY: MicroRNA regulatory networks and
human disease. Cell Mol Life Sci. 69:3529–3531. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Moridikia A, Mirzaei H, Sahebkar A and
Salimian J: MicroRNAs: Potential candidates for diagnosis and
treatment of colorectal cancer. J Cell Physiol. 233:901–913.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Slattery ML, Herrick JS, Pellatt DF,
Stevens JR, Mullany LE, Wolff E, Hoffman MD, Samowitz WS and Wolff
RK: MicroRNA profiles in colorectal carcinomas, adenomas and normal
colonic mucosa: Variations in miRNA expression and disease
progression. Carcinogenesis. 37:245–261. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
You C, Liang H, Sun W, Li J, Liu Y, Fan Q,
Zhang H, Yue X, Li J, Chen X and Ba Y: Deregulation of the
miR-16-KRAS axis promotes colorectal cancer. Sci Rep.
6(37459)2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Huang C, Liu J, Xu L, Hu W, Wang J, Wang M
and Yao X: MicroRNA-17 promotes cell proliferation and migration in
human colorectal cancer by downregulating SIK1. Cancer Manag Res.
11:3521–3534. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang X, Li X, Tan F, Yu N and Pei H:
STAT1 inhibits MiR-181a expression to suppress colorectal cancer
cell proliferation through PTEN/Akt. J Cell Biochem. 118:3435–3443.
2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Sun Y, Zhao X, Zhou Y and Hu Y: miR-124,
miR-137 and miR-340 regulate colorectal cancer growth via
inhibition of the Warburg effect. Oncol Rep. 28:1346–1352.
2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33(e179)2005.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Oparina NY, Snezhkina AV, Sadritdinova AF,
Veselovskii VA, Dmitriev AA, Senchenko VN, Mel'nikova NV,
Speranskaya AS, Darii MV, Stepanov OA, et al: Differential
expression of genes that encode glycolysis enzymes in kidney and
lung cancer in humans. Genetika. 49:814–823. 2013.PubMed/NCBI View Article : Google Scholar : (In Russian).
|
|
29
|
He Y, Deng F, Zhao S, Zhong S, Zhao J,
Wang D, Chen X, Zhang J, Hou J, Zhang W, et al: Analysis of
miRNA-mRNA network reveals miR-140-5p as a suppressor of breast
cancer glycolysis via targeting GLUT1. Epigenomics. 11:1021–1036.
2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Markopoulos GS, Roupakia E, Tokamani M,
Chavdoula E, Hatziapostolou M, Polytarchou C, Marcu KB,
Papavassiliou AG, Sandaltzopoulos R and Kolettas E: A step-by-step
microRNA guide to cancer development and metastasis. Cell Oncol
(Dordr). 40:303–339. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang X, Zhu J, Xing R, Tie Y, Fu H, Zheng
X and Yu B: miR-513a-3p sensitizes human lung adenocarcinoma cells
to chemotherapy by targeting GSTP1. Lung Cancer. 77:488–494.
2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
da Silveira MB, Lima KF, da Silva AR, Dos
Santos RAS and Moraes KCM: Mir-513a-3p contributes to the
controlling of cellular migration processes in the A549 lung tumor
cells by modulating integrin β-8 expression. Mol Cell Biochem.
444:43–52. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ju HQ, Zhan G, Huang A, Sun Y, Wen S, Yang
J, Lu WH, Xu RH, Li J, Li Y, et al: ITD mutation in FLT3 tyrosine
kinase promotes Warburg effect and renders therapeutic sensitivity
to glycolytic inhibition. Leukemia. 31:2143–2150. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Lee HG, Kim H, Son T, Jeong Y, Kim SU,
Dong SM, Park YN, Lee JD, Lee JM and Park JH: Regulation of HK2
expression through alterations in CpG methylation of the HK2
promoter during progression of hepatocellular carcinoma.
Oncotarget. 7:41798–41810. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Patra KC, Wang Q, Bhaskar PT, Miller L,
Wang Z, Wheaton W, Chandel N, Laakso M, Muller WJ, Allen EL, et al:
Hexokinase 2 is required for tumor initiation and maintenance and
its systemic deletion is therapeutic in mouse models of cancer.
Cancer Cell. 24:213–228. 2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Iwamoto M, Kawada K, Nakamoto Y, Itatani
Y, Inamoto S, Toda K, Kimura H, Sasazuki T, Shirasawa S, Okuyama H,
et al: Regulation of 18F-FDG accumulation in colorectal cancer
cells with mutated KRAS. J Nucl Med. 55:2038–2044. 2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhu W, Huang Y, Pan Q, Xiang P, Xie N and
Yu H: MicroRNA-98 suppress warburg effect by targeting HK2 in colon
cancer cells. Dig Dis Sci. 62:660–668. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Jin F, Wang Y, Zhu Y, Li S, Liu Y, Chen C,
Wang X, Zenz K and Li L: The miR-125a/HK2 axis regulates cancer
cell energy metabolism reprogramming in hepatocellular carcinoma.
Sci Rep. 7(3089)2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li Q, Qin Y, Wei P, Lian P, Li Y, Xu Y, Li
X, Li D and Cai S: Gas1 inhibits metastatic and metabolic
phenotypes in colorectal carcinoma. Mol Cancer Res. 14:830–840.
2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Hamabe A, Yamamoto H, Konno M, Uemura M,
Nishimura J, Hata T, Takemasa I, Mizushima T, Nishida N, Kawamoto
K, et al: Combined evaluation of hexokinase 2 and phosphorylated
pyruvate dehydrogenase-E1α in invasive front lesions of colorectal
tumors predicts cancer metabolism and patient prognosis. Cancer
Sci. 105:1100–1108. 2014.PubMed/NCBI View Article : Google Scholar
|